Submitted:
16 April 2024
Posted:
16 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Patients and Methods
2.1. Patients and Samples
2.2. MicroRNA Quantification
2.2.1. RNA Isolation
2.2.2. cDNA Synthesis
2.2.3. Serum microRNA Expression Quantification by Quantitative Realtime PCR (qRT-PCR)
2.3. Statistical Analysis
2.4. Ethical Approval
3. Results
3.1. Demographic, Clinical and Laboratory Characteristics
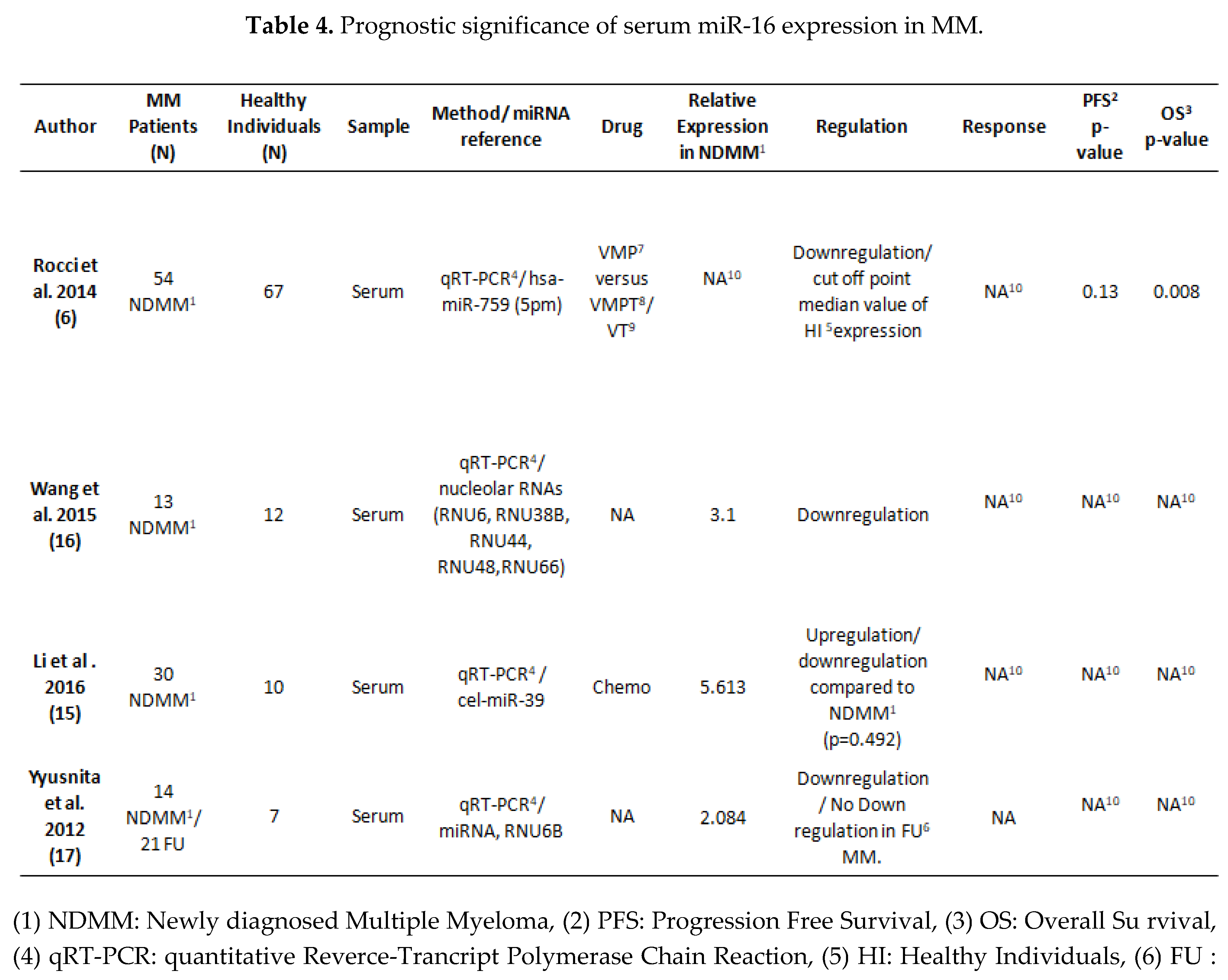
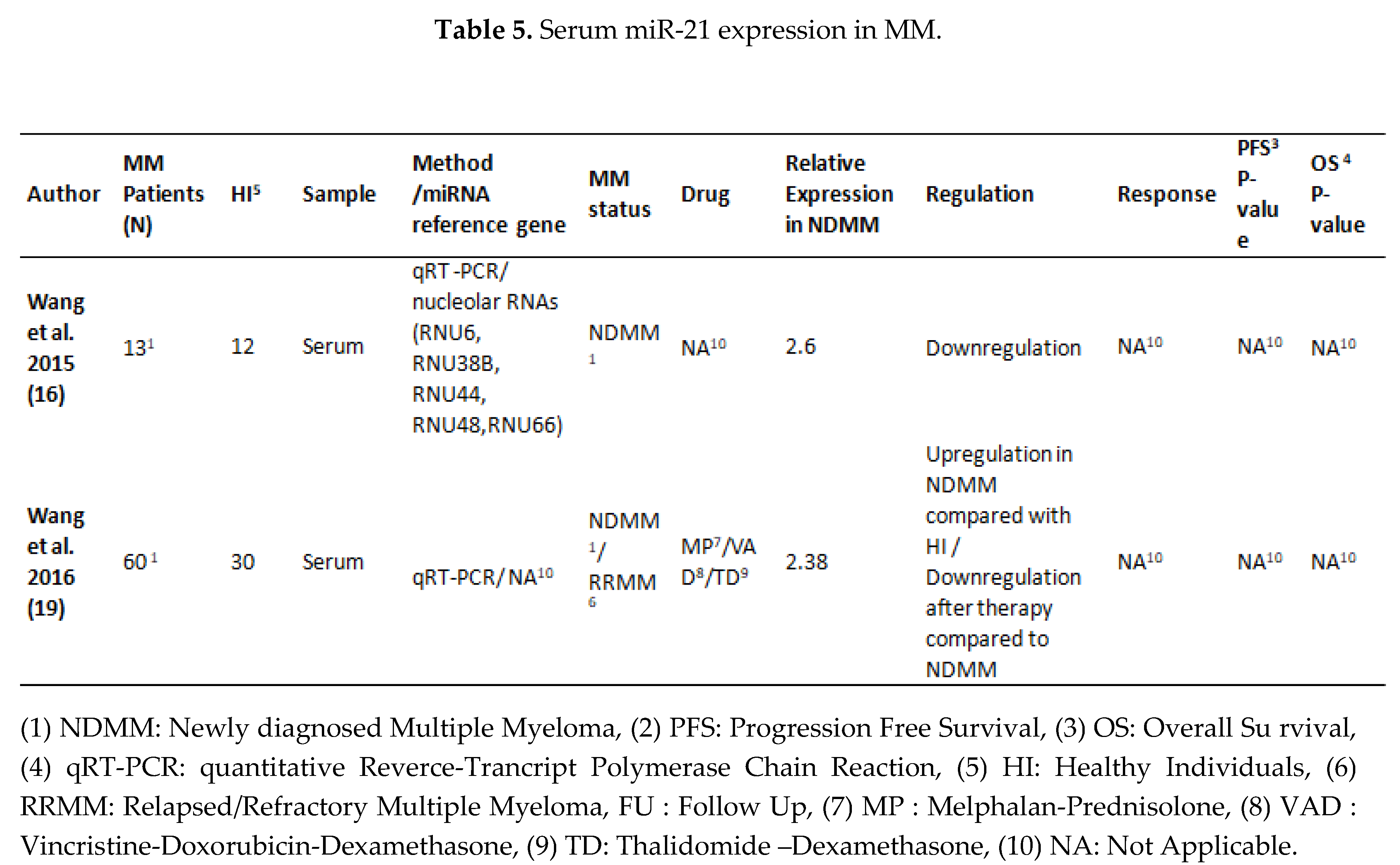
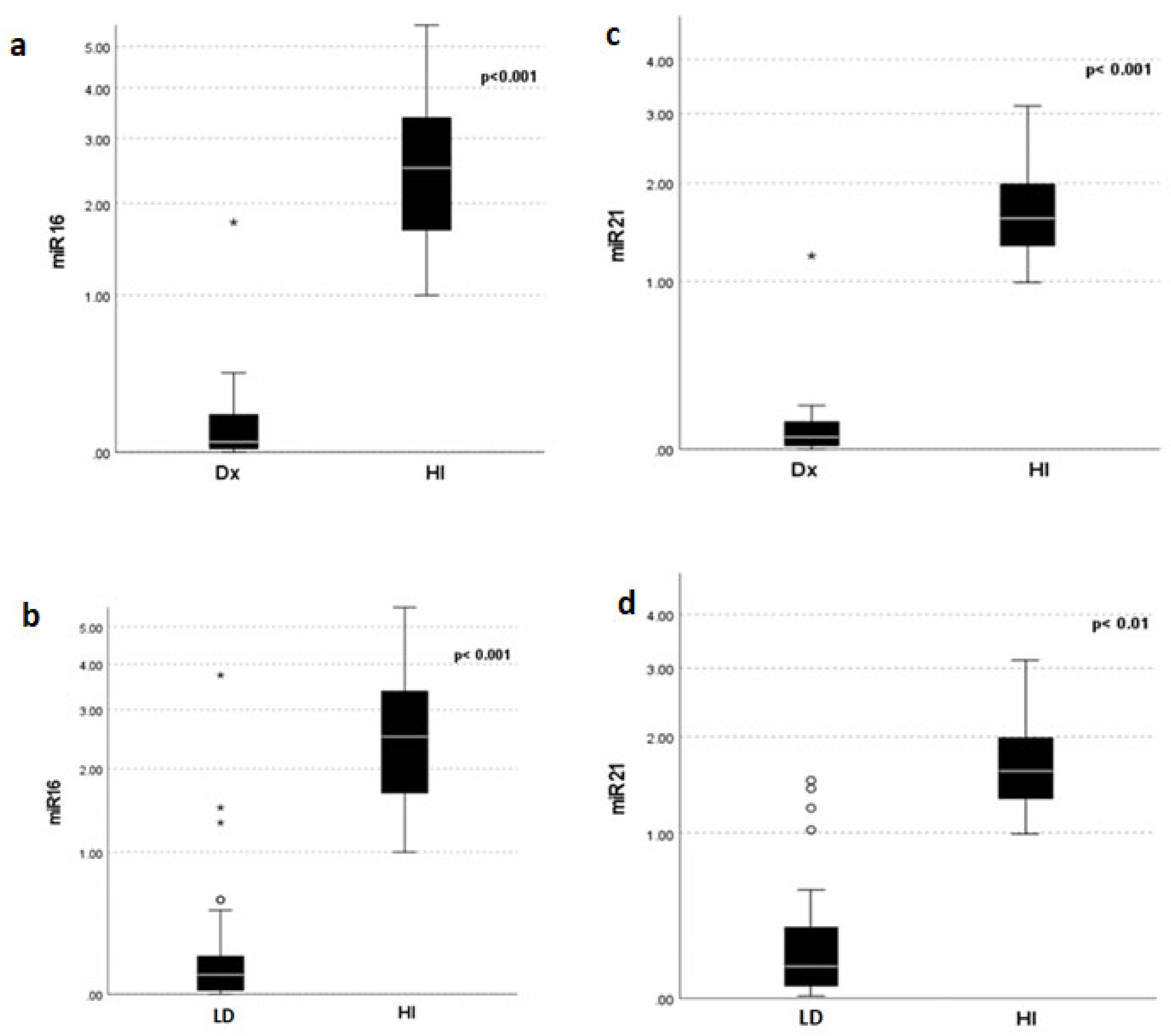
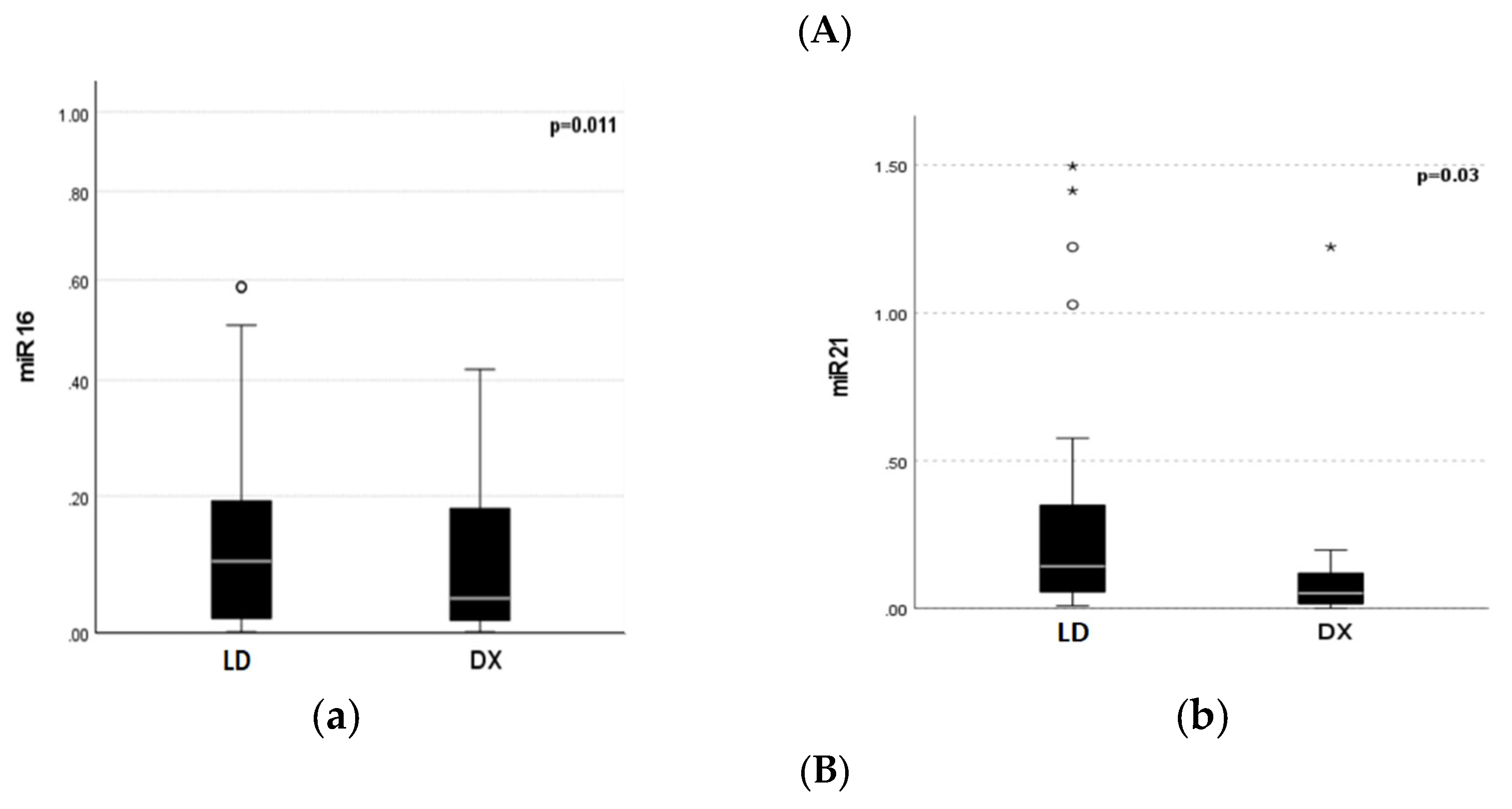
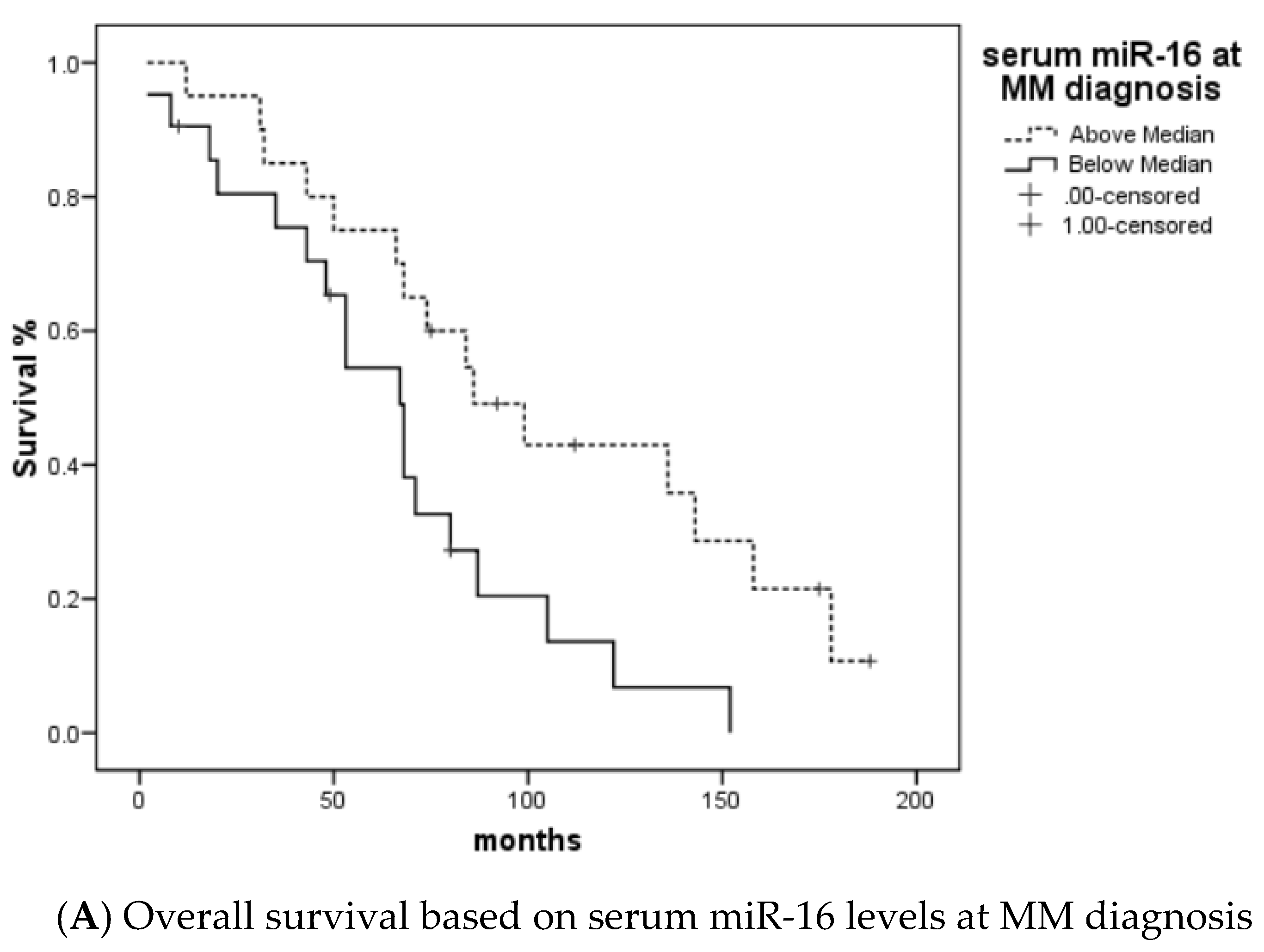
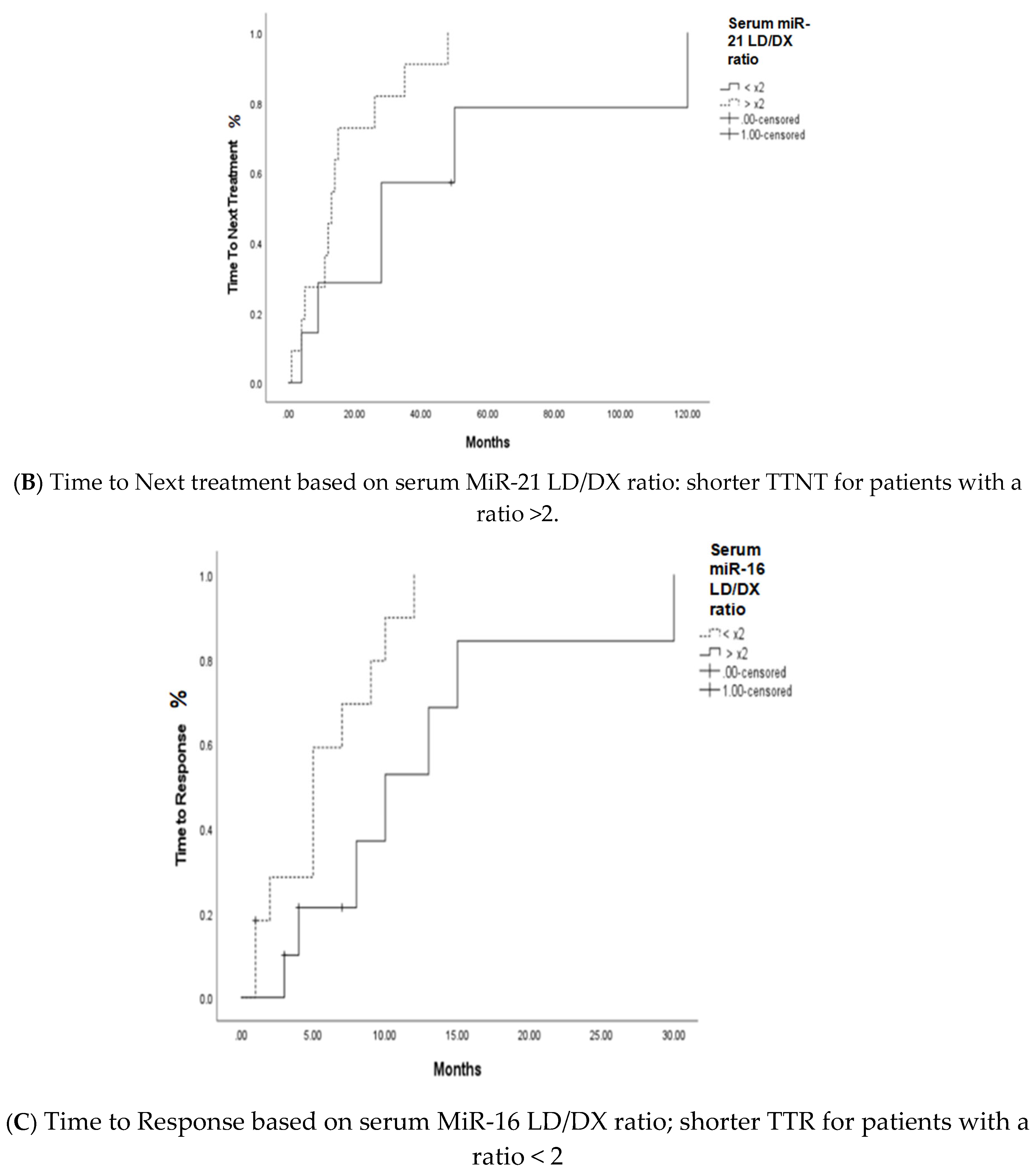
3.2. MiRNA 16 and MiRNA 21 Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cowan, A.J., D. J. Green, M. Kwok, S. Lee, D.G. Coffey, L.A. Holmberg, S. Tuazon, A.K. Gopal, and