Submitted:
10 May 2024
Posted:
13 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
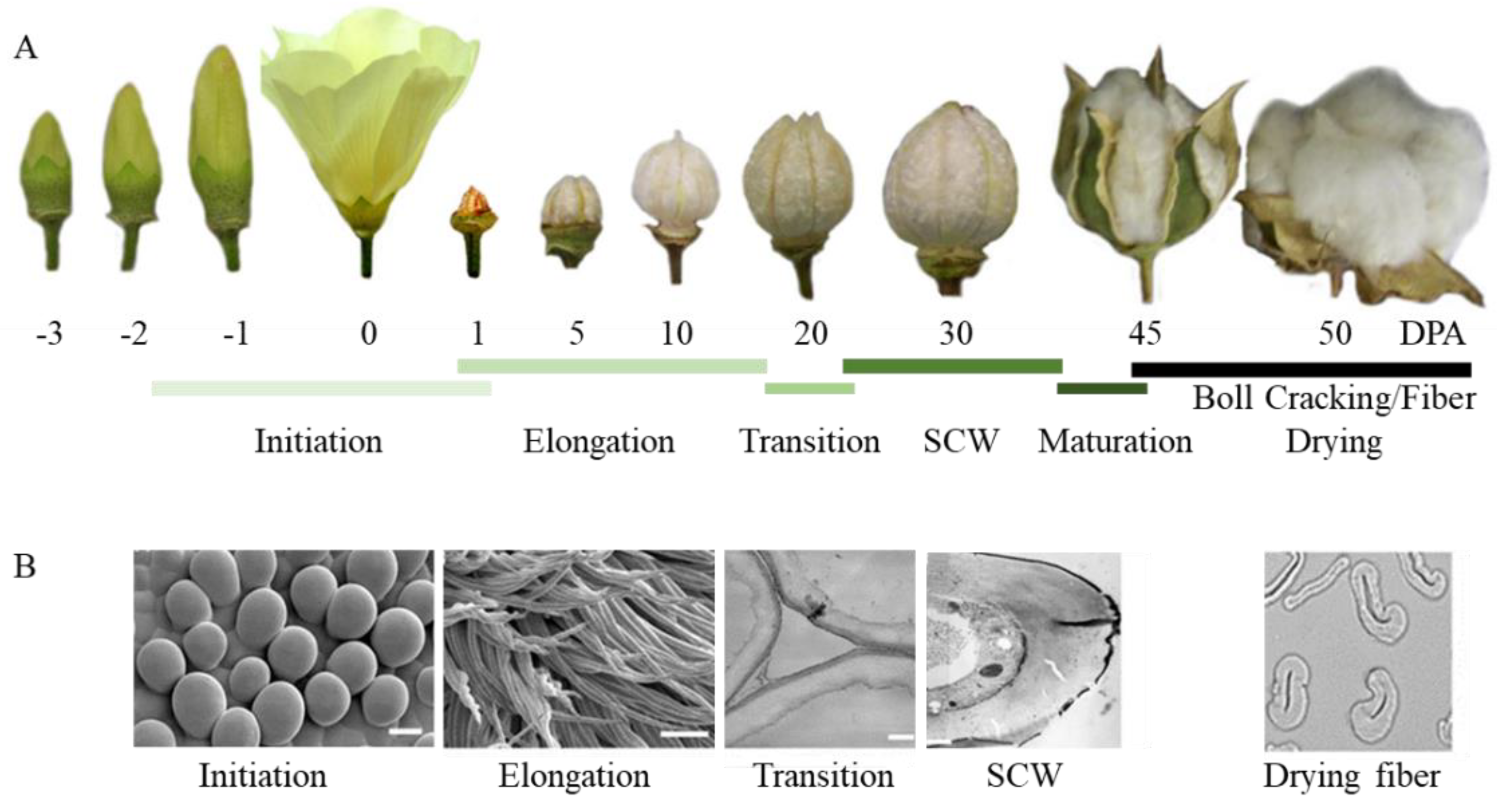
2. Fiberless Mutants Identification
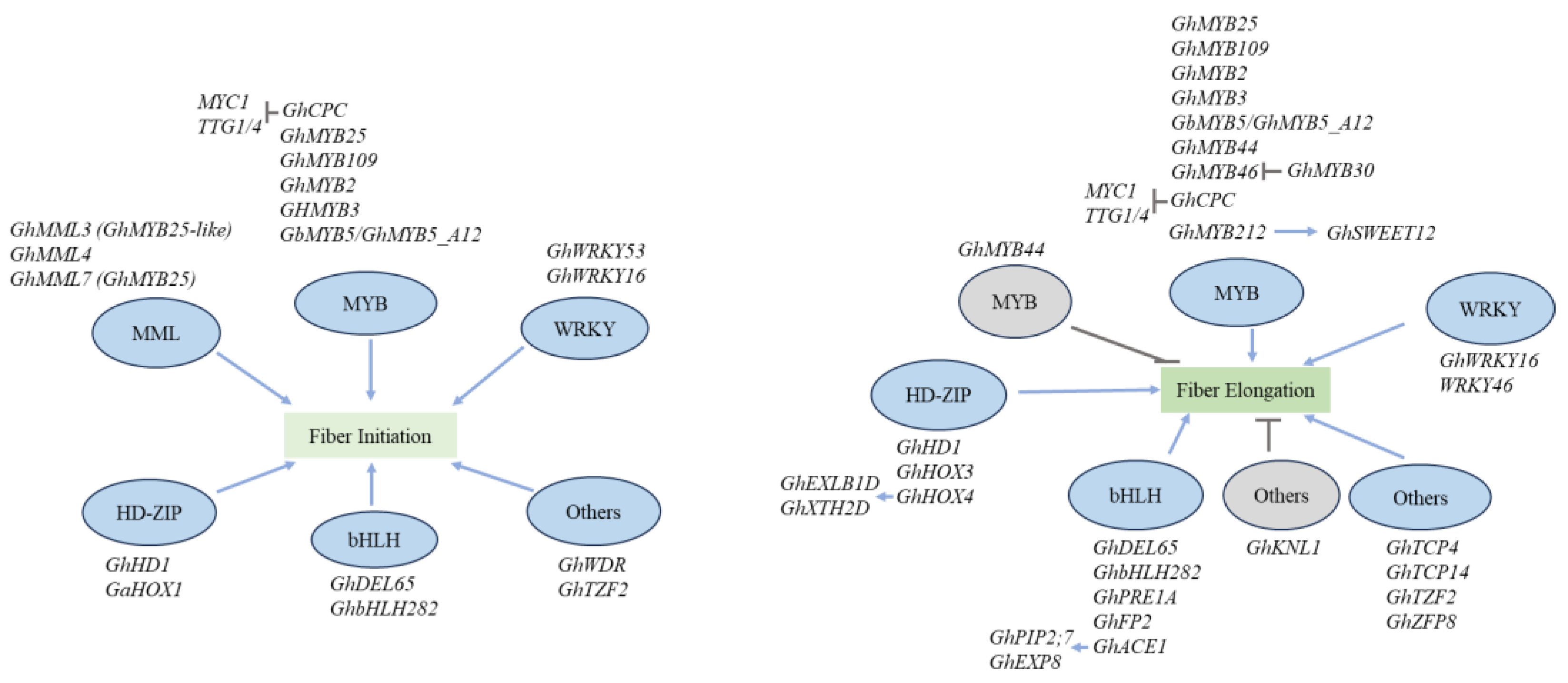
3. Transcriptional Regulation
3.1. MYB Transcription Factors
3.2. MYBMIXTA-like (MML) Transcription Factors
3.3. WRKY Transcription Factors
3.4. HD-ZIP Transcription Factors
3.5. bHLH Transcription Factors
3.6. Other Transcription Factors
