Introduction
Genomic Selection (GS)
The traditional breeding methods select excellent domestic animal by estimating breeding value (EBV) based on phenotypic information and genetic relationships. The development of sequencing technology, bioinformatics analysis and molecular breeding methods enabled the selection index method and the best linear unbiased prediction to combine with genomic information of animals for more accurate calculation of EBV. As the current state of art, the marker-assisted selection (Soller et al., 1983) and GS (Meuwissen et al., 2001) are the major breeding techniques using genomic information of animals to breeding. Marker-assisted selection (MAS) by adding the target trait affecting molecular markers, such as microsatellite, single nucleotide polymorphisms (SNPs), insertions and deletions, and quantitative trait loci (QTL), to improve the genetic evaluation (Freking et al., 2018; Imran et al., 2020). GS estimates the Genomic Estimated Breeding Value (GEBV) of individuals by whole-genome marker and individual phenotype information. Compared with MAS, GS uses almost all the markers from the whole-genome for EBV estimation, which provides more variation information and solves problems such as the difficulty in detecting genes or markers linked to QTL, it also greatly improves the accuracy of prediction. There are many research on GS of livestock and major progress has been made. Moreover, GS has become a routine breeding method in dairy cattle immediately. With the advancement and improvement of molecular breeding technology and the reference genomes of sheep and goat, GS has become more prevalent in their breeding programs. This will further drive the progress of genetic breeding for sheep and goat.
The Principle of GS
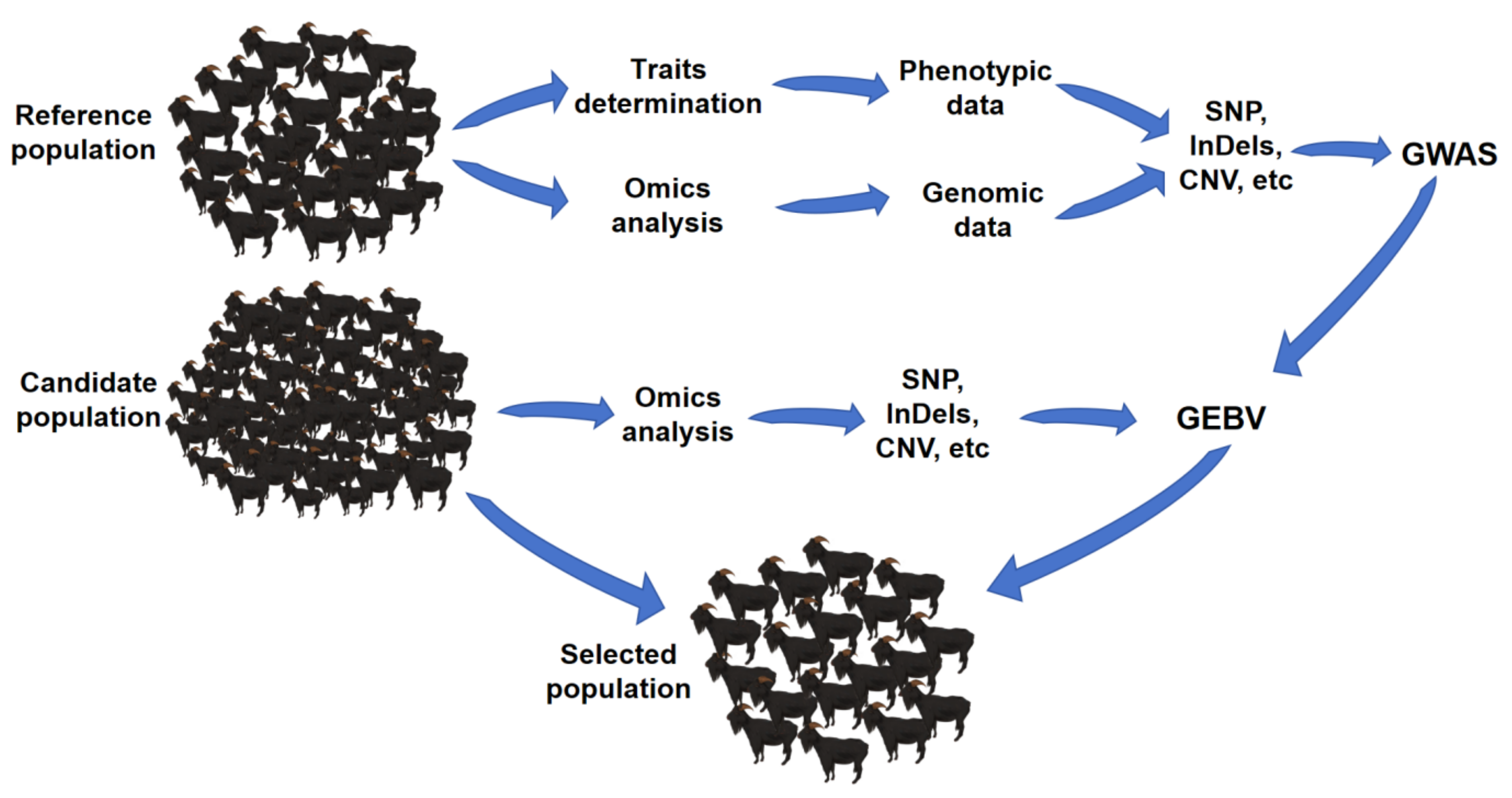
GS, which uses whole-genome information to select superior livestock, was first proposed in 2001 by Meuwissen (Meuwissen et al., 2001). GS estimates the effect value of each molecular marker or chromosome segment by using the whole-genome marker information and phenotype information of individuals. By integrating all effect values, the EBV of an individual with genomic information, also known as the GEBV, could be obtained. The specific procedure of GS include: first, a reference population with both genotype and phenotype information needs to be constructed; then, the genotype information is used to obtain whole-genome marker information; to find the best calculation model, the effective value of each marker was calculated by different models; following the best model is used to calculate the individual GEBV of the candidate population by its genomic information; finally, the excellent individuals with the genomic information and the higher GEBV are selected. The technical route of GS is shown in
Figure 1
Compared to other breeding methods, GS using genomic information is less dependent on phenotypic information, so it can use the genomic information of candidate populations for early selection to shorten the generation interval (Gore et al., 2021), and it is beneficial to the selection of limiting traits and low heritability traits, further reduce breeding costs to a certain extent. GS assumes that there is linkage disequilibrium (LD) between the quantitative trait locus QTLs and at least one molecular marker, then identifies all QTLs that affect the target trait to achieve more accurate predictions of individual breeding values. Selection of candidate individuals by estimating GEBV can distinguish the differences among individuals and improve the intensity of selection (Song et al., 2021). GS is based on high-throughput sequencing technologies, such as SNP chip, whole-genome sequencing and simplified genome sequencing and so on. With the development of sequencing technology, the accuracy of whole-genome sequencing and SNP chip has been improved, and the cost has been gradually reduced, which has been more applied in breeding.