E.N. Libby, Diagnosis and Management of Multiple Myeloma: A Review. Jama, 2022. 327(5): p. 464-477.
- Kyrtsonis, M.C., V. Bartzis, X. Papanikolaou, E. Koulieris, G. Georgiou, M. Dimou, T. Tzenou, and P. Panayiotidis, Genetic and molecular mechanisms in multiple myeloma: a route to better understand disease pathogenesis and heterogeneity. Appl Clin Genet, 2010. 3: p. 41-51.
- Cortez, M.A., C. Bueso-Ramos, J. Ferdin, G. Lopez-Berestein, A.K. Sood, and G.A. Calin, MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol, 2011. 8(8): p. 467-77.
- Manier, S., C. J. Liu, H. Avet-Loiseau, J. Park, J. Shi, F. Campigotto, K.Z. Salem, D. Huynh, S.V. Glavey, B. Rivotto, et al., Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood, 2017. 129(17): p. 2429-2436.
- Yao, Q., Y. Chen, and X. Zhou, The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol, 2019. 51: p. 11-17.
- Rocci, A., C. C. Hofmeister, S. Geyer, A. Stiff, M. Gambella, L. Cascione, J. Guan, D.M. Benson, Y.A. Efebera, T. Talabere, et al., Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia, 2014. 28(9): p. 1922-6.
- Casabonne, D., Y. Benavente, J. Seifert, L. Costas, M. Armesto, M. Arestin, C. Besson, F.S. Hosnijeh, E.J. Duell, E. Weiderpass, et al., Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int J Cancer, 2020. 147(5): p. 1315-1324.
- Masri, A.A., T. Price-Troska, M. Chesi, T.-H. Chung, S. Kim, J. Carpten, P.L. Bergsagel, and R. Fonseca, MicroRNA Expression Analysis in Multiple Myeloma. Blood, 2005. 106(11): p. 1554-1554.