3.7. Omic Tools for Studying the Fiber Initiation and Elongation
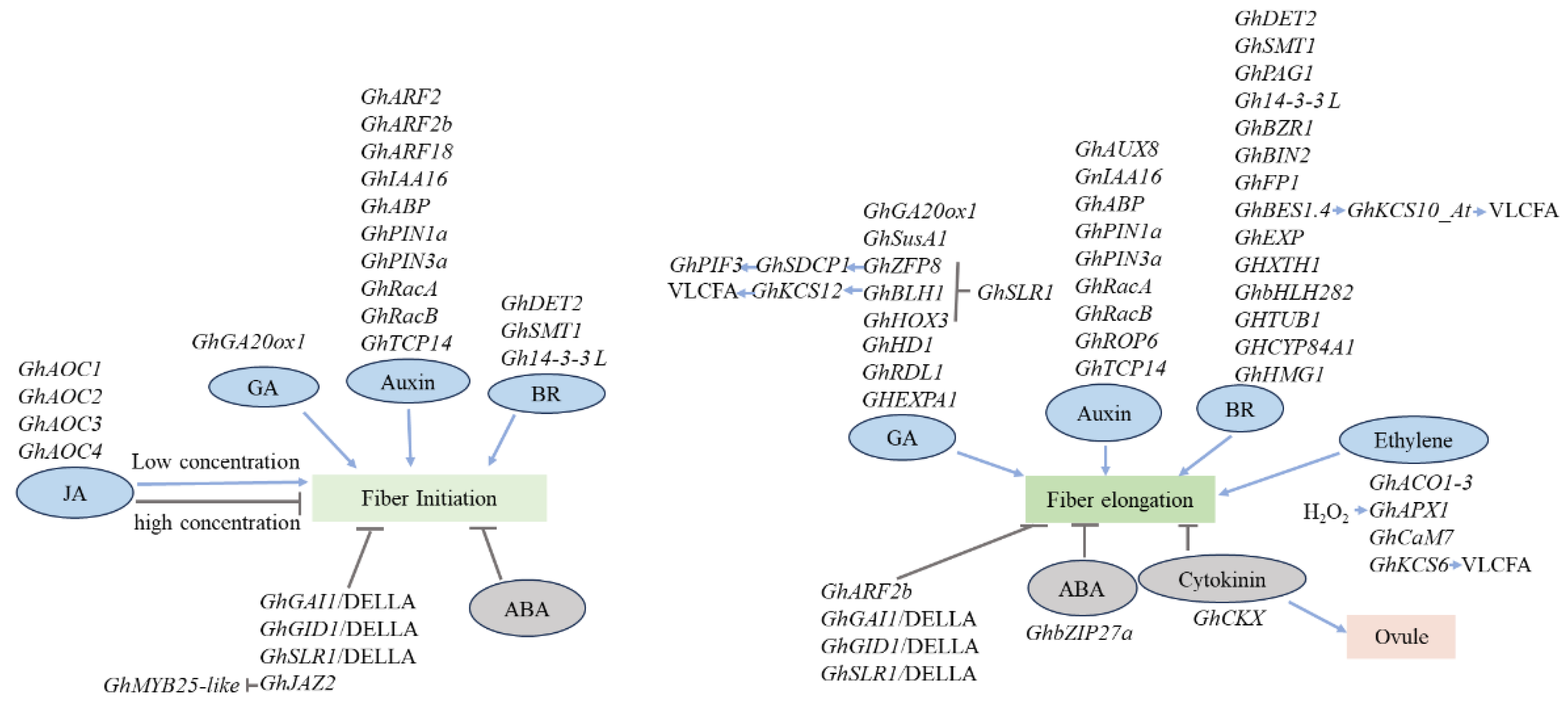
4. Phytohormonal Regulation
4.1. Auxin
4.2. Gibberellic Acid (GA)
4.3. Brassinosteroids (BR)
4.4. Jasmonic Acid (JA)
4.5. Ethylene
4.6. Abscisic Acid (ABA)
4.7. Cytokinin
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
EEO/Non-Discrimination
References
- Wendel, J.F.; Grover, C.E. Taxonomy and evolution of the cotton genus, Gossypium. Cotton 2015, 57, 25–44. [Google Scholar]
- Chen, Z.J.; et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Constable, G.; et al. Cotton breeding for fiber quality improvement. Industrial crops: Breeding for bioenergy and bioproducts 2015, 191–232. [Google Scholar]
- Basra, A.S.; C.P. Malik, Development of the Cotton Fiber, in International Review of Cytology, G.H. Bourne, J.F. Danielli, and K.W. Jeon, Editors. 1984, Academic Press. p. 65-113.