Methods for GEBV Estimation
The essence of GS lies in the estimation of genomic breeding value, which is commonly achieved through the methods of Best Linear Unbiased Prediction (BLUP) and Bayesian inference. BLUP assumes that all markers are micro-effect markers while all markers conform to a normal distribution with the same variance. The traditional BLUP uses the relatedness matrix (A) to predict the EBV of an individual, and methods using other relationship matrices for estimation have emerged based on BLUP. Ridge, Regression Best Linear Unbiased Prediction (RRBLUP) included individuals as random effects and calculated marker effects using matrix A which is constructed by genetic information from the reference and predicted populations. Genomic Best Linear Unbiased Prediction (GBLUP) replaces relatedness (matrix A) with genomic marker information (matrix G) and uses both genomic marker information and phenotypic information of reference population to estimate the individual GEBV of candidate group. Based on GBLUP, the single-step GBLUP (ssGBLUP) uses the integrated information of matrix A and matrix G to construct matrix H, it directly estimates the GEBV of individual of candidate group in the set of the mixed model system of equations. According to the different variances of the markers, the Bayes is divided into BayesA, BayesB, BayesC, BayesCpπ, Bayes R, and so on (Wu et al., 2021). Using Bayes is more difficult and time-consuming than BLUP. At present, GBLUP and ssGBULP are commonly used to estimate the GEBV of individual.
Influential Factors of the GS Accuracy
Although GS has certain advantages in various breeding methods, its accuracy is also affected by some factors:
Size of the reference population. The size of the reference population had a great impact on the accuracy of GS. With the larger size of the reference population, the GEBV of individuals in the candidate population will be more accurate (Song et al., 2019). For example, in GS of goat, Carillier (Carillier et al., 2014) constructed different levels of reference population (20, 705 bucks and 1946 does) for estimating the GEBV using Alpine dairy goats and Saanen dairy goats, respectively, in order to resolve the reference population size. The results showed that the accuracy of individual GEBV of the candidate population increased from 2% to 31% with the increase of the reference population size. Meanwhile, the structure of the reference population also has a certain impact on the accuracy of GEBV. Raoul (Raoul et al., 2021) explored the influence of reference population with different population structure on the accuracy of GEBV, and found that different sex ratio structure of reference population had certain influence of GEBV. Wei (Wei et al., 2022) investigated the effect of different population sizes on the accuracy of GS in Chinese pig and found that genome prediction accuracy was positively correlated with the size of reference population.
Marker density and the degree of LD between markers. Marker density affects the accuracy of GS to a large extent. Increasing marker density can improve the degree of LD between markers. Therefore, the higher marker density, the more accuracy of GS and better selection effect. In addition, some studies have shown that grouping SNPs based on regional LD can effectively reduce the adverse effect of the uneven distribution of LD on the genome, and greatly improve the efficiency of high-density SNP data for genome prediction and heritability estimation (Ren et al., 2022).
Generation intervals. With the increases of the generation interval between the reference population and the candidate population, LD between markers and QTLs can change. The smaller of the generation interval and the higher the correlation between individuals in the reference population and the candidate population, the higher accuracy of GS. After 3-4 generations, it is necessary to re-estimate the effect of markers using a new or improved reference population (Wu et al., 2021).
Heritability of the target traits. Compared with traditional breeding methods, GS is more effective of selecting traits with low heritability. However, previous studies have shown that the heritability of target traits also affects the accuracy of GS. The higher the heritability of target traits, the higher the accuracy of GS (Cleveland, et al., 2012). To explore the influence of trait heritability on GS effect, Zheng (Zheng et al., 2022) used different models to conduct GS on traits with different heritability (h2=0.1, 0.3, 0.5) of cattle. The results showed that the selection effect of traits with high heritability was higher than that of traits with low heritability under different models and conditions.
Methods and models for estimating GEBV. As mentioned above, the estimation methods of GEBV mainly include BLUP and Bayes. Different traits have their own applicable models, and the suitability of models can directly affect the accuracy of GS. Baloche (Baloche et al., 2014) compared the accuracy of three different models (ssGBLUP, pseudo-BLUP and pseudo-ssGBLUP) using SNP chip and a reference population of 2, 892 Lacaune sheep. The results found that the accuracy of GS in each model increased by 0.1-0.2 compared with that of their parents, and conventional ssGBLUP had the highest accuracy in this population. Yan (Yan et al., 2022) conducted a simulated study of GS on body weight and wool fiber diameter in Inner Mongolia Cashmere goat, and GBLUP, ssGBLUP, Bayes A, Bayes B, Bayesian R and Bayesian LASSO methods were used to calculate the GEBV, respectively. The results showed that the most suitable model needs to be selected in combination with the current conditions, and the most suitable model that enable GEBV with the highest accuracy.
Research Progress of GS in Sheep and Goat Breeding
Breeding is important content of animal husbandry. GS has good applications in livestock breeding, especially in cattle breeding (Wiggans et al., 2022). Although the studies of GS on the economic traits in sheep and goat are still under a developmental stage, there has been many attentions paid towards it, and the platform and the system of GS study has been gradually increasing.
