Submitted:
10 May 2024
Posted:
12 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
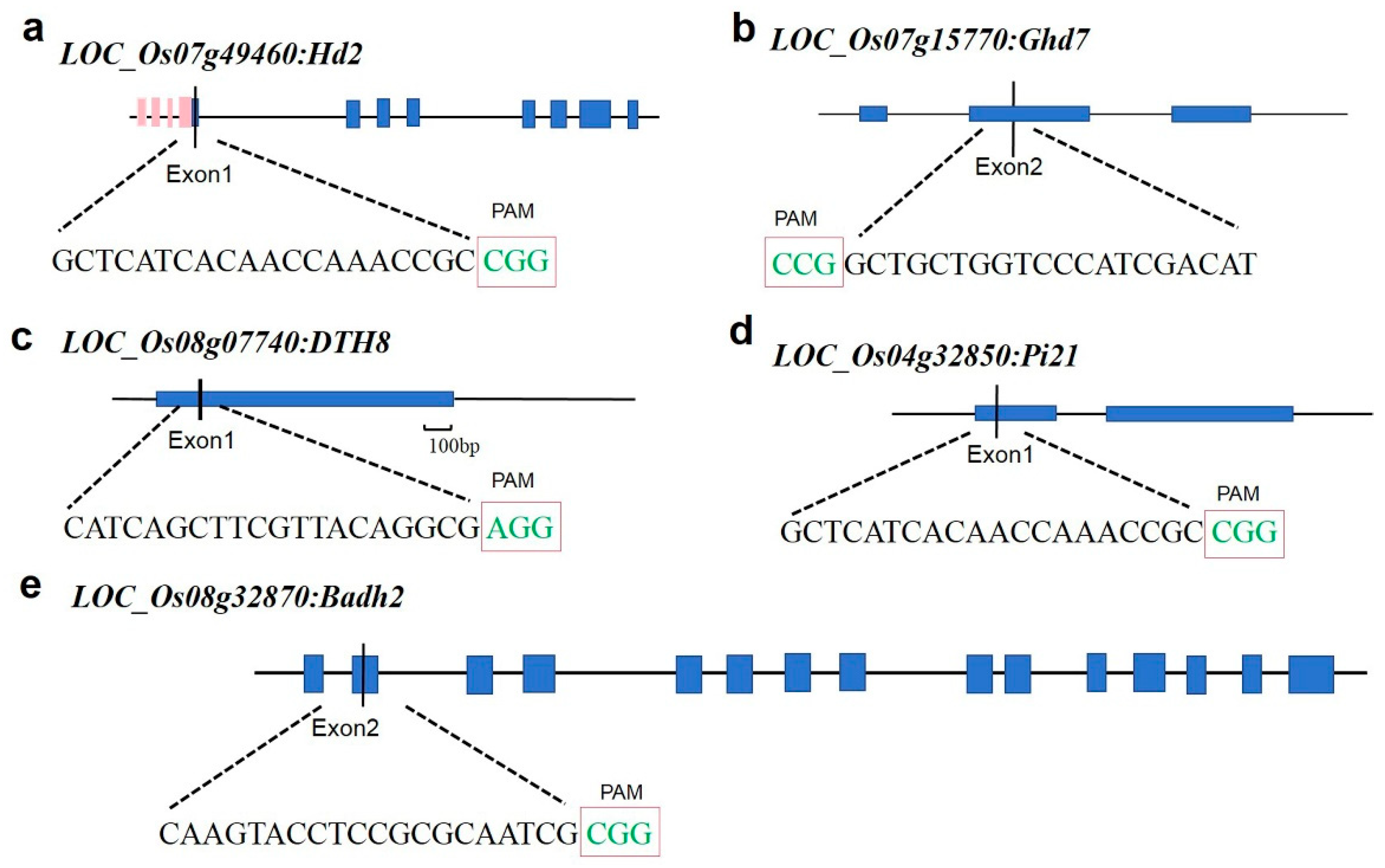
2.1. Target Design and Vector Construction
2.2. Target Design and Off-Target Analysis
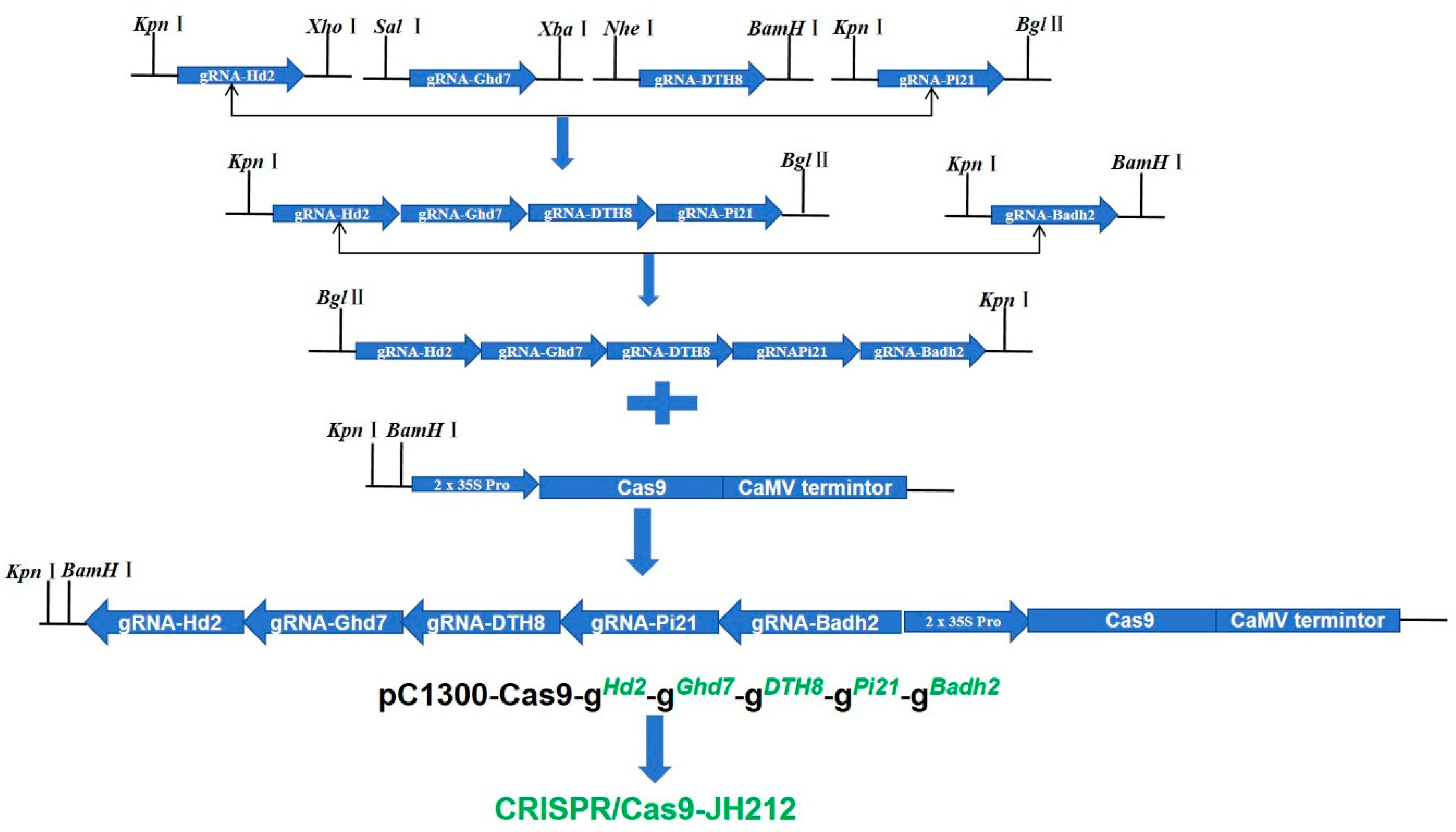
2.3. Vector Construction and Genetic Transformation
2.4. Acquisition of T0 Transgenic Positive Plants