- Haigler, C.H.; et al. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Frontiers in Plant Science 2012, 3. [Google Scholar] [CrossRef]
- Stiff, M.R.; Haigler, C.H. Recent advances in cotton fiber development. Flowering and fruiting in cotton. Tennessee: The Cotton Foundation 2012, 163–192. [Google Scholar]
- Fan, S.; et al. Evolution of pectin synthesis relevant galacturonosyltransferase gene family and its expression during cotton fiber development. Journal of Cotton Research 2021, 4, NA. [Google Scholar] [CrossRef]
- Taliercio, E.W.; Boykin, D. Analysis of gene expression in cotton fiber initials. BMC Plant Biol 2007, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; et al. Comprehensive analysis of cellulose content, crystallinity, and lateral packing in Gossypium hirsutum and Gossypium barbadense cotton fibers using sum frequency generation, infrared and Raman spectroscopy, and X-ray diffraction. Cellulose 2015, 22. [Google Scholar] [CrossRef]
- Kabir, N.; et al. Functional characterization of TBL genes revealed the role of GhTBL7 and GhTBL58 in cotton fiber elongation. Int J Biol Macromol 2023, 241, 124571. [Google Scholar] [CrossRef]
- Wu, C.; et al. Genome-wide analysis elucidates the roles of GhHMA genes in different abiotic stresses and fiber development in upland cotton. Plant Physiology and Biochemistry 2023, 194, 281–301. [Google Scholar] [CrossRef]
- Fu, G.; et al. Genome-wide analysis of the serine carboxypeptidase-like protein family reveals Ga09G1039 is involved in fiber elongation in cotton. Plant Physiology and Biochemistry 2023, 107759. [Google Scholar] [CrossRef] [PubMed]
- Xing, K.; et al. N(6) -Methyladenosine mRNA modification regulates transcripts stability associated with cotton fiber elongation. Plant J 2023, 115, 967–985. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; et al. Comparative phosphoproteomic analysis reveals that phosphorylation of sucrose synthase GhSUS2 by Ca2+-dependent protein kinases GhCPK84/93 affects cotton fiber development. Journal of Experimental Botany 2023, 74, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; et al. Large-fragment insertion activates gene GaFZ (Ga08G0121) and is associated with the fuzz and trichome reduction in cotton (Gossypium arboreum). Plant Biotechnology Journal 2021, 19, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Zuo, D.; et al. Genome-wide identification of cotton CrRLK1L family genes and potential function of GhCrRLK1L104 in cell elongation. Current Plant Biology 2024, 37, 100325. [Google Scholar] [CrossRef]
- Yu, K.; et al. Potassium ameliorates cotton (Gossypium hirsutum L.) fiber length by regulating osmotic and K+/Na+ homeostasis under salt stress. Physiologia Plantarum 2023, 175, e13842. [Google Scholar] [CrossRef] [PubMed]
- Sun, M., C. Zheng, and W. Feng, Low soil available phosphorus level reduces cotton fiber length via osmoregulation. Frontiers in Plant Science 2023, 14, 1254103. [CrossRef]
- Fang, S.; et al. A cell wall-localized beta-1,3-glucanase promotes fiber cell elongation and secondary cell wall deposition. Plant Physiol 2023, 194, 106–123. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; et al. GhIQD10 interacts with GhCaM7 to control cotton fiber elongation via calcium signaling. The Crop Journal 2023, 11, 447–456. [Google Scholar] [CrossRef]
