Submitted:
19 April 2024
Posted:
23 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plasmid Vectors and Inhibitors
2.2. Production of HIV-1 and HIV-2 Pseudovirions
2.3. Virus Production to Assay for Effects of Lenacapavir on HIV-2 Capsid Formation
2.4. MTT Cell Proliferation Assay
2.5. Inhibition Profiling in Jurkat Cells
2.6. Raltegravir Inhibition Assay against SARs-CoV-2
2.7. Molecular Docking
3. Results
3.1. Susceptibility of HIV-2 to INSTIs
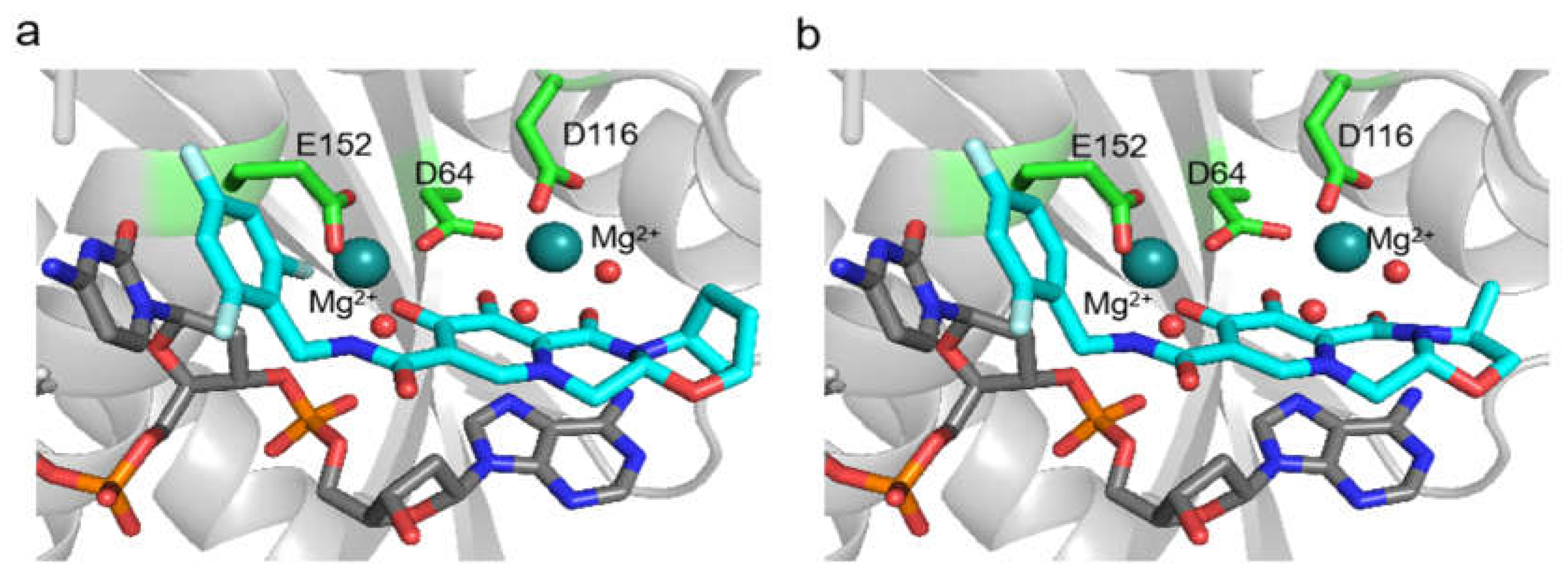
3.2. In Silico Analysis
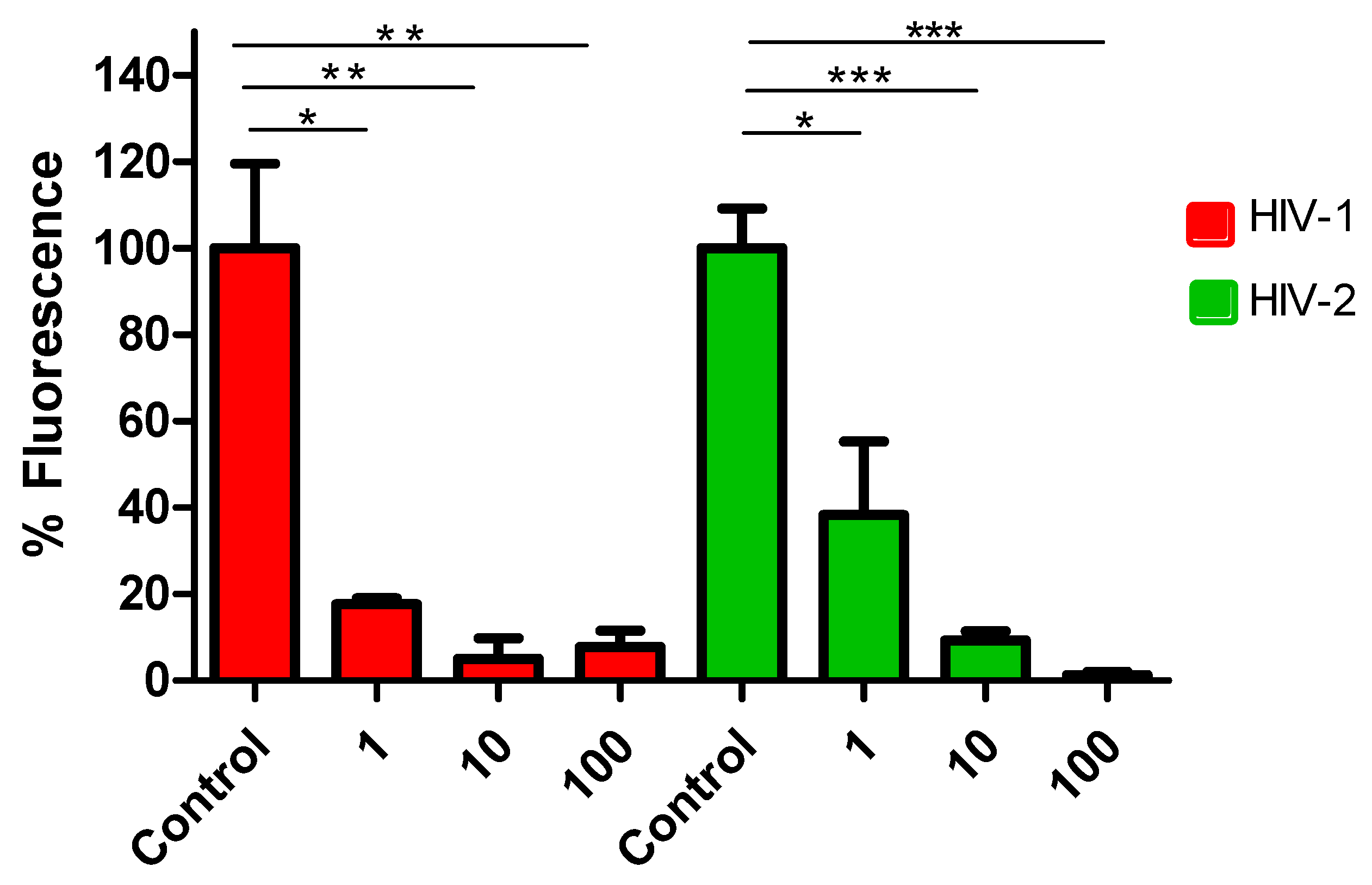