Research Progress of GS in Meat Traits of Sheep and Goat
China is a large producer and consumer of sheep and goat products. In 2022, the total production of sheep and goat meat in China reached 5.250 million tons, ranking first in the world for 33 consecutive years (National Bureau of Statistics of China. 2022). The improvement of meat quality are conducive to the development of sheep and goat industry. More GS studies were conducted on meat quality traits in sheep and goat than on other traits. Werf (Werf et al., 2009) conducted a simulation study on GS using a selection index method for three carcass traits of Merino sheep, which included growth rate, fat, and muscle conformation. The results showed that the application of GS in Merino sheep breeding could increase mutton yield index by 32%, and further promote the genetic research of sheep. Daetwyler (Daetwyler et al., 2010) estimated the GEBV of weight, loin-eye area and fat traits in Australian sheep using 7, 180 reference population of multi-breed sheep with phenotypic records (Merino sheep, etc.), and found that the accuracy of prediction for each trait ranged from 0.07 to 0.57. At the same time, the accuracy of prediction was improved by increasing the reference population. Slack-Smith (Slack-Smith et al., 2010) used the Bayes B to perform GS on three traits (carcass weight, loin-eye area and intramuscular fat) of meat purpose sheep and goat, and the prediction accuracy of GEBV of the three traits were 0.45, 0.72 and 0.32, respectively. Based on the Annual Genetic Gain (AGG) of the meat sheep breeding program, Shumbusho (Shumbusho et al., 2013.) optimized the GS strategy for mutton, milk and maternal traits. The accuracy of genome prediction for meat sheep, dairy sheep and dairy goat in a population of 2000 individuals was improved 17.9%, 51.7% and 26.2%, respectively, compared with the traditional breeding methods. Moghaddar (Moghaddar, et al., 2014) used a multi-species reference population to estimate GEBV of 26 traits, including muscle, weight, fat, wool traits and so on, in Australian sheep. The results showed that the accuracy of GEBV for body weight was 0.11-0.27 for Poll Dorset and White Suffolk sheep, and 0.25-0.63 for Border Leicester and Merino sheep. Brito (Brito et al., 2017) used a 15K SNP chip to estimate GEBV for many traits, such as carcass traits and mutton quality traits, in New Zealand sheep of a large reference population with 14, 845 individuals. The results showed that the accuracy of prediction of GEBV for carcass traits and mutton quality traits were 0.28±0.09-0.55±0.05 and 0.21±0.07-0.36±0.08, respectively. Lillehammer (Lillehammer et al., 2020) conducted GS simulation studies under different models for carcass traits, growth traits and maternal traits of Norwegian white sheep and found that GS increased the genetic benefits of these traits by 18% to 20%. Scholtens (Scholtens et al., 2020) used Bayes C to estimate the GEBV of milk yield, protein content, and somatic score of New Zealand dairy goat, and the average prediction accuracy was 0.34 to 0.43, which was more reliable than the traditional BLUP. Ashraf (Ashraf et al., 2022) used GBLUP and Bayes R, Bayes A, Bayes B, Bayes L to perform GS and compared eight different traits of Soay Sheep, such as body weight, length of front and rear legs, coat color, patterns and so on. The results showed that the prediction models based on genome had high accuracy and high correlation with each other. Combined with pedigree analysis, Hunter (Hunter et al., 2022) performed genomic prediction of adult body weight in Soay Sheep, and found that using genome prediction to study the microevolution of wild populations can eliminate the need for pedigree data, which is expected to open up new research systems. Oliveira (Oliveira et al., 2022) used ssGBLUP to predicted GEBV for the traits of birth weight, weaning weight, carcass weight and fat in Norwegian white sheep and New Zealand sheep. The results showed that the accuracy of the GEBV was 0.13-0.44 in just Norwegian white sheep, 0.06-0.15 in just New Zealand sheep, and 0.10-0.41 in the mixed population of both breeds. Moghaddar (Moghaddar et al., 2022) performed genomic prediction of body weight and mutton quality traits in Duper sheep by GBLUP, and the accuracy of genomic prediction for different traits ranged from 20.0 to 30.50. Massender (Massender et al., 2022) predicted the GEBV of eight traits, in 5, 158 Alpine dairy goat and 2, 342 Saanen dairy goat, and the results showed that the theoretical accuracy of GEBV for all eight traits increased 32%-37% and 40%-41% on average compared with the traditional genetic evaluation, and the accuracy of GEBV estimation by multiple breeds was significantly higher than that by single breeds. With the in-depth development of GS, more studies are utilizing multi-breed approaches to further estimate GEBV for meat quality traits in sheep and goat. These research of GS use multi-breed for analysis have demonstrated improved accuracy compared to single-breed approaches. The utilization of multi-breed joint predictions may potentially serve as a pathway for further enhancing the accuracy of GS in the future.
