1. Introduction
Rice
(Oryza sativa L.) is a vital staple food crop globally, yet its productivity is frequently compromised by various biotic stresses, including attacks by pathogens[
1]. Lesion mimic mutants (LMMs) in rice are characterized by spontaneous lesion formation on leaves, mirroring disease-like symptoms in the absence of pathogens. These mutants serve as a valuable tool for unraveling the molecular mechanisms underlying plant immunity and programmed cell death (PCD)[
2], and they hold promise for breeding disease-resistant rice varieties.
LMMs display a range of phenotypic traits, including the spontaneous emergence of necrotic lesions on leaves, which vary in size, shape, and distribution depending on the specific mutant. For instance, the
blm mutant features large, spreading lesions and is linked to enhanced resistance to blast pathogens, suggesting a potential role in defense[
3]. Temperature and light conditions can also influence lesion formation in LMMs. The
lrd mutant, for instance, shows increased lesion formation under high light intensity, highlighting the interaction between environmental factors and genetic predisposition in lesion development[
4].
Recent advancements in genetic mapping and molecular cloning have unveiled several genes responsible for lesion mimic phenotypes in rice. These genes play pivotal roles in a variety of biological processes, including PCD, defense signaling, and stress responses. The
SPL11 gene, which encodes a U-box/armadillo repeat protein with E3 ubiquitin ligase activity, negatively regulates cell death and defense mechanisms. This suggests its involvement in the ubiquitination-mediated degradation of proteins associated with defense[
5]. The
OsSPL1 gene, encoding a sphingosine-1-phosphate lyase, is a key player in sphingolipid metabolism and is associated with disease resistance responses[
6]. Furthermore, some LMMs genes are crucial for the regulation of reactive oxygen species (ROS) production and signaling. For instance, the
OsLMS gene, which encodes a protein with double-stranded RNA-binding motifs, is implicated in RNA metabolism and ROS accumulation. This leads to lesion formation and early senescence[
7]. The interaction between various hormonal pathways, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), is also vital in modulating the lesion mimic phenotype. The
OsEDR1 gene, for example, negatively regulates bacterial resistance by activating ethylene biosynthesis, demonstrating the intricate crosstalk between hormone signaling pathways in plant immunity[