3.3. Efficacy of Lenacapavir against HIV-2
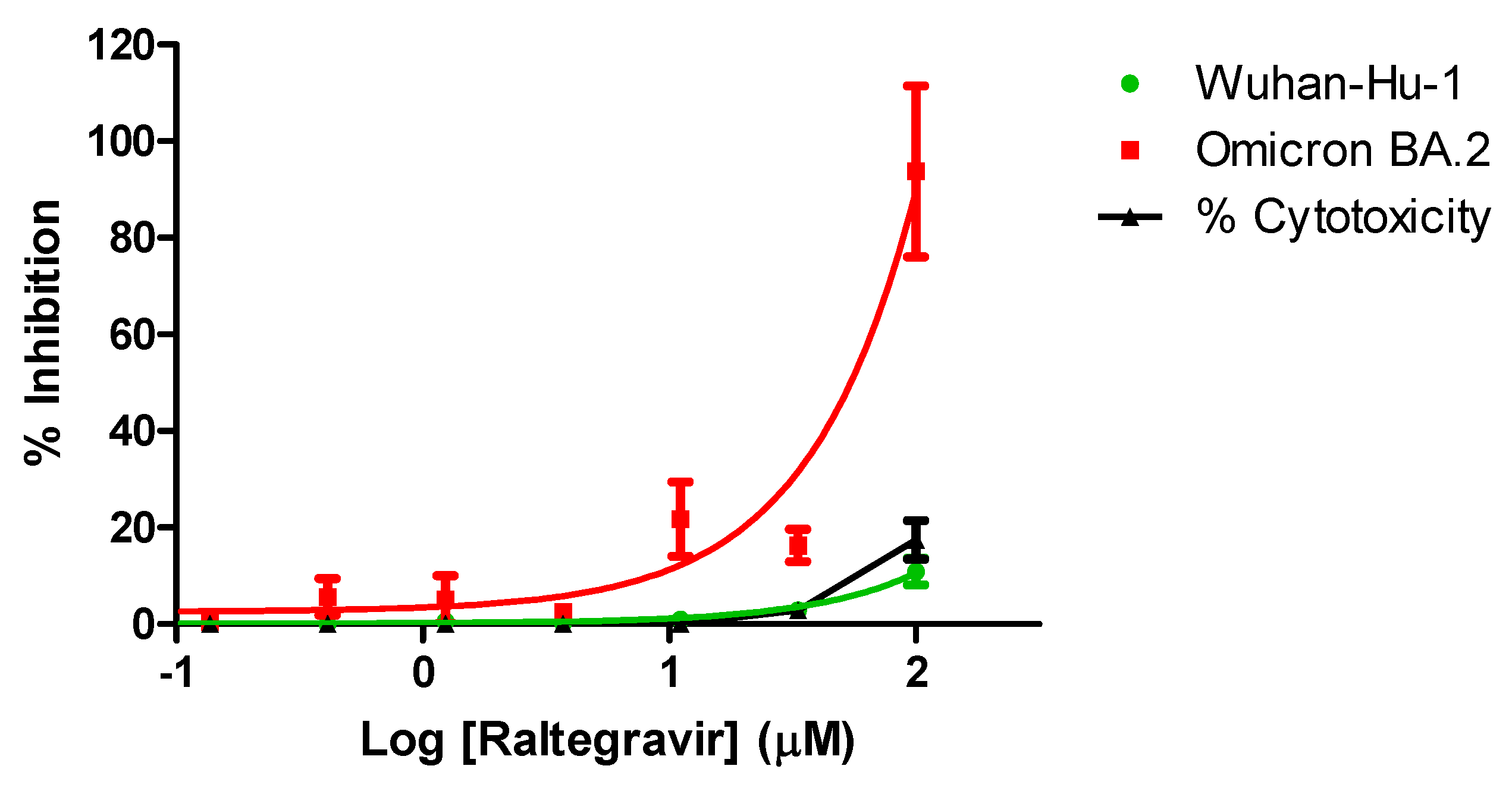
3.4. Anti-SARS-CoV-2 Activity of Raltegravir
4. Discussion
Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNAIDS. Global HIV/AIDS Data and Statistics. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on.