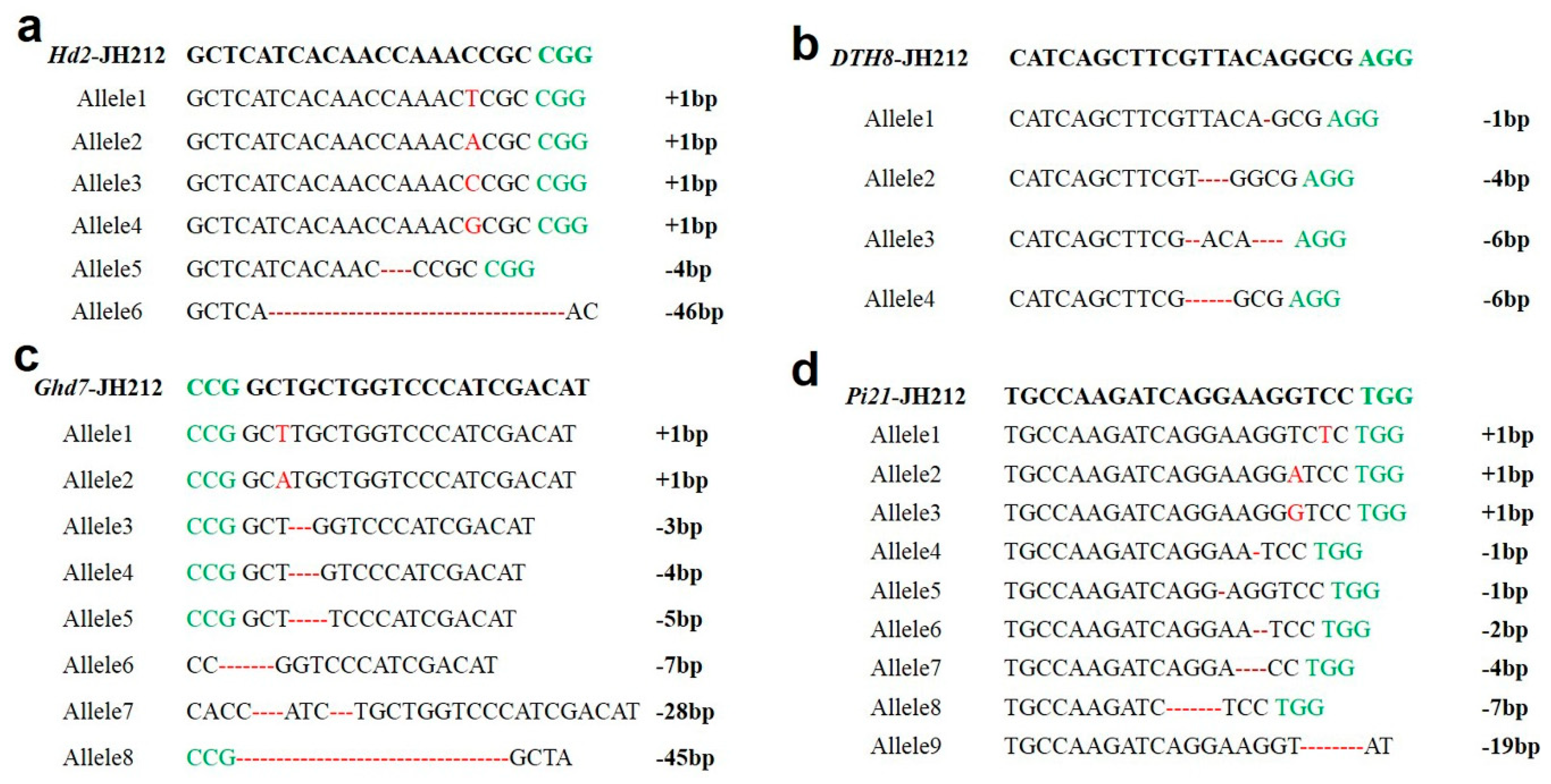
2.5. Mutation Analysis of Transgenic Seedlings in T0 Generation
2.6. Analysis of Mutation Sites in T1 Mutants
2.7. Analysis of Mutation Sites in Homozygous T1 Mutants
2.8. T1 Generation and T2 Generations of Cas9-Free Plants Were Obtained.
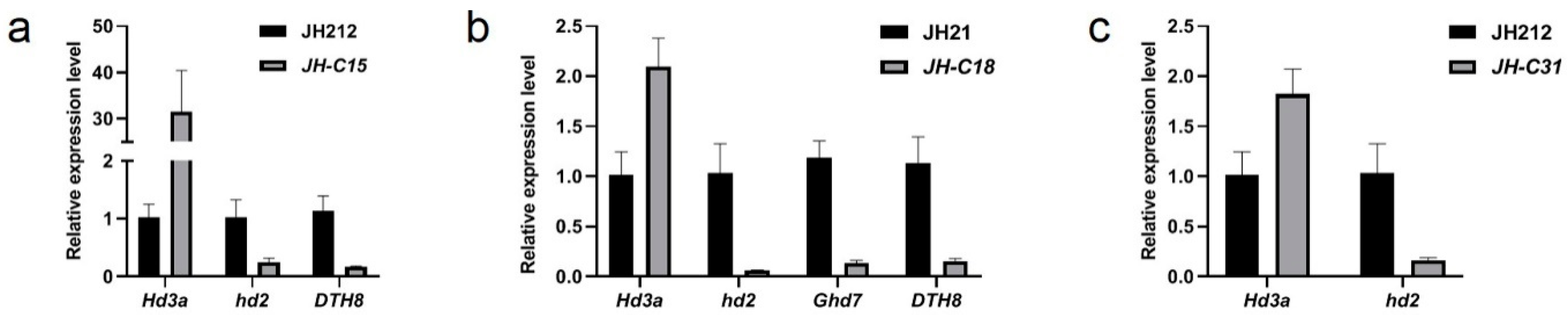
2.9. qRT-PCR Detection of Gene Editing in Homozygous Mutant Lines