8].
Investigations into rice lesion mimic mutants have shed light on the intricate molecular mechanisms governing plant immunity and PCD. Unraveling the genetic basis of LMMs paves the way for crafting rice varieties with enhanced disease resistance, thereby bolstering sustainable agriculture and ensuring food security. Here, we selected and characterized a spotted leaf mutant from the IR64 mutant library. This research endeavors aimed at the functional elucidation of OsRPT5A gene and its incorporation into breeding strategies will amplify the utility of the mutant in advancing crop improvement.
2. Results
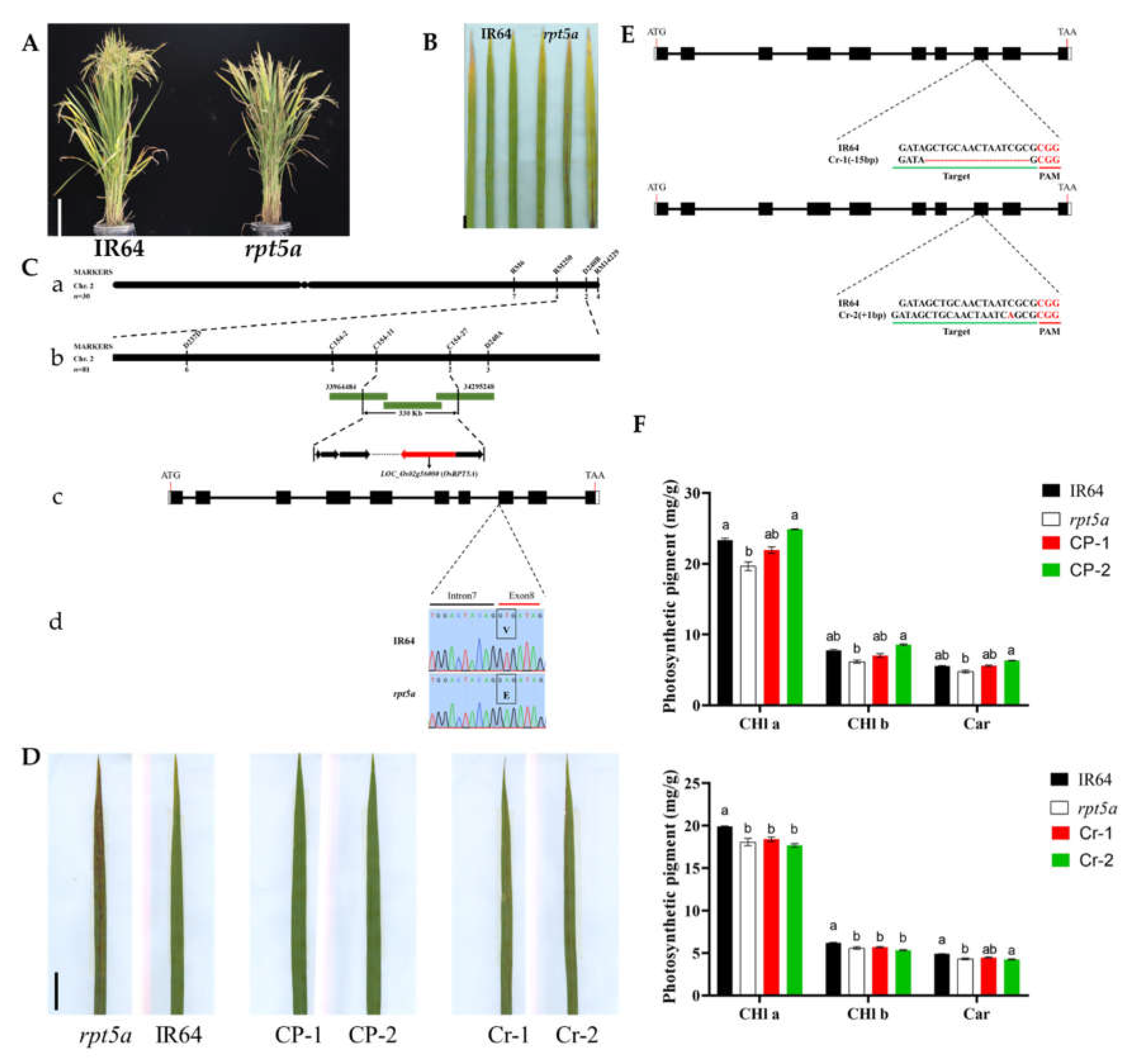
2.1. The Mutation of OsRPT5A Leads to the Spotted leaf Phenotype
In our rice spotted leaf mutant library, we focused particularly on a mutant named
rpt5a, which exhibited reddish-brown spots from the seedling stage, persisting throughout the plant's life cycle and ultimately leading to extensive leaf necrosis (
Figure 1A-B, Supplemental
Figure S1). To elucidate the genetic mechanism underlying this phenotype, we employed a map-based cloning approach and analyzed the progeny of a cross between
rpt5a and the japonica rice variety 80A90YR72. Our initial mapping efforts confined the locus to an interval between markers RM6 and RM14229 on chromosome 2. Further fine-mapping efforts narrowed this interval to a 330Kb region between markers C154-11 and C154-27.
Through a comprehensive analysis of related literature and targeted sequencing of candidate genes within the narrowed locus, we identified the target gene
LOC_Os02g56000. Given its high similarity to the Arabidopsis gene
RPT5A, we named it
OsRPT5A. This discovery was significant as a point mutation from thymine (T) to adenine (A) occurred in the eighth exon of
OsRPT5A, leading to the substitution of valine (Val) for glutamic acid (Glu) in the protein, which is likely a key factor causing the spotted leaf phenotype (
Figure 1C).
To further validate this hypothesis, we conducted functional complementation experiments in the
rpt5a mutant background and successfully obtained transgenic lines with restored phenotypes, strongly confirming the crucial role of
OsRPT5A in controlling the spotted leaf trait. Moreover, we generated
OsRPT5A knockout lines in the IR64 background, resulting in positive lines Cr-1 and Cr-2 that exhibited the spotted leaf phenotype, indicating that the loss of
OsRPT5A can lead to the spotted leaf phenotype (
Figure 1D-E).