- Campbell-Yesufu, O.T.; Gandhi, R.T. Update on human immunodeficiency virus (HIV)-2 infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 2011, 52, 780–787. [Google Scholar] [CrossRef]
- Gottlieb, G.S.; Raugi, D.N.; Smith, R.A. 90-90-90 for HIV-2? Ending the HIV-2 epidemic by enhancing care and clinical management of patients infected with HIV-2. The lancet. HIV 2018, 5, e390–e399. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Giovanetti, M.; Sagnelli, C.; Ciccozzi, A.; d’Ettorre, G.; Angeletti, S.; Borsetti, A.; Ciccozzi, M. Human Immunodeficiency Virus Type 2: The Neglected Threat. Pathogens 2021, 10. [Google Scholar] [CrossRef]
- Esbjornsson, J.; Mansson, F.; Kvist, A.; da Silva, Z.J.; Andersson, S.; Fenyo, E.M.; Isberg, P.E.; Biague, A.J.; Lindman, J.; Palm, A.A.; et al. Long-term follow-up of HIV-2-related AIDS and mortality in Guinea-Bissau: a prospective open cohort study. The lancet. HIV 2018. [Google Scholar] [CrossRef]
- Nachega, J.B.; Marconi, V.C.; van Zyl, G.U.; Gardner, E.M.; Preiser, W.; Hong, S.Y.; Mills, E.J.; Gross, R. HIV treatment adherence, drug resistance, virologic failure: evolving concepts. Infectious disorders drug targets 2011, 11, 167–174. [Google Scholar] [CrossRef]
- Slama, L.; Porcher, R.; Linard, F.; Chakvetadze, C.; Cros, A.; Carillon, S.; Gallardo, L.; Viard, J.P.; Molina, J.M. Injectable long acting antiretroviral for HIV treatment and prevention: perspectives of potential users. BMC infectious diseases 2023, 23, 98. [Google Scholar] [CrossRef]
- Witvrouw, M.; Pannecouque, C.; Van Laethem, K.; Desmyter, J.; De Clercq, E.; Vandamme, A.M. Activity of non-nucleoside reverse transcriptase inhibitors against HIV-2 and SIV. Aids 1999, 13, 1477–1483. [Google Scholar] [CrossRef]
- Desbois, D.; Roquebert, B.; Peytavin, G.; Damond, F.; Collin, G.; Benard, A.; Campa, P.; Matheron, S.; Chene, G.; Brun-Vezinet, F.; et al. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrobial agents and chemotherapy 2008, 52, 1545–1548. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Arias, L.; Alvarez, M. Antiretroviral therapy and drug resistance in human immunodeficiency virus type 2 infection. Antiviral research 2014, 102, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R. The molecular biology of HIV integrase. Future virology 2012, 7, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Jozwik, I.K.; Passos, D.O.; Lyumkis, D. Structural Biology of HIV Integrase Strand Transfer Inhibitors. Trends in pharmacological sciences 2020, 41, 611–626. [Google Scholar] [CrossRef]
- Trivedi, J.; Mahajan, D.; Jaffe, R.J.; Acharya, A.; Mitra, D.; Byrareddy, S.N. Recent Advances in the Development of Integrase Inhibitors for HIV Treatment. Current HIV/AIDS reports 2020, 17, 63–75. [Google Scholar] [CrossRef]
- Smith, R.A.; Raugi, D.N.; Pan, C.; Coyne, M.; Hernandez, A.; Church, B.; Parker, K.; Mullins, J.I.; Sow, P.S.; Gottlieb, G.S.; et al. Three main mutational pathways in HIV-2 lead to high-level raltegravir and elvitegravir resistance: implications for emerging HIV-2 treatment regimens. PloS one 2012, 7, e45372. [Google Scholar] [CrossRef]
- Descamps, D.; Peytavin, G.; Visseaux, B.; Tubiana, R.; Damond, F.; Campa, P.; Charpentier, C.; Khuong-Josses, M.A.; Duvivier, C.; Karmochkine, M.; et al. Dolutegravir in HIV-2-Infected Patients With Resistant Virus to First-line Integrase Inhibitors From the French Named Patient Program. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 2015, 60, 1521–1527. [Google Scholar] [CrossRef] [PubMed]
- Le Hingrat, Q.; Collin, G.; Le, M.; Peytavin, G.; Visseaux, B.; Bertine, M.; Tubiana, R.; Karmochkine, M.; Valin, N.; Collin, F.; et al. A New Mechanism of Resistance of Human Immunodeficiency Virus Type 2 to Integrase Inhibitors: A 5-Amino-Acid Insertion in the Integrase C-Terminal Domain. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 2019, 69, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Pujari, S.; Patel, A.; Gaikwad, S.; Patel, K.; Dabhade, D.; Chitalikar, A.; Joshi, K.; Bele, V. Effectiveness of dolutegravir-based antiretroviral treatment for HIV-2 infection: retrospective observational study from Western India. The Journal of antimicrobial chemotherapy 2020, 75, 1950–1954. [Google Scholar] [CrossRef]