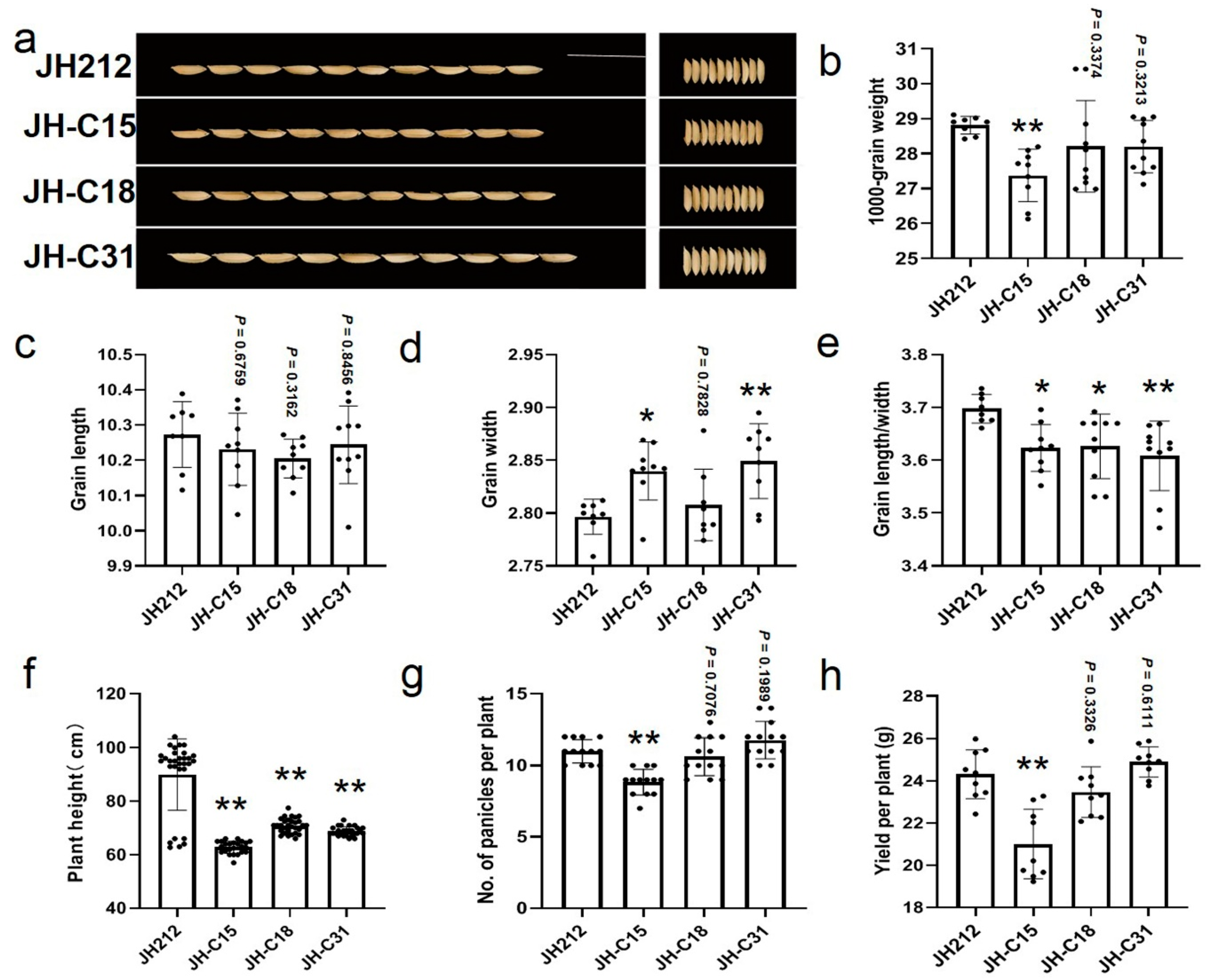
2.9. Phenotypic Analysis of T2 Generation Knockout Mutants
3. Discussion
4. Materials and Methods
4.1. Experimental Materials and Planting
4.2. gRNA Target Site Design
4.3. Construction of CRISPR/Cas9 Expression Vector
4.4. Acquisition of T0 Generation Positive Strains and Identification of Target Sites
4.5. Screening for GM-Free Components in T1 and T2 Generations
4.6. qRT-PCR Analysis of Transgenic Plants
4.7. Mutant Phenotype Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, S.; Zhu, S.; Cui, S.; Hou, H.; Wu, H.; Hao, B.; Cai, L.; Xu, Z.; Liu, L.; Jiang, L.; et al. Transcriptional and Post-Transcriptional Regulation of Flowering Date in Rice. New Phytologist 2021, 230, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Yuqi, S.U.N.; Qingyun, B.U.; Yunqing, C.; Xiaojie, T.; Zhenyu, W.; Yuekun, R.E.N.; Jiaqi, T.; Xiufeng, L.I. Research progress on the Flowering Date of rice in Northeast China. tryzw 2018, 7, 177–183. [Google Scholar] [CrossRef]
- Error: DOI Not Found Available online:. https://doi.org/10.11689/j.issn.2095-2961.2018.02.010 (accessed on 8 May 2024).