- Ma, J., S. Liu, and Y. Wang, MicroRNA-21 and multiple myeloma: small molecule and big function. Med Oncol, 2014. 31(8): p. 94.
- Dubaj, M., K. Bigosiński, A. Dembowska, R. Mlak, A. Szudy-Szczyrek, T. Małecka-Massalska, and I. Homa-Mlak, Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma. Cancers (Basel), 2024. 16(5).
- Pfeffer, S.R., C. H. Yang, and L.M. Pfeffer, The Role of miR-21 in Cancer. Drug Dev Res, 2015. 76(6): p. 270-7.
- Dimopoulos, M.A., P. Moreau, E. Terpos, M.V. Mateos, S. Zweegman, G. Cook, M. Delforge, R. Hájek, F. Schjesvold, M. Cavo, et al., Multiple myeloma: EHA-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up(†). Ann Oncol, 2021. 32(3): p. 309-322.
- Abdi, J., H. Jian, and H. Chang, Role of micro-RNAs in drug resistance of multiple myeloma. Oncotarget, 2016. 7(37): p. 60723-60735.
- Jiang, Y., Y. Luan, H. Chang, and G. Chen, The diagnostic and prognostic value of plasma microRNA-125b-5p in patients with multiple myeloma. Oncol Lett, 2018. 16(3): p. 4001-4007.
- Li, F., Y. Xu, S. Deng, Z. Li, D. Zou, S. Yi, W. Sui, M. Hao, and L. Qiu, MicroRNA-15a/16-1 cluster located at chromosome 13q14 is down-regulated but displays different expression pattern and prognostic significance in multiple myeloma. Oncotarget, 2015. 6(35): p. 38270-82.
- Wang, W., M. Corrigan-Cummins, E.A. Barber, L.M. Saleh, A. Zingone, A. Ghafoor, R. Costello, Y. Zhang, R.J. Kurlander, N. Korde, et al., Aberrant Levels of miRNAs in Bone Marrow Microenvironment and Peripheral Blood of Myeloma Patients and Disease Progression. J Mol Diagn, 2015. 17(6): p. 669-78.
- Xu, Y.N., C. R. Xiao, Y.D. Huang, and Q.Y. Lu, [Circulating Serum MicroRNA as Diagnostic Biomarkers for Multiple Myeloma]. Zhongguo Shi Yan Xue Ye Xue Za Zhi, 2017. 25(2): p. 471-475.
- Rhim, J., W. Baek, Y. Seo, and J.H. Kim, From Molecular Mechanisms to Therapeutics: Understanding MicroRNA-21 in Cancer. Cells, 2022. 11(18).
- Wang, X., C. Li, S. Ju, Y. Wang, H. Wang, and R. Zhong, Myeloma cell adhesion to bone marrow stromal cells confers drug resistance by microRNA-21 up-regulation. Leuk Lymphoma, 2011. 52(10): p. 1991-8.
- Zhou, L., X. L. Liu, Y.W. Li, L. Wu, G.Z. Wang, Z.F. Wang, L. Ma, J. Guan, and C.X. Han, [Clinical Study of miRNAs Derived from Serum Exosomes in Multiple Myeloma]. Zhongguo Shi Yan Xue Ye Xue Za Zhi, 2022. 30(5): p. 1490-1495.
- Moura, S.R., H. Abreu, C. Cunha, C. Ribeiro-Machado, C. Oliveira, M.A. Barbosa, H. Marques, and M.I. Almeida, Circulating microRNAs Correlate with Multiple Myeloma and Skeletal Osteolytic Lesions. Cancers, 2021. 13(21): p. 5258.
- Robak, P., I. Dróżdż, D. Jarych, D. Mikulski, E. Węgłowska, M. Siemieniuk-Ryś, M. Misiewicz, K. Stawiski, W. Fendler, J. Szemraj, et al., The Value of Serum MicroRNA Expression Signature in Predicting Refractoriness to Bortezomib-Based Therapy in Multiple Myeloma Patients. Cancers (Basel), 2020. 12(9).
- Hao, M., L. Zhang, G. An, W. Sui, Z. Yu, D. Zou, Y. Xu, H. Chang, and L. Qiu, Suppressing miRNA-15a/-16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J Hematol Oncol, 2011. 4: p. 37.
- Wang, J.H., W. W. Zhou, B.X. Liu, D.L. Man, Z.D. Yang, F.R. Liu, and H. Shang, Expression and significance of miR-21 in multiple myeloma patients. Genet Mol Res, 2016. 15(1).