Research Progress of GS in Fibre Traits of Sheep and Goat
Wool, as one of the most important raw materials in the textile industry, is also one of the first natural fiber to be used. The quality of wool has significant impact on the industry of sheep and goat and the textile industry, and is affected by the length, fineness, strength, crimp and so on (Henkel et ai., 2019). At present, some progress has been made in research of GS on wool. Daetwyler (Daetwyler et al., 2010) used a reference population of 7, 180 multi-breed sheep with phenotypic records (Merino sheep, etc.) to estimate GEBV for many traits, such as wool weight, fiber diameter, and staple fiber strength, in Australian sheep. The results showed that the predication accuracy of GBLUP, which is a representative method of GS, was slightly higher than that of BayesA in wool traits of sheep, and the prediction accuracy of GS in estimating the GEBV of Merino sheep wool traits was 0.15-0.79. Moghaddar (Moghaddar et al., 2014) used a multi-breed reference population to estimate the GEBV about the quantity of wool in Australia by GBLUP and found that the average accuracy of the GEBV about the quantity of wool was high, between 0.33 and 0.75. Dodds (Dodds et al., 2014) applied GS to a reference population of 13, 364 sheep of different breeds (Romney, Coperworth and Pollendale) and found that the accuracy of genomic prediction improved to 0.26. Bolormaa (Bolormaa, et al., 2017) used Bayes R and GBLUP to estimate the GEBV of wool quality, yield and turbid degree in 5, 726 Merino sheep and crossbreed Merino sheep, and found that the average accuracy of genomic prediction of wool traits was about 0.22. Moghaddar (Moghaddar et al., 2019) used GBLUP and Bayes RC model to estimate GEBV for eight economic traits related to meat quality and wool production in Australian sheep. In purebred and crossbred populations, the average genomic estimated breeding values of GBLUP for each trait were 0.083 and 0.073, while those of Bayes RC were 0.102 and 0.087. Wei (Wei et al., 2020) used PBLUP and ssGBLUP to estimate the EBV and GEBV of wool traits in Merino sheep, and the results showed that the average accuracy of GEBV increased by 3.016%-11.736%, compared with EBV. Wang (Wang. 2021) used a reference population of 1, 920 individuals to estimate the GEBV of villi yield, villi length, villi fine traits and villi length of Inner Mongolia Cashmere goat, and found the prediction accuracy of villi yield, villi length and villi fine was about 0.78, 0.79-0.80, and 0.55-0.56, respectively. The prediction accuracy of hair length was about 0.78-0.82, and the prediction accuracy of different models was follows the trend of ABLUP<GBLUP<ssGBLUP, and it follows that ssGBLUP has higher accuracy and advantage. Zhu (Zhu et al., 2021) used a reference population of 11, 500 Merino sheep from seven groups, and used GBLUP to estimate GEBV for six traits, including hair length, net hair weight, average coarse fiber density, coefficient of variation of average coarse fiber density, short-fiber strength and wool elongation. The prediction accuracy of the six traits ranged from 0.28 to 0.60. Yan (Yan et al., 2022) simulated GS study on wool fiber diameter of Inner Mongolia Cashmere goat, and found that GS was more accurate than conventional selection methods. Those studies on GS of wool traits have shown that the accuracy of GS is higher than that of traditional selection methods, and the accuracy of GBLUP in GS wool traits of sheep and goat is higher than that of Bayes A. Studies in cashmere goats also showed that ssGBLUP, which combines phenotypic, genealogical and genomic information, is more accurate than GBLUP (Daetwyler et al., 2010; Moghaddar et al., 2014). Moreover, in GS studies on wool traits of sheep and goat, reference population of multi-breed have been established for selection.
Research Progress of GS in Dairy Traits of Sheep and Goat
The milk of Goat and Sheep contains a variety of elements required by the human body, has a nutrients ratio that is more suitable for human absorption, and is easier to digest than cow’s milk, and is hypoallergenic (Hammam et al., 2022). Dairy trait is also an important economic trait of sheep and goat, and GS research for dairy traits, such as milk yield, milk protein quantity and percentage, and milk fat quantity and percentage, have also been studied to some extent. Duchemin (Duchemin et al., 2012) used different methods such as BLUP, Bayes Cπ, partial least squares (PLS), and sparse PLS to estimate the GEBV and compare their accuracies for 14 milk traits, such as milk yield, fat content and somatic cell score in sheep. Duchemin’s study found that there was no significant difference in the accuracy of prediction among different methods using genomic information, and the accuracy of genomic estimation was improved by 0.4-0.6 compared to conventional BLUP. Carillier (Carillier et al., 2013) performed a genomic assessment of milk production, somatic cell count, and udder traits in Alpine and Saanen dairy goat using SNP chip, with accuracy ranging from 36% to 53%. Carillier (Carillier et al., 2014) used ssGBLUP to estimate GEBV for breast traits in 677 French alpine and Saanen dairy goat, the selection accuracy improved from 22% to 37% by GS. Mucha (Mucha et al., 2015) used 50k SNP chip of goat to estimate the GEBV of 14, 453 hybrid dairy goats of British Togenberg, Alpine dairy goat and Saanen dairy goat, using SNPBLUP and ssGBLUP models to estimate their GEBV of milk yield, lactation duration and other lactation traits. The results showed that ssGBLUP was more accurate in this population, and the accuracy of GEBV estimated by ssGBLUP was 0.61. Antonio (Antonio et al., 2018) used BLUP and ssGBLUP separately to estimate the EBV and GEBV of 50, 649 Florida goat for dairy traits such as milk yield, and found that the reliability of ssGBLUP increased by 1.06% relative to BLUP. Teissier (Teissier et al., 2018) used the 50k SNP chip of goat to estimate the GEBV of protein content in the milk of 2, 955 French dairy goats by using ssGBLUP. The results showed that the accuracy of prediction of ssGBLUP using genomic information increased by 5%-7% compared to the traditional prediction method. The accuracy of three WssGBLUP was 6%, higher than that of the unweighted ssGBLUP. Cesarani (Cesarani et al., 2019) used BLUP, GBLUP, and ssGBLUP to estimate EBV and GEBV for milk fatty acid content in sheep. The study showed that the accuracy of prediction using genomic information was higher than that using traditional EBV estimation methods, and the accuracy of prediction was higher in the older group. Teissier (Teissier et al., 2020) used ssGBLUP and weighted ssGBLUP (WssGBLUP) of haplotypes to perform genomic prediction for five milk production traits, five breast shape and somatic score in French dairy goat, and found that haplotype-based models could improve the accuracy of genomic prediction of some traits. Sousa (Sousa et al., 2021) investigated the optimal GS model for milk production and composition traits in Brazilian Saanen dairy goat. Using GBLUP, Bayes Cπ, and Bayesian LASSO for estimated GEBV in the reference population, and found that the accuracy of the three methods was consistent, while GBLUP had better selection performance and lower cost in this reference population. Massender (Massender et al., 2022) used ssGBLUP to perform GS for protein and fat content in milk from 1, 707 Canadian Alpine and Saanen dairy goats, and the accuracy of GEBV increased by 12%-16% compared to traditional selection accuracy. Marina (Marina et al., 2022) used a low-density SNP chip to estimate EBV of milk coagulation properties and cheese yield-related traits in 2, 020 dairy goats using the models of BLUP, SNPBLUP, and ssGBLUP. The results showed that the prediction accuracy of ssGBLUP was improved by 0.19-0.44 after considering genomic information compared to BLUP. In the current studies on the GS for dairy traits of sheep and goat, several studies have shown that ssGBLUP has relatively superior prediction accuracy in groups with different numbers and structures (Cesarani et al., 2019; Carillier et al., 2014; Marina et al., 2022).