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol Plant Pathol 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Qu, R.; Zhang, P.; Liu, Q.; Wang, Y.; Guo, W.; Du, Z.; Li, X.; Yang, L.; Yan, S.; Gu, X. Genome-Edited ATP BINDING CASSETTE B1 Transporter SD8 Knockouts Show Optimized Rice Architecture without Yield Penalty. Plant Commun 2022, 3, 100347. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent Selection of a WD40 Protein That Enhances Grain Yield in Maize and Rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, S.; Huang, Y.; Ma, X.; Tan, L.; Liu, F.; Lv, Q.; Zhu, Z.; Hu, M.; Fu, Y.; et al. OsMADS17 Simultaneously Increases Grain Number and Grain Weight in Rice. Nat Commun 2023, 14, 3098. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid Improvement of Grain Weight via Highly Efficient CRISPR/Cas9-Mediated Multiplex Genome Editing in Rice. J Genet Genomics 2016, 43, 529–532. [Google Scholar] [CrossRef]
- Huang, Y.; Dong, H.; Mou, C.; Wang, P.; Hao, Q.; Zhang, M.; Wu, H.; Zhang, F.; Ma, T.; Miao, R.; et al. Ribonuclease H-like Gene SMALL GRAIN2 Regulates Grain Size in Rice through Brassinosteroid Signaling Pathway. J Integr Plant Biol 2022, 64, 1883–1900. [Google Scholar] [CrossRef]
- ZHANG, H.; LIU, X.; XUAN, N.; ZHANG, H.; GAO, R.; ZHAO, Q.; YAO, F. Editing DTH8 Gene Using CRISPR /Cas9 Technology to Improve Heading Date of Rice 99-25. Acta Agriculturae Boreali-Sinica 2020, 35, 58–66. [Google Scholar]
- Huang, L.; Xiao, Y.; Zhao, W.; Rao, Y.; Shen, H.; Gu, Z.; Fan, X.; Li, Q.; Zhang, C.; Liu, Q. Creating High-Resistant Starch Rice by Simultaneous Editing of SS3a and SS3b. Plant Biotechnol J 2024, 22, 787–789. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal Seeds from Hybrid Rice by Simultaneous Genome Engineering of Meiosis and Fertilization Genes. Nat Biotechnol 2019, 37, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Wenjia, Z.; Xiaojie, T.; Yuekun, R.E.N.; Xiangjin, W.E.I.; Yang G. a., O.; Lihong, X.I.E.; Huazhao, L.I.U.; Qingyun, B.U.; Xiufeng, L.I. Breeding of Early-maturatity and Fragrant Rice via CRISPR/Cas9 Mediated Genome Editing. tryzw 2017, 6, 146–152. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of Rice Varieties with Enhanced Resistances to Both Blast and Bacterial Blight Diseases via CRISPR/Cas9. Plant Biotechnol J 2022, 20, 876–885. [Google Scholar] [CrossRef]
- Sun, K.; Huang, M.; Zong, W.; Xiao, D.; Lei, C.; Luo, Y.; Song, Y.; Li, S.; Hao, Y.; Luo, W.; et al. Hd1, Ghd7, and DTH8 Synergistically Determine the Rice Flowering Date and Yield-Related Agronomic Traits. J Genet Genomics 2022, 49, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qu, X.; Zhou, Y.; Song, G.; Abiri, N.; Xiao, Y.; Liang, F.; Jiang, D.; Hu, Z.; Yang, D. OsPRR37 Confers an Expanded Regulation of the Diurnal Rhythms of the Transcriptome and Photoperiodic Flowering Pathways in Rice. Plant Cell Environ 2018, 41, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural Variation in Ghd7 Is an Important Regulator of Flowering Date and Yield Potential in Rice. Nat Genet 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Chen, S.; Wu, M.; Zheng, T.; Zhou, L.; Li, C.; Zhang, H.; Wang, J.; Xu, X.; Chai, J.; et al. Early Flowering 7 Interacts with DTH8, and Regulates Flowering Time in Rice. Plant Cell Rep 2019, 38, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zheng, X.-M.; Chen, D.; Zhang, Y.; Ma, W.; Zhang, H.; Sun, L.; Yang, Z.; Zhao, C.; Zhan, X.; et al. OsCOL16, Encoding a CONSTANS-like Protein, Represses Flowering by up-Regulating Ghd7 Expression in Rice. Plant Sci 2017, 260, 60–69. [Google Scholar] [CrossRef]