Additionally, we measured the chlorophyll content of the transgenic plants and found that the chlorophyll content of the complementation lines was close to that of the wild type, while the knockout lines were similar to the mutant (
Figure 1F). The agronomic traits of the knockout lines were close to
rpt5a, and some agronomic traits of the complementation lines were restored to the wild type level (Supplemental
Figure S2). Additionally, we observed the chloroplast structure of
rpt5a using transmission electron microscopy and noted that its arrangement was sparser, and exhibited more severe degradation compared to IR64, potentially contributing to the reduced photosynthetic capacity (Supplemental
Figure S3).
2.2. OsRPT5A Is a Constitutively Expressed Gene
To elucidate the spatiotemporal expression pattern of
OsRPT5A, total RNA was extracted from various tissues at different developmental stages of IR64 and
rpt5a, and its relative expression was quantified using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The results revealed that
OsRPT5A is ubiquitously expressed across all examined tissues, including roots, stems, leaf sheaths, and panicles, with its lowest expression observed in stems at 2 weeks (
Figure 2A). Furthermore, a GUS reporter construct containing the
OsRPT5A promoter was generated, and the GUS activity pattern corroborated the qRT-PCR findings, collectively indicating that OsRPT5A is constitutively expressed in rice (
Figure 2B).
To ascertain whether mutations in OsRPT5A protein influence its subcellular localization, we expressed GFP-tagged OsRPT5A and its mutant proteins in rice protoplasts. The results revealed that both the OsRPT5A-GFP fusion protein and OsRPT5A
V318E-GFP were localized in the cytoplasm, suggesting that the mutation in the OsRPT5A does not alter its cellular localization (
Figure 2C).
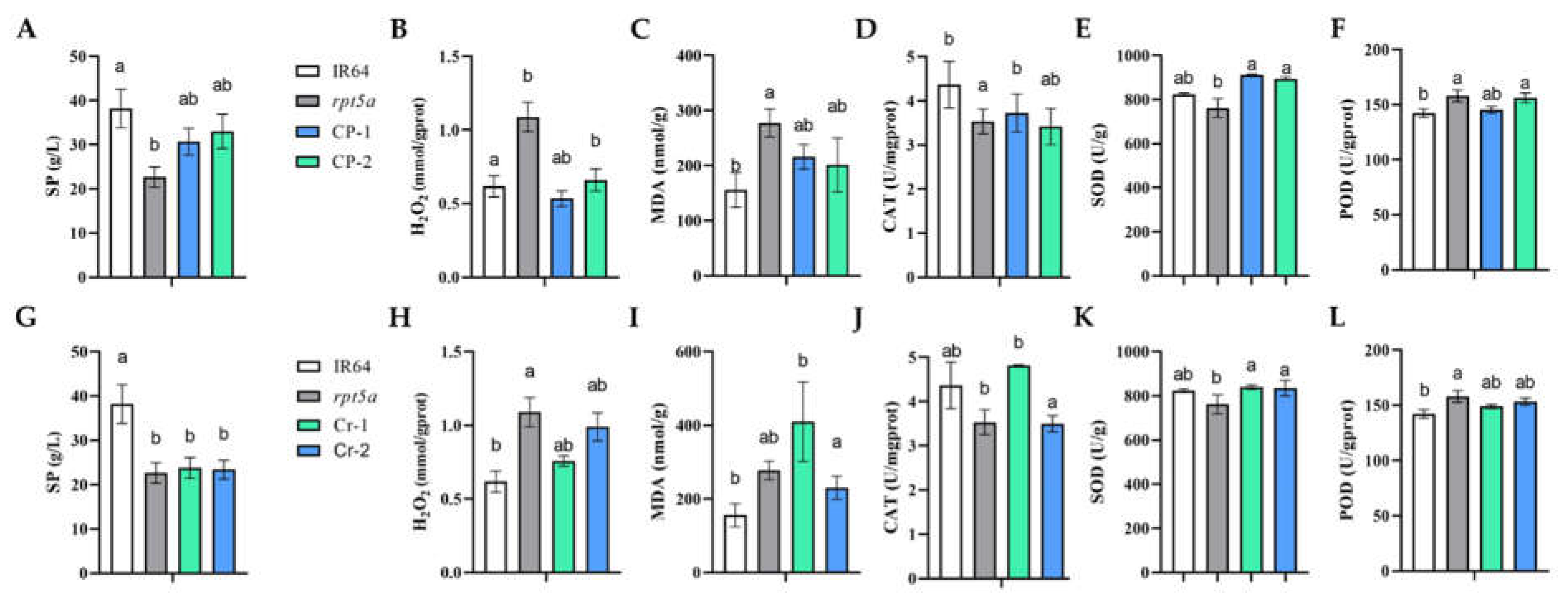
2.3. ROS Is Accumulated in rpt5a