- Smith, R.A.; Raugi, D.N.; Pan, C.; Sow, P.S.; Seydi, M.; Mullins, J.I.; Gottlieb, G.S.; University of Washington-Dakar, H.I.V.S.G. In vitro activity of dolutegravir against wild-type and integrase inhibitor-resistant HIV-2. Retrovirology 2015, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Raugi, D.N.; Wu, V.H.; Zavala, C.G.; Song, J.; Diallo, K.M.; Seydi, M.; Gottlieb, G.S.; University of Washington-Dakar, H.I.V.S.G. Comparison of the Antiviral Activity of Bictegravir against HIV-1 and HIV-2 Isolates and Integrase Inhibitor-Resistant HIV-2 Mutants. Antimicrobial agents and chemotherapy 2019, 63. [Google Scholar] [CrossRef]
- Smith, R.A.; Wu, V.H.; Zavala, C.G.; Raugi, D.N.; Ba, S.; Seydi, M.; Gottlieb, G.S.; University of Washington-Dakar, H.I.V.S.G. In Vitro Antiviral Activity of Cabotegravir against HIV-2. Antimicrobial agents and chemotherapy 2018, 62. [Google Scholar] [CrossRef]
- Gilead Sciences, I. Sunlenca® (lenacapavir) Receives FDA Approval as a First-in-Class, Twice-Yearly Treatment Option for People Living With Multi-Drug Resistant HIV. Available online: . (accessed on 22.12.2022).
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 2020, 584, 614–618. [Google Scholar] [CrossRef]
- Mahdi, M.; Motyan, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tozser, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2’s main protease. Virology journal 2020, 17, 190. [Google Scholar] [CrossRef]
- Indu, P.; Rameshkumar, M.R.; Arunagirinathan, N.; Al-Dhabi, N.A.; Valan Arasu, M.; Ignacimuthu, S. Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: A molecular docking and drug repurposing approach. Journal of infection and public health 2020, 13, 1856–1861. [Google Scholar] [CrossRef] [PubMed]
- Cavaco-Silva, J.; Abecasis, A.; Miranda, A.C.; Pocas, J.; Narciso, J.; Aguas, M.J.; Maltez, F.; Almeida, I.; Germano, I.; Diniz, A.; et al. HIV-2 integrase polymorphisms and longitudinal genotypic analysis of HIV-2 infected patients failing a raltegravir-containing regimen. PloS one 2014, 9, e92747. [Google Scholar] [CrossRef] [PubMed]
- Hutapea, H.M.L.; Maladan, Y.; Widodo. Relationship between HIV integrase polymorphisms and integrase inhibitor susceptibility: An in silico analysis. Heliyon 2018, 4, e00956. [Google Scholar] [CrossRef] [PubMed]
- Bercoff, D.P.; Triqueneaux, P.; Lambert, C.; Oumar, A.A.; Ternes, A.M.; Dao, S.; Goubau, P.; Schmit, J.C.; Ruelle, J. Polymorphisms of HIV-2 integrase and selection of resistance to raltegravir. Retrovirology 2010, 7, 98. [Google Scholar] [CrossRef]
- Miklossy, G.; Tozser, J.; Kadas, J.; Ishima, R.; Louis, J.M.; Bagossi, P. Novel macromolecular inhibitors of human immunodeficiency virus-1 protease. Protein engineering, design & selection : PEDS 2008, 21, 453–461. [Google Scholar] [CrossRef]
- Mahdi, M.; Matuz, K.; Toth, F.; Tozser, J. A modular system to evaluate the efficacy of protease inhibitors against HIV-2. PloS one 2014, 9, e113221. [Google Scholar] [CrossRef] [PubMed]
- Jochmans, D.; Leyssen, P.; Neyts, J. A novel method for high-throughput screening to quantify antiviral activity against viruses that induce limited CPE. Journal of virological methods 2012, 183, 176–179. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic acids research 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Sondergaard, C.R.; Olsson, M.H.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. Journal of chemical theory and computation 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. Journal of chemical theory and computation 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- D.A. Case, R.M.B., D. D.A. Case, R.M.B., D.S. Cerutti, T.E. Cheatham, III, T.A. Darden, R.E. Duke, T.J. Giese, H. Gohlke, A.W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T.S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K.M. Merz, G. Monard, H. Nguyen, H.T. Nguyen, I. Omelyan, A. Onufriev, D.R. Roe, A. Roitberg, C. Sagui, C.L. Simmerling, W.M. Botello-Smith, J. Swails, R.C. Walker, J. Wang, R.M. Wolf, X. Wu, L. Xiao and P.A. Kollman AMBER 2016, University of California, San Francisco.