Research Progress of GS in Reproductive Traits of Sheep and Goat
Reproductive traits are very important economic traits in sheep and goat, and their performance directly affects the development of the sheep and goat industry. Most reproductive traits have low heritability and are difficult to assess, however, GS can improve the selection efficiency for traits with low heritability. Pickering (Pickering et al., 2013) performed GS on 4, 237 Romney sheep and estimated the GEBV for reproductive traits. The results showed that the prediction accuracy of GEBV of reproductive traits ranged from 0.16 to 0.52. The application of GS shortened the generation interval, and the estimated genetic gain had the potential to increase up to 84%. Granleese (Granleese et al., 2015) simulated GS in sheep under optimal contribution selection, and found that the MOET and MOET+JIVET programs increased the genetic gain by 38%-76% and 51%-81%, respectively. Newton (Newton et al., 2017) used a genomic information-based simulation to model the impact of different age structures of sheep populations on reproductive performance in Australian sheep. The results showed that the availability of ram and ewe age as well as male genomic information significantly affected the predicted genetic gain, and genetic progress was unlikely to be adversely affected at fertility rates of 10% or more. Bolormaa (Bolormaa et al., 2017) used GBLUP to conduct GS on reproductive traits such as lambing number, litter size, and weaning weight in 5, 340 Merino sheep, and resulted with an average accuracy of 43.0 based on parent-offspring trait deviations and 17.0-61.0 for random cross-validation of GEBV. The highest accuracy of prediction based on lambing number and litter size among the reproductive traits studied were also obtained to be between 41.0 and 54.0. Moghaddar (Moghaddar et al., 2017) using GBLUP and Bayes R to estimate GEBV for reproductive traits in Australian Merino sheep, showed that the accuracy of genomic prediction increased by 1.0%-12.0% in population with distant genetic relationships, with an average improvement of 5.2%, and the two models had similar predictive performance. This was done to improve the accuracy of genomic prediction for reproductive traits in Australian Merino sheep. Lillehammer (Lillehammer et al., 2020) conducted a simulation experiment on maternal traits of Norwegian White Sheep under different GS models. The results showed that the implementation of GS increased the genetic gain of maternal traits in Norwegian White Sheep by 65%-77%. Araujo (Araujo et al., 2023) used ssGBLUP based on SNPs and haplotypes to estimate GEBV for some traits, such as birth weight, weaning weight, adult weight, and litter size, in Lacaune sheep. And Araujo found that using genomic information provided similar or higher levels of predictive and theoretical accuracy compared to traditional methods across all traits, and reduced the dispersion of GEBV for Lacaune sheep reproductive traits. Due to the complexity of reproductive traits, there is currently relatively limited research on GS of sheep and goat reproductive traits, and further in-depth research is needed. Compared with other traits, there are relatively few studies on the reproductive traits of GS in sheep and goat, which may be due to the low heritability of the reproductive traits.
Research Progress of GS in Other Economic Traits of Sheep and Goat
In addition to the above traits, GS has also been used in studies of other traits of sheep and goat. Phua (Phua et al., 2014) conducted GS research on New Zealand sheep and dairy industry to reduce the impact of facial eczema (Pithomycotoxicosis), and the best genome prediction accuracy obtained from the study was 0.38. Rowe (Rowe et al., 2014) performed a GS study on enteric methane emissions in 1, 872 New Zealand sheep with GBLUP, and the genomic prediction accuracy was 0.37. Duijvesteijn (Duijvesteijn et al., 2018) conducted a prediction of horn type in Merino sheep using traditional pedigree estimation and genomic information. The accuracy of prediction of horn type was 0.323 for females and 0.725 for males using pedigree, and 0.713 for females and 0.620 for males using genomic information. In order to increase the genetic resistance of sheep to gastrointestinal nematode infection, Dos (Dos et al., 2021) evaluated the genetic resistance using both traditional evaluation methods and GS. The results showed that genomic models provided more accurate estimates of EBV compared to pedigree-based models. Carracelas (Carracelas et al., 2022) used ssGBLUP to obtain GEBV for increasing the genetic resistance of sheep to gastrointestinal nematodes, and selected relevant genomic regions associated with resistance in the Coopworth breed.
Conclusions
Currently, GS has been widely applied in the livestock industry, particularly in dairy cattle and pigs, and achieved significant improvement on the accuracy of predictions compared to traditional methods. However, the research on GS in sheep and goat is not yet complete and requires further investigation. Currently, the sheep and goat industry is facing the bottleneck of slow growth and low reproductive efficiency. Compared to other livestock, the foundation of the sheep and goat breeding is relatively weak, with poor breeding infrastructure, outdated breeding methods, and inadequate fundamentals, such as performance testing and genetic evaluation. Therefore, it is necessary to continue to improve the ability of breeding independent innovations. Improvements in the speed and accuracy of genome sequence as well as reductions in cost, have made GS is expected to become a routine method in sheep and goat breeding. GS has become a routine method in cattle breeding, with official genomic evaluation platforms established in many countries and regions for regular genomic evaluation (Wiggans et al., 2017). However, the application of GS in sheep and goat breeding still needs further development. Compared to other livestock, sheep and goat breeding has a relatively small scale, and integrating GS into sheep and goat breeding also faces some difficulties and challenges, such as the reduction of accuracy due to long-term selection, incomplete data, and the lack of large and excellent reference population. It is necessary to gain a deep understanding of the potential factors and regularly update the prediction models.
In addition, recent studies have also shown an increasing trend in conducting GS research on sheep and goat by utilizing joint analysis of multi-breed as multiple beneficial factors have been shown (
Table 1). Multiple studies have indicated that utilizing joint GS analysis with multi-breed significantly improves the accuracy of GEBV compared to studies with merely single breed. The substantial number of multi-breed GS studies conducted for both meat and fiber traits of sheep and goat shows a clear evidence of such trend. Particularly, it was observed that the improvement in GEBV estimation by incorporating multiple breeds is more significant compared to single-breed GS for meat quality traits in sheep and goats (Moghaddar, et al., 2014; Massender et al., 2022; Oliveira et al., 2022). This finding holds significant guidance for future GS research, suggesting that combining multiple breeds for the same trait in GS analysis could potentially elevate the accuracy of GS to a higher level, especially for meat quality traits Furthermore, the enhancement of precision for GS by combining multiple varieties of breeds for the same trait shows promising and exciting prospects for advancing selective breeding programs in the sheep and goat industry. In the context of diverse population structures and calculation methods, the accuracy of GS may vary considerably.