- Davière, J.M.; P. Achard, A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Molecular Plant 2016, 9, 10–20. [CrossRef]
- Wen, Y.-z.; et al. Strigolactones modulate cotton fiber elongation and secondary cell wall thickening. Journal of Integrative Agriculture. 2023. [Google Scholar]
- Zeng, J.; et al. Fiber-specific increase of carotenoid content promotes cotton fiber elongation by increasing abscisic acid and ethylene biosynthesis. The Crop Journal. 2023. [Google Scholar]
- Stewart, J.M. Fiber initiation on the cotton ovule (Gossypium hirsutum). American Journal of Botany 1975, 62, 723–730. [Google Scholar] [CrossRef]
- Wu, Y.; et al. Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 2006, 47, 107–127. [Google Scholar] [CrossRef] [PubMed]
- Kohel, R.J., E.V. Narbuth, and C.R. Benedict, Fiber Development of Ligon Lintless-2 Mutant of Cotton. Crop science 1992, 32, 733–735. [CrossRef]
- Du, X.M.; et al. Genetic analysis of presence and absence of lint and fuzz in cotton. Plant Breeding 2001, 120, 519–522. [Google Scholar] [CrossRef]
- Karaca, M.; et al. Simple sequence repeat (SSR) markers linked to the Ligon lintless (Li 1) mutant in cotton. Journal of Heredity 2002, 93, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Turley, R.B.; R.H. Kloth, The inheritance model for the fiberless trait in upland cotton (Gossypium hirsutum L.) line SL1-7-1: variation on a theme. Euphytica 2008, 164, 123–132. [CrossRef]
- Bechere, E., D.L. Auld, and E. Hequet, Development of ‘naked-tufted’ seed coat mutants for potential use in cotton production. Euphytica 2009, 167, 333–339. [CrossRef]
- Patel, J.D.; et al. The Ligon lintless-2 short fiber mutation is located within a terminal deletion of chromosome 18 in cotton. Plant Physiology 2020, 183, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.; et al. A deletion/duplication in the Ligon lintless-2 locus induces siRNAs that inhibit cotton fiber cell elongation. Plant Physiology 2022, 190, 1792–1805. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; et al. A Novel Tandem Zinc Finger Protein in Gossypium hirsutum, GhTZF2, Interacts with GhMORF8 to Regulate Cotton Fiber Cell Development. Agronomy 2023, 13, 519. [Google Scholar] [CrossRef]
- Wan, Q.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol 2016, 210, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; et al. Cotton microtubule-associated protein GhMAP20L5 mediates fiber elongation through the interaction with the tubulin GhTUB13. Plant Science 2023, 327, 111545. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; et al. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 2003, 1630, 25–34. [Google Scholar] [CrossRef]
- Lee, J.J.; et al. Developmental and gene expression analyses of a cotton naked seed mutant. Planta 2006, 223, 418–432. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; et al. Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta 2007, 226, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Walford, S.A.; et al. GhMYB25-like: a key factor in early cotton fibre development. Plant J 2011, 65, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Pu, L.; et al. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 2008, 180, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Liu, B., Y. Zhu, and T. Zhang, The R3-MYB gene GhCPC negatively regulates cotton fiber elongation. PLoS One 2015, 10, e0116272.