- Scott Le Grand, A.W.G. , Ross C. Walker. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Computer Physics Communications 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. Journal of chemical theory and computation 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. Journal of chemical theory and computation 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbova, M.; Sponer, J.; Otyepka, M.; Jurecka, P.; Cheatham, T.E. , 3rd. Assessing the Current State of Amber Force Field Modifications for DNA. Journal of chemical theory and computation 2016, 12, 4114–4127. [Google Scholar] [CrossRef]
- Li, P.; Merz, K.M., Jr. Taking into Account the Ion-induced Dipole Interaction in the Nonbonded Model of Ions. Journal of chemical theory and computation 2014, 10, 289–297. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. Journal of chemical theory and computation 2020, 16, 1367–1372. [Google Scholar] [CrossRef]
- M. J. Frisch, G.W.T., H. M. J. Frisch, G.W.T., H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, and D. J. Fox Gaussian 09, Gaussian, Inc., Wallingford CT: 2016.
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theoretical Chemistry Accounts 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. The Journal of Physical Chemistry 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. Journal of molecular graphics & modelling 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stützle, T.; Exner, T.E. An ant colony optimization approach to flexible protein–ligand docking. Swarm Intelligence 2007, 1, 115–134. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In Proceedings of the Ant Colony Optimization and Swarm Intelligence, Berlin, Heidelberg, 2006; 2006//; pp. 247–258. [Google Scholar]