Table 1 presents a overview of studies involving the selection of phenotypic traits in sheep and goat, using in multi-breed or different methods. These selection methods using genomic information show little difference in many studies, but for those studies about different selection methods, ssGBLUP shows superior prediction compared with other methods. And in some studies the prediction accuracy of WssGBLUP is higher than that of ssGBLUP, thereby providing valuable guidance for prospective GS studies in sheep and goat. Furthermore, it is noteworthy that gender ratios and group structures have also been identified as potential factors influencing the accuracy of GS predictions. This insight opens up new avenues of exploration for future GS studies in this field. As such, understanding and considering these various factors can significantly contribute to the refinement of genomic selection techniques for sheep and goat.
GS, as it becomes a popular choice to estimate GEBV, can also obtain benefits from other interdisciplinary development, such as the advancement of data analysis tools, the development of machine learning and deep learning for optimizing models, gene editing, transgenics, and phenomics. This interdisciplinary cooperation can enable GS to be more effective in improving selection efficiency and breeding progress. By utilizing and developing of GS, it is promising to find the best model and configuration for various traits, which will strengthen the efficiency and accuracy of selection, accelerate the genetic progress, and provide a foundation for breeding selection, purification and expansion of sheep and goat.
List of Abbreviations
The full name of abbreviations were listed in
Table 2.
Funding
This work was financially supported by the National Key Research and Development Program of China (No.2022YFD1300202), The National Natural Science Foundation of China (No. 32372834), the Collection, Utilization and Innovation of Germplasm Resources by Research Institutes and Enterprises of Chongqing, China (cqnyncw-kqlhtxm) and Chongqing Modern Agricultural Industry Technology System (CQMAITS202313), the Fundamental Research Funds for the Central Universities (7110100870).
Authors Contributions
Linyun Zhang performed the data curation and writing; Yixin Duan performed the data curation; Naiyi Xu and Yongju Zhao performed the wrting-review and editing.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (No.2022YFD1300202), The National Natural Science Foundation of China (No. 32372834), the Collection, Utilization and Innovation of Germplasm Resources by Research Institutes and Enterprises of Chongqing, China (cqnyncw-kqlhtxm) and Chongqing Modern Agricultural Industry Technology System (CQMAITS202313), the Fundamental Research Funds for the Central Universities (7110100870).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of Interest
The authors declare that they have no conflicts of interest to this work.
References
- Antonio Molina, Eva Muñoz, Clara Díaz, Alberto Menéndez-Buxadera, Manuel Ramón, Manuel Sánchez, María J. Carabaño, Juan M. Serradilla (2018) Goat genomic selection: Impact of the integration of genomic information in the genetic evaluations of the Spanish Florida goats. Small Ruminant Research: 163.
- Araujo AC, Carneiro PLS, Oliveira HR, Lewis RM, Brito LF. SNP- and haplotype-based single-step genomic predictions for body weight, wool, and reproductive traits in North American Rambouillet sheep (2023) Anim Breed Genet: 140(2):216-234. [CrossRef] [PubMed]
- Ashraf B, Hunter DC, Bérénos C, Ellis PA, Johnston SE, Pilkington JG, Pemberton JM, Slate J (2022) Genomic prediction in the wild: A case study in Soay sheep. Mol Ecol: 31(24):6541-6555. [CrossRef] [PubMed]
- Bolormaa S, Swan AA, Brown DJ, Hatcher S, Moghaddar N, van der Werf JH, Goddard ME, Daetwyler HD (2017) Multiple-trait QTL mapping and genomic prediction for wool traits in sheep. Genet Sel Evol: 49(1): 62.
- Bolormaa S, Brown D J, Swan A A, van der Werf J H J, Hayes B J, Daetwyler H D (2017) Genomic prediction of reproduction traits for Merino sheep. Animal genetics: 48(3).
- Brito Luiz F, Clarke Shannon M, McEwan John C, Miller Stephen P, Pickering Natalie K, Bain Wendy E, Dodds Ken G, Sargolzaei Mehdi, Schenkel Flávio S (2017) Prediction of genomic breeding values for growth, carcass and meat quality traits in a multi-breed sheep population using a HD SNP chip. BMC genetics: 18(1).
- C. Carillier, H. Larroque, I. Palhière, V. Clément, R. Rupp, C. Robert-Granié (2013) A first step toward genomic selection in the multi-breed French dairy goat population. Journal of Dairy Science: 96(11).
- Carillier C, Larroque H, Robert-Granié C (2014) Comparison of joint versus purebred genomic evaluation in the French multi-breed dairy goat population. Genet Sel Evol:29;46(1):67. [CrossRef] [PubMed]
- Carracelas B, Navajas EA, Vera B, Ciappesoni G (2022) Genome-Wide Association Study of Parasite Resistance to Gastrointestinal Nematodes in Corriedale Sheep. Genes (Basel): 27;13(9):1548. [CrossRef] [PubMed]
- Cesarani A, Gaspa G, Correddu F, Cellesi M, Dimauro C, Macciotta NPP (2019) Genomic selection of milk fatty acid composition in Sarda dairy sheep: Effect of different phenotypes and relationship matrices on heritability and breeding value accuracy. Dairy Sci: 102(4):3189-3203. [CrossRef] [PubMed]
- Cleveland MA, Hickey JM, Forni S (2012) A common dataset for genomic analysis of livestock populations. G3 (Bethesda): 2(4):429-35. [CrossRef] [PubMed]
- Daetwyler H. D., Hickey J. M., Henshall J. M., Dominik S., Gredler B., van der Werf J. H. J., Hayes B. J. (2010) Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Animal Production Science: 50(12).
- de Sousa DR, do Nascimento AV, Lôbo RNB (2021) Prediction of genomic breeding values of milk traits in Brazilian Saanen goats. Anim Breed Genet: 138(5):541-551. [CrossRef] [PubMed]
- Dodds K G, Auvray B, Lee M, et al. (2014) Genomic Selection in New Zealand Dual Purpose Sheep. 10 Th World Congress of Genetics Applied to Livestock Production.