- Guan, X.; et al. Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS One 2011, 6, e21301. [Google Scholar] [CrossRef]
- Wang, N.; et al. Genetic variation in MYB5_A12 is associated with fibre initiation and elongation in tetraploid cotton. Plant Biotechnology Journal 2021, 19, 1892. [Google Scholar] [CrossRef]
- Wu, A.; et al. GhMYB30-GhMUR3 affects fiber elongation and secondary wall thickening in cotton. Plant J 2024, 117, 694–712. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; et al. Jasmonic Acid Signaling Pathway in Plants. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; et al. Cotton fiber elongation requires the transcription factor Gh MYB 212 to regulate sucrose transportation into expanding fibers. New Phytologist 2019, 222, 864–881. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; et al. Recent Advances and Future Perspectives in Cotton Research. Annu Rev Plant Biol 2021, 72, 437–462. [Google Scholar] [CrossRef]
- Wu, H.; et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol 2018, 217, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; et al. Genetic dissection of the fuzzless seed trait in. Journal of Experimental Botany 2018, 69, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; et al. Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol Genet Genomics 2015, 290, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; et al. Identification of the Group III WRKY Subfamily and the Functional Analysis of GhWRKY53 in Gossypium hirsutum L. Plants 2021, 10, 1235. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.-N.; et al. Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum). The Plant Cell 2021, 33, 2736–2752. [Google Scholar] [CrossRef]
- Chen, E.; et al. Genome-wide analysis of the HD-ZIP IV transcription factor family in Gossypium arboreum and GaHDG11 involved in osmotic tolerance in transgenic Arabidopsis. Mol Genet Genomics 2017, 292, 593–609. [Google Scholar] [CrossRef]
- Guan, X.Y.; et al. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiologia plantarum 2008, 134, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.M.; et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun 2014, 5, 5519. [Google Scholar] [CrossRef] [PubMed]
- Walford, S.A.; et al. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J 2012, 71, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; et al. Phosphatidic acid interacts with an HD-ZIP transcription factor GhHOX4 to influence its function in fiber elongation of cotton (Gossypium hirsutum). Plant J 2024. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, X.X.; et al. Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol Plant 2016, 158, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytologist 2018, 218, 1061–1075. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; et al. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC plant biology 2018, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; et al. The bHLH/HLH transcription factors GhFP2 and GhACE1 antagonistically regulate fiber elongation in cotton. Plant Physiology 2022, 189, 628–643. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.F.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef]
- Tian, Y.; et al. The transcription factor MML4_D12 regulates fiber development through interplay with the WD40-repeat protein WDR in cotton. Journal of Experimental Botany 2020, 71, 3499–3511. [Google Scholar] [CrossRef]
- Wang, Y.; et al. GhKNL1 controls fiber elongation and secondary cell wall synthesis by repressing its downstream genes in cotton (Gossypium hirsutum). J Integr Plant Biol 2022, 64, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; et al. Zinc Finger Protein8 (GhZFP8) Regulates the Initiation of Trichomes in Arabidopsis and the Development of Fiber in Cotton. Plants 2024, 13, 492. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-W.; et al. Comparative proteomic analysis provides new insights into the fiber elongating process in cotton. Journal of Proteome Research 2008, 7, 4623–4637. [Google Scholar] [CrossRef]
- Ho, M.H.; et al. Characterization and promoter analysis of a cotton RING-type ubiquitin ligase (E3) gene. Mol Biotechnol 2010, 46, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; et al. Characterization of the early fiber development gene, Ligon-lintless 1 (Li1), using microarray. Plant Gene 2016, 6, 59–66. [Google Scholar] [CrossRef]