- Xu, Q., Y. X. Hou, P. Langlais, P. Erickson, J. Zhu, C.X. Shi, M. Luo, Y. Zhu, Y. Xu, L.J. Mandarino, et al., Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer, 2016. 16: p. 297.
- Jung, S.H., S. E. Lee, M. Lee, S.H. Kim, S.H. Yim, T.W. Kim, C.K. Min, and Y.J. Chung, Circulating microRNA expressions can predict the outcome of lenalidomide plus low-dose dexamethasone treatment in patients with refractory/relapsed multiple myeloma. Haematologica, 2017. 102(11): p. e456-e459.
- Majithia, N., S. V. Rajkumar, M.Q. Lacy, F.K. Buadi, A. Dispenzieri, M.A. Gertz, S.R. Hayman, D. Dingli, P. Kapoor, L. Hwa, et al., Early relapse following initial therapy for multiple myeloma predicts poor outcomes in the era of novel agents. Leukemia, 2016. 30(11): p. 2208-2213.
| Variable (N=48) |
Number (%) / Median (Range) |
|---|---|
| Age | 67 (40-85) |
| Sex | |
| Male | 28 (58%) |
| Female | 20 (42%) |
| Ig Type | |
| IgG | 22 (46%) |
| IgA | 16 (33%) |
| Light Chain | 8 (17%) |
| Biclonal | 2 (4%) |
| Light Chain Restriction | |
| Kappa | 31 (64 %) |
| Lamda | 17 (36 %) |
| ISS1 | |
| I | 15 (33%) |
| II | 13 (28%) |
| III | 18 (39%) |
| Variable (N=48) | Median (Range) |
|---|---|
| Hb1g/dl | 11.1 (6.7-15) |
| PLTs2 109/μL | 212 (52-375) |
| Cr3mg/dl | 1 (0.5-4.6) |
| Hypercalcemia | 10 (22 %) |
| Alb4 | 4.08 (2.5-5) |
| Bone Disease (determined by Imaging) | 36 (80 %) |
| IgG mg/L | 3360 (1850-8730) |
| IgA mg/L | 3255 (944-6450) |
| IgM mg/L | 17.3 (16.8-99.8) |
| FLCk5 mg/L | 1398 (1.98-1510) |
| FLCλ6mg/L | 3.36 (0.08-2690) |
| FLCR7 | 42 (0.9-52750) |
| B2M8 mg/L | 3.8 (2-16) |
| BMINF9 % | 50 (10-95) |
| LDH UNL10 | 3 (7 %) |
| Neu11 109/μL | 3455 (500-17880) |
| Lymph12 109/μL | 1610 (780-4340) |
| Neu/Lymph13 | 2 (0.52-17.5) |
| CRP14 mg/L | 3.45 (0.470-86.5) |
| Variables | miR-16 Dx * | miR-21 Dx * | ||
|---|---|---|---|---|
| R | P value | r | P value | |
| B2M1 | -0.370 | 0.021 | NS | NS |
| B2M < 3.5 | NS | NS | -0.372 | 0.017 |
| CRP2 | -0.356 | 0.022 | -0.285 | 0.068 |
| NEU3/LYMPH4 | -0.307 | 0.069 | -0.332 | 0.045 |
| Renal Failure | -0.410 | 0.008 | -0.301 | 0.05 |
| GFR5 | 0.419 | 0.012 | NS | NS |
| Hypercalcemia | -0.460 | 0.003 | NS | NS |
| Variables | miR-16 LD v | miR-21 LD v | miR-16 LD v /DX* | miR-21 LD v /DX* | ||||
|---|---|---|---|---|---|---|---|---|
| R | P-value | r | P-value | R | P-value | r | P-value | |
| Biochemical Relapse | NS | NS | -0.371 | 0.068 | NS | NS | 0.547 | 0.015 |
| ISS1 LD | NS | NS | 0.468 | 0.018 | NS | NS | NS | NS |
| Relapse > 24 months | NS | NS | NS | NS | 0.709 | 0.0001 | 0.461 | 0.035 |
| Response ≥ VGPR2 | NS | NS | 0.453 | 0.034 | NS | NS | NS | NS |
| Response ≥PR3 | NS | NS | NS | NS | -0.413 | 0.05 | NS | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