- Dos Santos GV, Santos NPDS, Figueiredo Filho LAS, Britto FB, Sena LS, Torres TS, Carneiro PLS, Sarmento JLR (2021) Comparison of genetic parameters and estimated breeding values for worm resistance in meat sheep obtained using traditional and genomic models. Trop Anim Health Prod: 23;53(2):283. [CrossRef] [PubMed]
- Duijvesteijn N, Bolormaa S, Daetwyler HD, van der Werf JHJ (2018) Genomic prediction of the polled and horned phenotypes in Merino sheep. Genet Sel Evol: 22;50(1):28. [CrossRef] [PubMed]
- Freking Brad A, King David A, Shackelford Steven D, Wheeler Tommy L, Smith Tim P L (2018) Effects and interactions of myostatin and callipyge mutations: I. Growth and carcass traits. Journal of animal science:96(2).
- G. Baloche, A. Legarra, G. Sallé, H. Larroque, J.-M. Astruc, C. Robert-Granié, F. Barillet (2014) Assessment of accuracy of genomic prediction for French Lacaune dairy sheep. Journal of Dairy Science: 97(2).
- Gore D.L.M., Okeno T.O., Muasya T.K., Mburu J.N. (2021) Improved response to selection in dairy goat breeding programme through reproductive technology and genomic selection in the tropics. Small Ruminant Research (prepublish).
- Granleese T, Clark SA, Swan AA, van der Werf JH (2015) Increased genetic gains in sheep, beef and dairy breeding programs from using female reproductive technologies combined with optimal contribution selection and genomic breeding values. Genet Sel Evol: 14;47(1):70. [CrossRef] [PubMed]
- Hammam Ahmed R.A., et al. (2022) “Goat Milk: Compositional, Technological, Nutritional and Therapeutic Aspects: A Review.” Asian Journal of Dairy and Food Research: 41.4. [CrossRef]
- Henkel Jan, Saif Rashid, Jagannathan Vidhya, Schmocker Corinne, Zeindler Flurina, Bangerter Erika, Herren Ursula, Posantzis Dimitris, Bulut Zafer, Ammann Philippe, Drögemüller Cord, Flury Christine, Leeb Tosso (2019) Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS genetics: 15(12).
- Hunter DC, Ashraf B, Bérénos C, Ellis PA, Johnston SE, Wilson AJ, Pilkington JG, Pemberton JM, Slate J (2022) Using genomic prediction to detect microevolutionary change of a quantitative trait. Proc Biol Sci: 11;289(1974):20220330. [CrossRef] [PubMed]
- Imran FS, Al-Thuwaini TM, Al-Shuhaib MBS, Lepretre F (2021) A Novel Missense Single Nucleotide Polymorphism in the GREM1 Gene is Highly Associated with Higher Reproductive Traits in Awassi Sheep. Biochem Genet: 59(2):422-436. [CrossRef] [PubMed]
- Lillehammer Marie, Sonesson Anna K, Klemetsdal Gunnar, Blichfeldt Thor, Meuwissen Theo H E (2020) Genomic selection strategies to improve maternal traits in Norwegian White Sheep. Journal of animal breeding and genetics = Zeitschrift fur Tierzuchtung und Zuchtungsbiologie: 137(4).
- Marina H, Pelayo R, Gutiérrez-Gil B, Suárez-Vega A, Esteban-Blanco C, Reverter A, Arranz JJ (2022) Low-density SNP panel for efficient imputation and genomic selection of milk production and technological traits in dairy sheep. J Dairy Sci: 105(10):8199-8217. [CrossRef] [PubMed]
- Massender E, Brito LF, Maignel L, Oliveira HR, Jafarikia M, Baes CF, Sullivan B, Schenkel FS (2022) Single- and multiple-breed genomic evaluations for conformation traits in Canadian Alpine and Saanen dairy goats. J Dairy Sci: 105(7):5985-6000. [CrossRef] [PubMed]
- Massender E, Brito LF, Maignel L, Oliveira HR, Jafarikia M, Baes CF, Sullivan B, Schenkel FS (2022) Single-step genomic evaluation of milk production traits in Canadian Alpine and Saanen dairy goats. J Dairy Sci: 105(3):2393-2407. [CrossRef] [PubMed]
- Meuwissen T H, Hayes B J, Goddard M E (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics: 157(4).
- Moghaddar N, Brown DJ, Swan AA, Gurman PM, Li L, van der Werf JH (2022) Genomic prediction in a numerically small breed population using prioritized genetic markers from whole-genome sequence data. J Anim Breed Genet: 139(1):71-83. [CrossRef] [PubMed]
- Moghaddar N, Khansefid M, van der Werf JHJ, Bolormaa S, Duijvesteijn N, Clark SA, Swan AA, Daetwyler HD, MacLeod IM (2019) Genomic prediction based on selected variants from imputed whole-genome sequence data in Australian sheep populations. Genet Sel Evol: 5;51(1):72. [CrossRef] [PubMed]
- Moghaddar N, Swan AA, van der Werf JHJ (2017) Genomic prediction from observed and imputed high-density ovine genotypes. Genet Sel Evol:20;49(1):40. [CrossRef] [PubMed]
- Moghaddar N., Swan A. A., van der Werf J. H. J. (2014) Genomic prediction of weight and wool traits in a multi-breed sheep population. Animal Production Science:54(5).
- National Bureau of Statistics of China (2022) China Statistical Yearbook. Beijing: China Statistics Press.
- Newton J. E., Brown D. J., Dominik S., van der Werf J. H. J. (2017) Impact of young ewe fertility rate on risk and genetic gain in sheep-breeding programs using genomic selection. Animal Production Science: 57(8).
- Oliveira HR, McEwan JC, Jakobsen JH, Blichfeldt T, Meuwissen THE, Pickering NK, Clarke SM, Brito LF (2022) Across-country genomic predictions in Norwegian and New Zealand Composite sheep populations with similar development history. J Anim Breed Genet: 139(1):1-12. [CrossRef] [PubMed]
- Phua S H, Hyndman D L, Baird H J, Auvray B, McEwan J C, Lee M A, Dodds K G (2014) Towards genomic selection for facial eczema disease tolerance in the New Zealand sheep industry. Animal genetics: 45(4).