Lesion mimic mutants in rice are frequently associated with abnormal reactive oxygen species (ROS) accumulation. In this context, we evaluated physiological parameters in the rpt5a mutant and transgenic plants. Our analysis demonstrated a significant elevation in H2O2 levels in rpt5a compared to the wild type, accompanied by a substantial reduction in catalase (CAT) activity, indicating a compromise in the ROS detoxification system. Additionally, the activities of superoxide dismutase (SOD) and peroxidase (POD) were markedly enhanced in rpt5a, suggesting that the mutation in OsRPT5A disrupts the balance between ROS production and scavenging, leading to an excessive ROS build-up in rpt5a.
Conversely, the trends in soluble protein (SP), H
2O
2, and malondialdehyde (MDA) contents in the complemented plants closely resembled those in the IR64 wild type, signifying a partial restoration of ROS homeostasis in these plants (
Figure 3G-L)
. The data from the knockout plants mirrored the mutant phenotype, confirming that the deletion of this gene triggers ROS accumulation. These findings highlight the pivotal role of
OsRPT5A stability in maintaining the integrity of the ROS scavenging system (
Figure 3A-F).
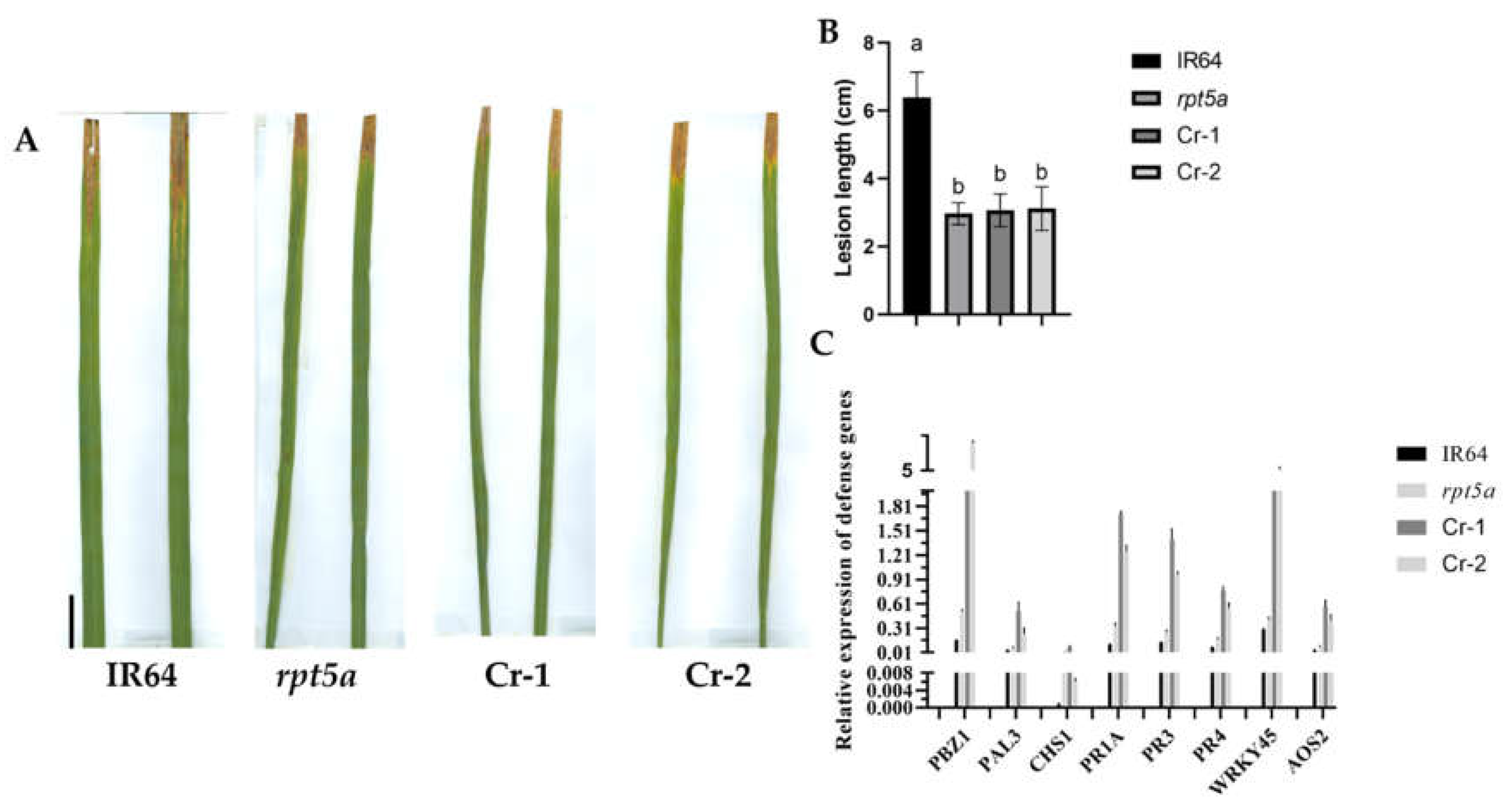
2.4. The Resistance of rpt5a Is Enhanced
In majority of rice lesion mimic mutants, alterations in resistance to pathogen infection are observed, accompanied by either upregulation or downregulation of defense response gene expression. Preliminary investigations have demonstrated that the
rpt5a mutant exhibits enhanced resistance to the
Xoo race C2. To ascertain if this enhanced resistance is attributable to the
OsRPT5A mutation, transgenic lines of
OsRPT5A were inoculated with C2. The lesion lengths of the knockout lines were found to be similar to those of the
rpt5a mutant, suggesting that
OsRPT5A plays a pivotal role in conferring resistance in rice (
Figure 4A-B).
After inoculation, we examined the expression levels of some defense genes and found that they were upregulated in the knockout lines, further validating the inoculation results (