- Tan, J.; Jin, M.; Wang, J.; Wu, F.; Sheng, P.; Cheng, Z.; Wang, J.; Zheng, X.; Chen, L.; Wang, M.; et al. OsCOL10, a CONSTANS-Like Gene, Functions as a Flowering Time Repressor Downstream of Ghd7 in Rice. Plant Cell Physiol 2016, 57, 798–812. [Google Scholar] [CrossRef]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H.; et al. Strong Photoperiod Sensitivity Is Controlled by Cooperation and Competition among Hd1, Ghd7 and DTH8 in Rice Flowering. New Phytol 2021, 229, 1635–1649. [Google Scholar] [CrossRef]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 Module Mediates Day-Length-Dependent Regulation of Rice Flowering. Mol Plant 2017, 10, 948–961. [Google Scholar] [CrossRef]
- Fukuoka, S.; Okuno, K. QTL Analysis and Mapping of Pi21, a Recessive Gene for Field Resistance to Rice Blast in Japanese Upland Rice. Theor Appl Genet 2001, 103, 185–190. [Google Scholar] [CrossRef]
- Tao, H.; Shi, X.; He, F.; Wang, D.; Xiao, N.; Fang, H.; Wang, R.; Zhang, F.; Wang, M.; Li, A.; et al. Engineering Broad-Spectrum Disease-Resistant Rice by Editing Multiple Susceptibility Genes. J Integr Plant Biol 2021, 63, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Liu, X.; Tang, W. Research Progress of Photoperiod Regulation in Rice Flowering. Chinese Journal OF Rice Science 2021, 35, 207. [Google Scholar] [CrossRef]
- Lorenzo, C.D.; Debray, K.; Herwegh, D.; Develtere, W.; Impens, L.; Schaumont, D.; Vandeputte, W.; Aesaert, S.; Coussens, G.; De Boe, Y.; et al. BREEDIT: A Multiplex Genome Editing Strategy to Improve Complex Quantitative Traits in Maize. Plant Cell 2023, 35, 218–238. [Google Scholar] [CrossRef] [PubMed]
- Romero, F.M.; Gatica-Arias, A. CRISPR/Cas9: Development and Application in Rice Breeding. Rice Science 2019, 26, 265–281. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLOS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, W.; Ren, Y.; Tian, X.; Lv, T.; Wang, Z.; Fang, J.; Chu, C.; Yang, J.; Bu, Q. High-Efficiency Breeding of Early-Maturing Rice Cultivars via CRISPR/Cas9-Mediated Genome Editing. J Genet Genomics 2017, 44, 175–178. [Google Scholar] [CrossRef]
- Xu, P.; Wang, H.; Tu, R .; Liu, Q.; Wu, W.; Fu, X.; Cao, L.; SHEN, X. Orientation Improvement of Blast Resistance in Rice via CRISPR/Cas9 System. Chinese Journal of Rice Science 2019, 33, 313–322. [Google Scholar] [CrossRef]
- LIANG, M.; ZHANG, H.; CHEN, J.; DAI, D.; DU, C.; WANG, H.; MA, L. Developing Fragrant Early indica TGMS Line with Blast Resistance by Using CRISPR/Cas9 Technology. Chinese Journal of Rice Science 2022, 36, 248–258. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Z.; Li, Y.; Xu, C.; Xia, H.; Zhuang, H.; Sun, S.; Guo, M.; Yan, C. Rapid Improvement of Rice Eating and Cooking Quality through Gene Editing toward Glutelin as Target. J Integr Plant Biol 2022, 64, 1860–1865. [Google Scholar] [CrossRef]