- Prasad, P.; et al. Transcriptional Landscape of Cotton Fiber Development and Its Alliance With Fiber-Associated Traits. Front Plant Sci 2022, 13, 811655. [Google Scholar] [CrossRef]
- Wang, D.; et al. Cell-specific clock-controlled gene expression program regulates rhythmic fiber cell growth in cotton. Genome Biol 2023, 24, 49. [Google Scholar] [CrossRef]
- Bao, Y.; et al. Genome-wide chromatin accessibility landscape and dynamics of transcription factor networks during ovule and fiber development in cotton. BMC Biol 2023, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.A.; I.P. Ting, Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. American Journal of Botany 1974, 61, 188–194. [CrossRef]
- Guinn, G.; D.L. Brummett, Changes in abscisic acid and indoleacetic acid before and after anthesis relative to changes in abscission rates of cotton fruiting forms. Plant physiology 1988, 87, 629–631. [CrossRef]
- Chen, Z.J.; X. Guan, Auxin boost for cotton. Nat Biotechnol 2011, 29, 407–409. [CrossRef] [PubMed]
- Zhang, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 2011, 29, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; et al. Auxin Regulates Cotton Fiber Initiation via GhPIN-Mediated Auxin Transport. Plant Cell Physiol 2017, 58, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Gokani, S.J.; V.S. Thaker, Role of gibberellic acid in cotton fibre development. The Journal of Agricultural Science 2002, 138, 255–260. [CrossRef]
- Xiao, G.; et al. Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. Journal of experimental botany 2018, 69, 4323–4337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; et al. Characterization of cotton ARF factors and the role of GhARF2b in fiber development. BMC Genomics 2021, 22, 202. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; et al. Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 2012, 503, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-G.; et al. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes & development 2001, 15, 902–911. [Google Scholar]
- Tao, L.Z., A.Y. Cheung, and H.M. Wu, Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 2002, 14, 2745–2760. [CrossRef]
- Wang, M.Y.; et al. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol 2013, 162, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Li, I.X.; et al. [Cloning and expression analysis of two Rac genes from cotton (Gossypium hirsutum L)]. Yi Chuan Xue Bao 2005, 32, 72–78. [Google Scholar]
- Zhang, M.; et al. PIN-formed protein, a door to reveal the mechanism for auxin-triggered initiation of cotton fiber. Plant Signal Behav 2017, 12, e1319031. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; et al. GhROP6 GTPase modulates auxin accumulation in cotton fibers by regulating cell-specific GhPIN3a localization. J Exp Bot 2023, 74, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; B.A. Triplett, Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 2001, 127, 1361–1366. [CrossRef]
- Gialvalis, S.; R.W. Seagull, Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. J. Cotton Sci 2001, 5, 252–258.
- Seagull, R.W.; S. Giavalis, Pre-and post-anthesis application of exogenous hormones alters fiber production in Gossypium hirsutum L. cultivar Maxxa GTO. 2004.
- Liao, W.-b.; et al. Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum. Plant growth regulation 2009, 58, 35–45. [Google Scholar] [CrossRef]
- Aleman, L.; et al. Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol Biol 2008, 68, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 2010, 167, 829–837. [Google Scholar] [CrossRef]
- Bai, W.Q.; et al. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS One 2014, 9, e96537. [Google Scholar] [CrossRef]
- Daviere, J.M.; P. Achard, Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [CrossRef]
- Hu, M.-y.; et al. Brassinosteroids and auxin down-regulate DELLA genes in fiber initiation and elongation of cotton. Agricultural Sciences in China 2011, 10, 1168–1176. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 2008, 6, e0103. [Google Scholar] [CrossRef]
- Jing, D.; et al. Cloning and Expression Profile of Gibberellin Insensitive Dwarf GID1 Homologous Genes from Cotton. Acta Agronomica Sinica 2009, 35, 1822–1830. [Google Scholar]
- He, P.; et al. Gibberellin acid promotes single-celled fiber elongation through the activation of two signaling cascades in cotton. Developmental Cell, 2024.