Figure 4C).
3. Discussion
In our study, we successfully isolated the target gene responsible for the lesion mimic mutant phenotype through map-based cloning. Although the segregation of the positioning group does not comply with Mendel's Law of Segregation, we then validated its role in controlling this phenotype using transgenic lines. The complemented lines effectively restored the wild-type phenotype, as the knockout lines exhibited lesion mimic phenotypes similar to the original mutant. This mutation induced significant differences in the major agronomic traits of the mutant compared to the wild type, highlighting its impact on plant development and yield. Notably, the chlorophyll content of the knockout lines remained similar to that of the mutant throughout the growth period. This suggests that the necrotic spots caused by the gene mutation adversely affected photosynthesis, thereby potentially impacting the plant's energy production and overall health. Additionally, the knockout lines showed a reduction in 1000-grain weight and seed-setting rate, indicating a negative effect on reproductive success and yield, implying that the absence of the functional gene RPT5A led to abnormalities in plant agronomic traits.
The restoration of hydrogen peroxide levels to wild-type levels in the complemented lines suggests that the excessive accumulation of hydrogen peroxide in the mutant disrupts ROS scavenging system homeostasis and is closely related to the occurrence of lesion mimic spots. The high levels of hydrogen peroxide in the knockout lines enhance their resistance to C2, as confirmed by the high expression levels of defense genes 72 hours after inoculation. Possible mechanisms include altering the strength of plant cell walls, activating other defense signaling pathways, or enhancing plant cell recognition of pathogens [9, 10]. Abnormal RPT5A protein may interfere with normal plant growth and response pathways, leading to the formation of lesion mimic spots.