- Shen, L.; Hua, Y.; Fu, Y.; Li, J.; Liu, Q.; Jiao, X.; Xin, G.; Wang, J.; Wang, X.; Yan, C.; et al. Rapid Generation of Genetic Diversity by Multiplex CRISPR/Cas9 Genome Editing in Rice. Sci China Life Sci 2017, 60, 506–515. [Google Scholar] [CrossRef]
- Zhou, W.; Tian, X.; Ren, Y.; Wei, X.; Gao, Y.; Xie, L.; Liu, H.; Bu, Q.; Li, X. Breeding of Early-maturatity and Fragrant Rice via CRISPR/Cas9 Mediated Genome Editing[J]. Soils and Crops, 2017; 6, 146–152. [Google Scholar] [CrossRef]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.-L.; et al. OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas Nuclease Specificity Using Truncated Guide RNAs. Nat Biotechnol 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-Throughput Profiling of off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat Biotechnol 2013, 31, 839–843. [Google Scholar] [CrossRef]


| Target | Position | Strand | GC(%) | Region | Potential off-target sites (Max score) |
Pairing with sg RNA (>= 8 nt) |
|---|---|---|---|---|---|---|
| Hd2 | 738 - 757 | + | 55 | CDS1 | 0.074 | None |
| Ghd7 | 366 - 385 | + | 60 | CDS2 | 0.25 | None |
| DTH8 | 986 - 1005 | + | 55 | CDS1 | 0.238 | None |
| Pi21 | 503 - 522 | + | 55 | CDS1 | 0.133 | None |
| Badh2 | 499 - 518 | + | 60 | CDS2 | 0.083 | None |
| Gene | No. of plants | Number of mutant strains | Mutation rate |
|---|---|---|---|
| Hd2 | 30 | 26/30 | 86.66% |
| Ghd7 | 30 | 25/30 | 83.33% |
| DTH8 | 30 | 25/30 | 83.33% |
| Pi21 | 30 | 26/30 | 86.66% |
| Line No.\Gene | Hd2 | Ghd7 | DTH8 | Pi21 | Badh2 | Flowering after sowing (Days) |
|---|---|---|---|---|---|---|
| JH212 | WT | WT | WT | WT | WT | 88.8±3.5 |
| JH-C15 | Allele1: +1bp | WT | Allele1: -4bp | Allele3: +1bp | WT | 57.38±2.75** |
| JH-C17 | Allele1: +1bp | Allele1: +1bp | WT | Allele6: -2bp | WT | 53.22±2.75** |
| JH-C18 | Allele6: -46bp | Allele1: +1bp | Allele1: -1bp | Allele1: +1bp | WT | 61.4±2.25** |
| JH-C19 | Allele2: +1bp | Allele7: -28bp | WT | Allele2: +1bp | WT | 56.55±3.25** |
| JH-C22 | Allele2: +1bp | Allele1: +1bp | WT | Allele9: -19bp | WT | 56.55±4.25** |
| JH-C23 | Allele3: +1bp | Allele5: -5bp | WT | Allele2: +1bp | WT | 50.5±2.75** |
| JH-C24 | Allele3: +1bp | Allele4: -4bp | WT | Allele9: -19bp | WT | 55.5±3.25** |
| JH-C26 | Allele1: +1bp | Allele5: +1bp | WT | Allele6: -2bp | WT | 50.5±2.25** |
| JH-C27-5 | Allele2: +1bp | WT | WT | Allele7: -4bp | WT | 51.55±4.25** |
| JH-C27-2 | WT | Allele7: -28bp | WT | Allele7: -4bp | WT | 52.55±3.25** |
| JH-C28-5 | Allele4: +1bp | Allele2: +1bp | WT | Allele8: -7bp | WT | 60.55±2.75** |
| JH-C31 | Allele5: -5bp | WT | WT | Allele4: -1bp | WT | 59.07±2.75** |
| JH-C32 | Allele2: +1bp | WT | WT | Allele4: -1bp | WT | 60.55±3.25** |
| JH-C34 | Allele5: -5bp | Allele5: -5bp | WT | Allele5: -1bp | WT | 63.55±2.75** |
| JH-C37 | Allele3: +1bp | Allele3: -3bp | WT | Allele2: +1bp | WT | 59.55±3.25** |
| JH-C38 | Allele2: +1bp | Allele8: -45bp | WT | Allele9: -19bp | WT | 55.5±3.75** |
| JH-C40 | Allele4: +1bp | Allele3: -3bp | WT | Allele4: -1bp | WT | 60.55±2.75** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