- Sun, Y.; et al. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol 2005, 46, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; et al. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J 2007, 51, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.H.; et al. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006, 18, 651–664. [Google Scholar] [CrossRef]
- Yang, Z.; et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol 2014, 203, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; et al. Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol J 2015, 13, 269–280. [Google Scholar] [CrossRef]
- Liu, Z.H.; et al. A basic helix-loop-helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling. New Phytol 2020, 225, 2439–2452. [Google Scholar] [CrossRef]
- Liu, L.; et al. A brassinosteroid transcriptional regulatory network participates in regulating fiber elongation in cotton. Plant Physiol 2023, 191, 1985–2000. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; et al. Brassinosteroids regulate cotton fiber elongation by modulating very-long-chain fatty acid biosynthesis. Plant Cell 2023, 35, 2114–2131. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; et al. Exogenous jasmonic acid inhibits cotton fiber elongation. Journal of Plant Growth Regulation 2012, 31, 599–605. [Google Scholar] [CrossRef]
- Wang, L.; et al. Comparative Transcriptomics Reveals Jasmonic Acid-Associated Metabolism Related to Cotton Fiber Initiation. PLoS One 2015, 10, e0129854. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; et al. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J 2016, 88, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.-M., C.-Y. Hu, and Y.-X. Zhu, The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant signaling & behavior 2008, 3, 194–196.
- Jiang, L.; et al. A role for APX1 gene in lead tolerance in Arabidopsis thaliana. Plant Sci 2017, 256, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; et al. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytologist 2007, 175, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.-M.; Y.-X. Zhu, How cotton fibers elongate: a tale of linear cell-growth mode. Current opinion in plant biology 2011, 14, 106–111. [CrossRef] [PubMed]
- Tang, W.; et al. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 2014, 202, 509–520. [Google Scholar] [CrossRef]
- Qin, Y.M.; et al. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.A.; et al. A quantitative procedure for estimating cotton fiber growth. Stain Technol 1974, 49, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; et al. Comparative proteomic analysis identified proteins and the phenylpropanoid biosynthesis pathway involved in the response to ABA treatment in cotton fiber development. Sci Rep 2023, 13, 1488. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.A.; F.T. Addicott, Abscisic Acid: correlations with abscission and with development in the cotton fruit. Plant Physiol 1972, 49, 644–648. [CrossRef] [PubMed]
- Gokani, S.J., R. Kumar, and V.S. Thaker, Potential role of abscisic acid in cotton fiber and ovule development. Journal of plant growth regulation 1998, 17, 1–5. [CrossRef]
- Nayyar, H.; et al. Hormonal regulation of cotton fibre elongation in Gossypium arboreum L. in vitro and in vivo. Biochemie und Physiologie der Pflanzen 1989, 185, 415–421. [Google Scholar] [CrossRef]
- Ma, Q., P. Hedden, and Q. Zhang, Heterosis in rice seedlings: its relationship to gibberellin content and expression of gibberellin metabolism and signaling genes. Plant Physiol 2011, 156, 1905–1920. [CrossRef] [PubMed]
- Gilbert, M.K.; et al. A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of Ligon lintless-2 mutant of cotton (Gossypium hirsutum L). PLoS One 2013, 8, e75268. [Google Scholar] [CrossRef]
- Zhang, H.; et al. Effect of phytohormones on fiber initiation of cotton ovule. Acta physiologiae plantarum 2009, 31, 979–986. [Google Scholar] [CrossRef]
- Dasani, S.H.; V.S. Thaker, Role of abscisic acid in cotton fiber development. Russian Journal of Plant Physiology 2006, 53, 62–67. [CrossRef]
- Chen, J.G.; et al. Levels of cytokinins in the ovules of cotton mutants with altered fiber development. Journal of Plant Growth Regulation 1997, 16, 181–185. [Google Scholar] [CrossRef]
- Spallek, T.; et al. Interspecies hormonal control of host root morphology by parasitic plants. Proc Natl Acad Sci U S A 2017, 114, 5283–5288. [Google Scholar] [CrossRef] [PubMed]
- Niemann, M.C.E.; et al. The cytokinin oxidase/dehydrogenase CKX1 is a membrane-bound protein requiring homooligomerization in the endoplasmic reticulum for its cellular activity. Plant Physiology 2018, 176, 2024–2039. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.J.; B.M.N. Schreiber, Role and function of cytokinin oxidase in plants. Plant Growth Regulation 1997, 23, 123–134. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