OsRPT5A encodes a subunit of the 26S proteasome, playing a pivotal role in diverse plant species, particularly in leaf development, DNA damage repair, and responses to environmental stress. In rice, the
rpt5a mutant demonstrates abnormal leaf development under zinc deficiency, highlighting the critical role of DNA damage mitigation in normal leaf development [
11]. This suggests that RPT5A may underpin normal leaf morphology by engaging in DNA repair and ensuring genomic stability. In Arabidopsis, the
NAC103 gene mutation alleviates DNA damage in mutants sensitive to excess boron, indicating that
RPT5A might be instrumental in the plant's response to environmental stress, such as excess boron, particularly in the DNA damage repair process[
12]. Concurrently, both RPT2A and RPT5A are essential for zinc deficiency tolerance, suggesting that RPT5A is key in the plant's adaptation to micronutrient deficiency, potentially by modulating protein degradation and signal transduction pathways[
13]. In papaya, proteomic analysis suggests a link between RPT5A and typical sticky disease symptoms, implying that RPT5A may contribute to the plant's disease response, possibly by regulating the degradation of immune-related proteins to participate in disease defense mechanisms[
14]. Coupled with existing experimental evidence, we hypothesize that RPT5A may be regulated by upstream transcription factors, thereby playing a role in the regulation of ROS in plants, although the potential interacting transcription factors are still under investigation. Concurrently, our screening of its interacting proteins (unpublished data) reveals interactions with protein kinases, indicating that RPT5A is not only tightly regulated upstream but also involved in the plant's response to various stresses downstream through serine/threonine protein kinases, such as salt stress, drought stress, and low-temperature stress. The mutation site in the OsRPT5A protein is the 318th amino acid, where valine is mutated to glutamate, located in the SMART AAA domain. Given that valine is hydrophobic and glutamate is a negatively charged polar amino acid, many proteins with the AAA domain interact with other proteins through this domain. The mutation-induced change in the protein surface charge or hydrophobicity can impact protein-protein interactions, thereby affecting the assembly and stability of the protein complex and interfering with various cellular activities, including protein folding, membrane fusion, and DNA replication. In summary, the role of OsRPT5A in ROS regulation and plant resistance regulation remains an area for further investigation.
4. Materials and methods
4.1. Plant Materials and Growth Conditions
Ethyl methanesulfonate (EMS) was utilized to induce mutagenesis in the indica rice variety IR64, leading to the development of a comprehensive mutant library. Within this library, a specific leaf spot mutant, named as rpt5a, was isolated. Genetic analyses and gene mapping were performed on an F2 population derived from a cross between rpt5a and the japonica rice variety 80A90YR72. The cultivation of these experimental materials took place at the Fu Yang experimental base of the China National Rice Research Institute (CNRRI), under the auspices of the Chinese Academy of Agricultural Sciences.
4.2. The Construction of Vectors and the Acquisition of Transgenic Lines
For the complementation lines, the allelic gene OsRPT5A from the wild-type IR64, which includes a 3000 bp sequence upstream of the transcription start site, the complete 4319 bp genomic DNA, and a 2500 bp sequence downstream of the termination site, was cloned into the vector pCAMBIA1300, resulting in the construction of pCAMBIA1300-RPT5A. For the knockout lines, we employed CRISPR/Cas9 technology to excise specific genomic sequences, constructing the knockout vector prpt5a. Utilizing the GUS reporter vector pCambia1381Z, we cloned the 3000 bp promoter region of OsRPT5A, leading to the creation of the vector pCambia1381Z-RPT5A-GUS. For subcellular localization assays, the coding sequences of both the wild-type OsRPT5A and its variant, RPT5AV318E, were integrated into pYBA1132, yielding the vectors GFP-RPT5A and GFP-RPT5AV318E. All engineered constructs were transformed by BIORUN BIOSCIENCES CO., LTD, and the plant lines were cultivated at the Fu Yang experimental base of the China National Rice Research Institute, affiliated with the Chinese Academy of Agricultural Sciences.