- Pickering N K, Dodds K G, Auvray B, et al. (2013) The impact of genomic selection on genetic gain in the New Zealand sheep dual purpose selection index. [CrossRef]
- Raoul J, Elsen JM (2021) The levels of artificial insemination and missing sire information make genomic selection not always beneficial in meat sheep. Animal: 15(2):100040. [CrossRef] [PubMed]
- Ren D, Cai X, Lin Q, Ye H, Teng J, Li J, Ding X, Zhang Z (2022) Impact of linkage disequilibrium heterogeneity along the genome on genomic prediction and heritability estimation. Genet Sel Evol: 54(1):47. [CrossRef] [PubMed]
- Rowe S J, Mcewan J C, Hickey S M, et al. (2014) Genomic selection as a tool to decrease greenhouse gas emission from dual purpose New Zealand sheep. Proceedings of the World Congress on Genetics Applied to Livestock Production. World Congress on Genetics Applied to Livestock Production.
- S. Mucha, R. Mrode, I. MacLaren-Lee, M. Coffey, J. Conington (2015) Estimation of genomic breeding values for milk yield in UK dairy goats. Journal of Dairy Science: 98(11).
- S.I. Duchemin, C. Colombani, A. Legarra, G. Baloche, H. Larroque, J.-M. Astruc, F. Barillet, C. Robert-Granié, E. Manfredi (2012) Genomic selection in the French Lacaune dairy sheep breed. Journal of Dairy Science: 95(5).
- Scholtens M, Lopez-Villalobos N, Lehnert K, Snell R, Garrick D, Blair HT (2020) Advantage of including Genomic Information to Predict Breeding Values for Lactation Yields of Milk, Fat, and Protein or Somatic Cell Score in a New Zealand Dairy Goat Herd. Animals (Basel): 25;11(1):24. [CrossRef] [PubMed]
- Shumbusho F, Raoul J, Astruc J M, Palhiere I, Elsen J M (2013) Potential benefits of genomic selection on genetic gain of small ruminant breeding programs. Journal of animal science: 91(8).
- SLACK-SMITH A, KINGHORN B P, VAN DER WERF J H J (2010) Accuracy of genomic selection in redicting carcass traits in meat sheep. Anim Prod Sci: 50(11-12): XIII.
- Soller M, and Beckmann J S (1983) “Genetic polymorphism in varietal identification and genetic improvement…” TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 67.1. [CrossRef]
- Song Hailiang, Hu Hongxia (2021) Strategies to improve the accuracy and reduce costs of genomic prediction in aquaculture species. Evolutionary Applications: 15(4).
- Song Hailiang, Ye Shaopan, Jiang Yifan, Zhang Zhe, Zhang Qin, Ding Xiangdong (2019) Using imputation-based whole-genome sequencing data to improve the accuracy of genomic prediction for combined populations in pigs. Genetics, selection, evolution: GSE: 51(1).
- Teissier M, Larroque H, Brito LF, Rupp R, Schenkel FS, Robert-Granié C (2020) Genomic predictions based on haplotypes fitted as pseudo-SNP for milk production and udder type traits and SCS in French dairy goats. J Dairy Sci: 103(12):11559-11573. [CrossRef] [PubMed]
- Teissier M, Larroque H, Robert-Granié C (2018) Weighted single-step genomic BLUP improves accuracy of genomic breeding values for protein content in French dairy goats: A quantitative trait influenced by a major gene. Genet Sel Evol: 15;50(1):31. [CrossRef] [PubMed]
- Wang fengxiang (2021) Design of Goat SNP Chip with Applications in Genome-wide Association Study and Genomic Selection of Important Economic Traits in Inner Mongolia Cashmere Goat. Inner Mongolia Agricultural University. [CrossRef]
- Wei C, Luo H, Zhao B, Tian K, Huang X, Wang Y, Fu X, Tian Y, Di J, Xu X, Wu W, Tulafu H, Yasen M, Zhang Y, Zhao W (2020) The Effect of Integrating Genomic Information into Genetic Evaluations of Chinese Merino Sheep. Animals (Basel): 28;10(4):569. [CrossRef] [PubMed]
- Wei X, Zhang T, Wang L, Zhang L, Hou X, Yan H, Wang L (2022) Optimizing the Construction and Update Strategies for the Genomic Selection of Pig Reference and Candidate Populations in China. Front Genet: 13:938947. [CrossRef] [PubMed]
- Werf J (2009) Potential benefit of genomic selection in sheep.
- Wiggans GR, Carrillo JA (2022) Genomic selection in United States dairy cattle. Front Genet: 13:994466. [CrossRef] [PubMed]
- Wiggans GR, Cole JB, Hubbard SM, Sonstegard TS (2017) Genomic Selection in Dairy Cattle: The USDA Experience. Annu Rev Anim Biosci: 5:309-327. [CrossRef] [PubMed]
- Wu Changxin, et al. (2021) Comparative animals breeding. Beijing: Published by China Agricultural University: 2.
- Yan X, Zhang T, Liu L, Yu Y, Yang G, Han Y, Gong G, Wang F, Zhang L, Liu H, Li W, Yan X, Mao H, Li Y, Du C, Li J, Zhang Y, Wang R, Lv Q, Wang Z, Zhang J, Liu Z, Wang Z, Su R (2022) Accuracy of Genomic Selection for Important Economic Traits of Cashmere and Meat Goats Assessed by Simulation Study. Front Vet Sci: 9:770539. [CrossRef] [PubMed]
- Zheng Xu, Zhang Tianliu, Wang Tianzhen, Niu Qunhao, Wu Jiayuan, Wang Zezhao, Gao Huijiang, Li Junya, Xu Lingyang (2022) Long-Term Impact of Genomic Selection on Genetic Gain Using Different SNP Density. Agriculture: 12(9).
- Zhu S, Guo T, Yuan C, et al. (2021) Evaluation of GBLUP and Bayes-Alphabet Based on Different Marker Density For Genomic Prediction in Alpine Merino Sheep. G3-Genes Genomes Genetics.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).