- Cook, N.J.; Li, W.; Berta, D.; Badaoui, M.; Ballandras-Colas, A.; Nans, A.; Kotecha, A.; Rosta, E.; Engelman, A.N.; Cherepanov, P. Structural basis of second-generation HIV integrase inhibitor action and viral resistance. Science 2020, 367, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lambert, C.; Arendt, V.; Seguin-Devaux, C. Virological and immunological outcomes of elvitegravir-based regimen in a treatment-naive HIV-2-infected patient. Aids 2014, 28, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Prather, C.; Lee, A.; Yen, C. Lenacapavir: A first-in-class capsid inhibitor for the treatment of highly treatment-resistant HIV. American journal of health-system pharmacy: AJHP: official journal of the American Society of Health-System Pharmacists. [CrossRef]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364. [Google Scholar] [CrossRef]
- Roquebert, B.; Damond, F.; Collin, G.; Matheron, S.; Peytavin, G.; Benard, A.; Campa, P.; Chene, G.; Brun-Vezinet, F.; Descamps, D.; et al. HIV-2 integrase gene polymorphism and phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitors raltegravir and elvitegravir in vitro. The Journal of antimicrobial chemotherapy 2008, 62, 914–920. [Google Scholar] [CrossRef]
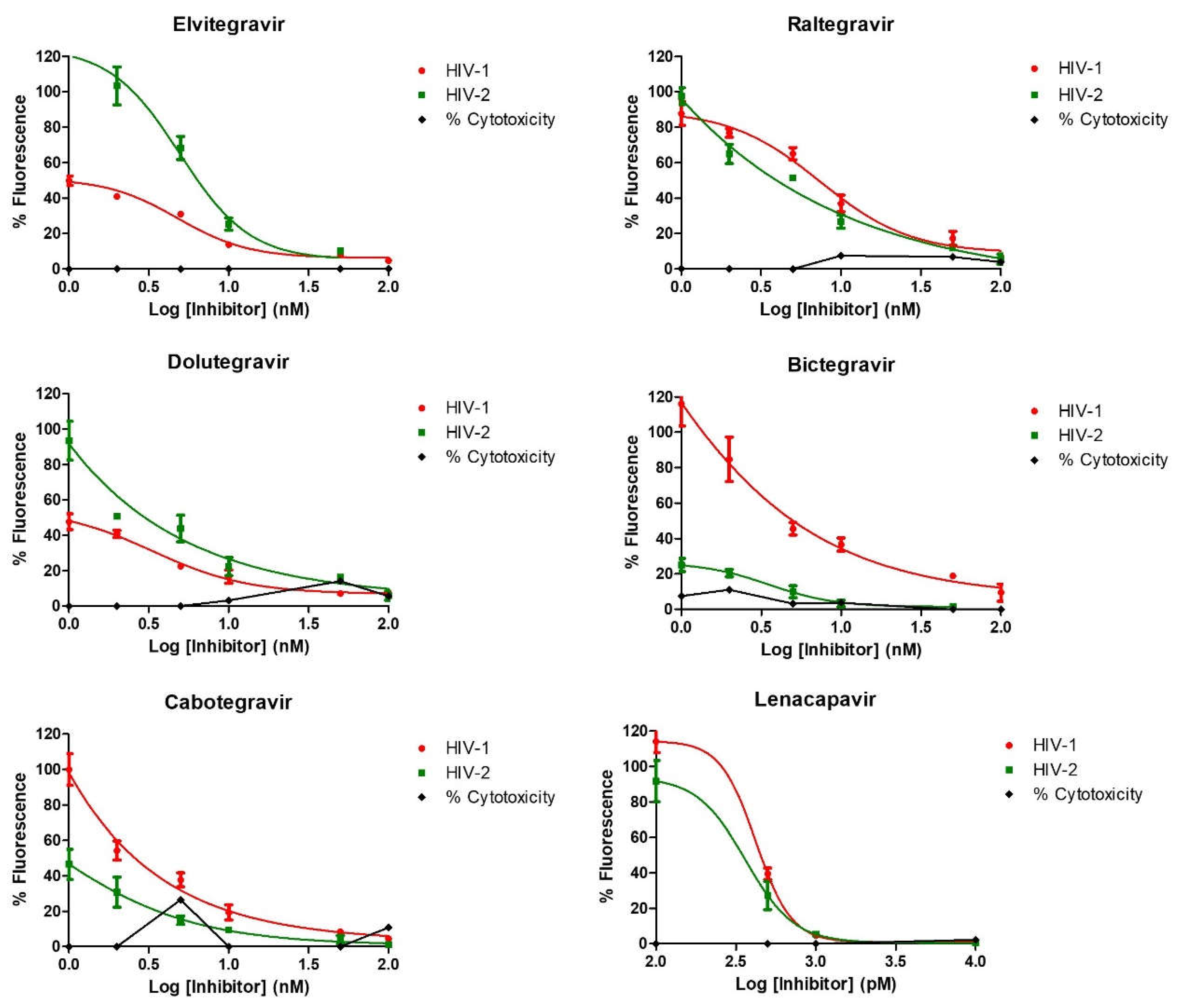
| IC50 (nM) | ||
|---|---|---|
| INSTI | HIV-1 | HIV-2 |
| Elvitegravir | 2.5 (±0.1) | 2.6 (±0.4) |
| Raltegravir | 6.9 (±0.1) | 2.1 (±0.1) |
| Dolutegravir | 2.2 (±0.1) | 1.1 (±0.3) |
| Bictegravir | 1.2 (±0.2) | 1.8 (±0.3) |
| Cabotegravir | 0.4 (±0.3) | 0.8 (±0.4) |
| Capsid Inhibitor | IC50 (pM) | |
| Lenacapavir | 89 (±0.2) | 50.6 (±0.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