4.3. Agronomic Trait Evaluation
Three individual plants were randomly selected from each of the IR64, rpt5a, its complementation lines, and knockout lines for evaluation of their agronomic traits at full maturity. These traits included panicle length, thousand grain weight, and seed setting rate. Three biological replicates were performed for all measurements.
4.4. Measurement of Physiological Indexes
According to the protocols provided by the Nanjing JianCheng Bioengineering Institute's assay kits, leaves of IR64, rpt5a, its complementation lines, and knockout lines at the tillering stage were homogenized to prepare a uniform slurry. The concentrations of stress biomarkers including soluble proteins (SP), malondialdehyde (MDA), and hydrogen peroxide (H2O2), as well as the enzymatic activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) were quantitatively assessed. Three biological replicates were performed for all measurements.
4.5. Gene Mapping
DNA was extracted utilizing the Cetrimonium Bromide (CTAB) method[
15]. Rice genomic sequences were retrieved from the GRAMENE (
http://ensembl.gramene.org/) and the Chinese Rice Genome Database (
http://rice.genomics.org.cn/). Primers were specifically designed to amplify genomic regions, based on the comparative genomic analysis between the japonica variety 'Nipponbare' and the indica variety '9311'. Primer designs and sequence analyses were facilitated by utilizing resources from the GRAMENE and MSU Rice Genome Annotation Project databases (
http://rice.uga.edu/). PCR amplifications were performed according to the protocols specified by Vazyme (P222-03).
4.6. qRT-PCR Analysis
RNA was extracted using the Invitrogen TRIzol reagent (ThermoFisher) according to the manufacturer's instructions. To assess the impact of inoculation with
Xanthomonas oryzae pv.
oryzae on the expression levels of defense genes, RNA was isolated from the leaves of IR64,
rpt5a, CR-1, and CR-2 lines at 72 hours post-inoculation. The qRT-PCR analyses were conducted using the SYBR Green qPCR Master Mix from Vazyme (Q511-02), with each experimental condition replicated three times. The primers utilized for qRT-PCR are detailed in
Supplementary Table S1.
4.7. Inoculation with the Bacterial Blight Pathogen
Based on the method[
16], it was conducted on the tillering stage leaves of IR64,
rpt5a, CR-1, and CR-2 lines using the race C2 of
Xanthomonas oryzae pv.
oryzae. Lesion lengths were measured 14 days after inoculation. For each group, six fully expanded leaves from different plants were inoculated. Six replicates were measured to analyze the data.
4.8. Chloroplast Structure Observation
For the TEM observation of chloroplast structure, the leaves of IR64, rpt5a at the tillering stage were first soaked in 2.5% glutaraldehyde for 24 h. Then, the samples were examined in the Instrument and Equipment Sharing Service Technology Platform of CNRRI.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1: Primers used for
OsRPT5A gene and vector construction in this study; Supplemental Figure S1: Phenotypes of IR64 and
rpt5a at 8 weeks, 12 weeks and 16 weeks; Supplemental Figure S2: Agronomic traits of IR64,
rpt5a and complementary lines (CP-1, CP-2), knockout lines (Cr-1, Cr-2); Supplemental Figure S3: TEM observation of the chloroplasts in IR64 and
rpt5a at the tillering stage.
Author Contributions
Conceptualization, Chen Wang and Jian-li Wu; Data curation, Xinwei Liao; Formal analysis, XiaoBo Zhang; Funding acquisition, Junyi Gong; Investigation, Chen Wang and Wenjun Liu; Project administration, Xia Xu; Software, Shihua Yang; Supervision, Hai Zhou and Chuxiong Zhuang; Writing – original draft, Chen Wang; Writing – review & editing, Jian-li Wu.
Funding
This research was supported by the National Key Research and Development Program of China (2022YFF1003301), and Zhejiang Provincial Natural Science Foundation of China (LZ24C130004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
Authors declare that there are no conflicts of interest.
References
- Liu, W.D.; Liu, J.L.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel Insights into Rice Innate Immunity Against Bacterial and Fungal Pathogens. Annual Review of Phytopathology 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Lee, J.H.; Agrawal, G.K.; Rakwal, R.; Kim, J.A.; Shim, J.K.; Lee, S.K.; Jeon, J.S.; Koh, H.J.; Lee, Y.H.; Iwahashi, H.; Jwa, N. S The rice (Oryza sativa) Blast Lesion Mimic Mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiology and Biochemistry 2005, 43, 397–406. [Google Scholar] [CrossRef]
- Wang, J.J.; Zhang, L.X.; Wang, L.Y.; Zhang, L.H.; Zhu, C.N.; He, Z.H.; JIin, Q.S.; Fan, H.H.; Yu, X. Response to Illumination Induction and Effect of Temperature on Lesion formation of lrd (Lesion Resembling Disease) in Rice. Scientia Agricultura Sinica 2010, 43, 2039–2044. [Google Scholar]
- Zeng, L.R.; Qu, S.H.; Bordeos, A.; Yang, C.W.; Baraoidan, M.; Yan, H.Y.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf11, a Negative Regulator of Plant Cell Death and Defense, Encodes a U-Box/Armadillo Repeat Protein Endowed with E3 Ubiquitin Ligase Activityw⃞. The Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.J.; Jin, X.Y.; Lei, H.; Hong, Y.B.; Zhang, Y.F.; Ouyang, Z.G.; Li, X.H.; Song, F.M.; Li, D.Y. Molecular characterization of rice sphingosine-1-phosphate lyase gene OsSPL1 and functional analysis of its role in disease resistance response. Plant Cell Reports 2014, 33, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Undan, J.R.; Tamiru, M.; Abe, A.; Yoshida, K.; Kosugi, S.; Takagi, H.; Yoshida, K.; Kanzaki, H.; Saitoh, H.; Fekih, R.; Sharma, S.; Undan, J.; Yano, M.; Terauchi, R. Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.). Genes & Genetic Systems 2012, 87, 169–179. [Google Scholar]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant, Cell & Environment 2011, 34, 179–191. [Google Scholar]
- Ballini, E.; Morel, J.B.; Droc, G.; Price, A.; Courtois, B.; Notteghem, J.L.; Tharreau, D. A Genome-Wide Meta-Analysis of Rice Blast Resistance Genes and Quantitative Trait Loci Provides New Insights into Partial and Complete Resistance. MPMI 2008, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Frick, M.; Huel, R.; Nykiforuk, C.L.; Wang, X.M.; Gaudet, D.A.; Eudes, F.; Conner, R.L.; Kuzyk, A.; Chen, Q.; Kang, Z.S.; Laroche, A. Stripe Rust Resistance Gene Yr10 Encodes an Evolutionary-conserved and Unique CC-NBS-LRR Sequence in Wheat. Molecular plant 2014, 7, 1740–1755. [Google Scholar] [CrossRef] [PubMed]
- Sotta, N.; Sakamoto, T.; Matsunaga, S.; Fujiwara, T. Abnormal leaf development of rpt5a mutant under zinc deficiency reveals important role of DNA damage alleviation for normal leaf development. Scientific Reports 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sotta, N.; Sakamoto, T.; Kamiya, T.; Tabata, R.; Yamaguchi, K.; Shigenobu, S.; Yamada, M.; Hasebe, M.; Sawa, S.; Fujiwara, T. NAC103 mutation alleviates DNA damage in an Arabidopsis thaliana mutant sensitive to excess boron. Frontiers in Plant Science 2023, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kamiya, T.; Sako, K.; Yamaguchi, J.; Yamagami, M.; Fujiwara, T. Arabidopsis thaliana 26S Proteasome Subunits RPT2a and RPT5a Are Crucial for Zinc Deficiency-Tolerance. Bioscience, biotechnology, and biochemistry 2011, 75, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.L.; Guyon-Debast, A.; Lécureuil, A.; Vezon, D.; Carpentier, V.; Bonhomme, S.; Guerche, P. The Arabidopsis proteasome RPT5 subunits are essential for gametophyte development and show accession-dependent redundancy. Plant Cell 2009, 21, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology 1985, 5, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Bordeos, A.; Madamba, M.R.S.; Baraoidan, M.; Ramos, M.; Wang, G.L.; Leach, J.E.; Leung, H. Rice lesion mimic mutants with enhanced resistance to diseases. Molecular Genetics and Genomics 2008, 279, 605–619. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).