Submitted:
12 April 2024
Posted:
15 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. A Historical Perspective on Legume Seed Coat Pigmentation
3. Mechanisms of Partner Selection in Symbiotic Interactions
4. Genetic Determinants of Legume Seed Coat Pigmentation
5. Genetic Determinants of Host Plants in Symbiotic Interactions with Rhizobia
6. Seed Coat Pigmentation and N2 Fixation in Legumes
7. Effect of Seed Coat Pigmentation on Microbial Colonization
8. Evolutionary Dynamics of the Legume-Rhizobia Partnership
9. Adapting to Environmental Changes: The Role of Seed Coat Pigmentation
10. Grain legume Seed Coats Are a Natural Source of Nutraceuticals and Anthocyanins
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Molefe, R.R.; Amoo, A.E.; Babalola, O.O. Communication between plant roots and the soil microbiome; involvement in plant growth and development. Symbiosis 2023, 90, 231–239. [Google Scholar] [CrossRef]
- Barbieri, P.; Starck, T.; Voisin, A.-S.; Nesme, T. Biological nitrogen fixation of legumes crops under organic farming as driven by cropping management: A review. Agric. Syst. 2023, 205, 103579. [Google Scholar] [CrossRef]
- Dimkpa, C.; Adzawla, W.; Pandey, R.; Atakora, W.K.; Kouame, A.K.; Jemo, M.; Bindraban, P.S. Fertilizers for food and nutrition security in sub-Saharan Africa: an overview of soil health implications. Front. Soil Sci. 2023, 3. [Google Scholar] [CrossRef]
- Nag, P.; Shriti, S.; Das, S. Microbiological strategies for enhancing biological nitrogen fixation in nonlegumes. J. Appl. Microbiol. 2020, 129, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Ngwenya, Z.D.; Mohammed, M.; Dakora, F.D. Monocropping and Intercropping of Maize with Six Food Legumes at Malkerns in Eswatini: Their Effects on Plant Growth, Grain Yield and N2 Fixation, Measured using the 15N Natural Abundance and Ureide Techniques. Symbiosis 2024, 1–13. [Google Scholar] [CrossRef]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. Black seedcoat pigmentation is a marker for enhanced nodulation and N2 fixation in Bambara groundnut (Vigna subterranea L. Verdc.) landraces. Front. Agron. 2021, 3, 692238. [Google Scholar] [CrossRef]
- Bopape, F.L.; Beukes, C.W.; Katlego, K.; Hassen, A.I.; Steenkamp, E.T.; Gwata, E.T. Symbiotic performance and characterization of Pigeonpea (Cajanus cajan L. Millsp.) Rhizobia occurring in South African soils. Agriculture 2022, 13, 30. [Google Scholar] [CrossRef]
- Nyemba, R.C.; Dakora, F.D. Evaluating N 2 fixation by food grain legumes in farmers ’ fields in three agro-ecological zones of Zambia, using 15 N natural abundance. Biol Fertil Soils 2010, 461–470. [Google Scholar] [CrossRef]
- Bohra, A.; Tiwari, A.; Kaur, P.; Ganie, S.A.; Raza, A.; Roorkiwal, M.; Mir, R.R.; Fernie, A.R.; Sm\`ykal, P.; Varshney, R.K. The key to the future lies in the past: insights from grain legume domestication and improvement should inform future breeding strategies. Plant Cell Physiol. 2022, 63, 1554–1572. [Google Scholar] [CrossRef] [PubMed]
- Konzen, E.R.; Tsai, S.M. Genetic variation of landraces of common bean varying for seed coat glossiness and disease resistance: valuable resources for conservation and breeding. Rediscovery Landrac. as a Resour. Futur. London IntechOpen 2018, 177–193. [Google Scholar]
- Smýkal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar] [CrossRef] [PubMed]
- Paauw, M.; Koes, R.; Quattrocchio, F.M. Alteration of flavonoid pigmentation patterns during domestication of food crops. J. Exp. Bot. 2019, 70, 3719–3735. [Google Scholar] [CrossRef] [PubMed]
- Von Wettberg, E.J.B.; Chang, P.L.; Ba\csdemir, F.; Carrasquila-Garcia, N.; Korbu, L.B.; Moenga, S.M.; Bedada, G.; Greenlon, A.; Moriuchi, K.S.; Singh, V.; et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 2018, 9, 649. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.-S.; Contador, C.A.; Ng, M.-S.; Yu, J.; Chung, G.; Lam, H.-M. The effects of domestication on secondary metabolite composition in legumes. Front. Genet. 2020, 11, 581357. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, U. a; Maxwell, C. a; Joseph, C.M.; Phillips, D. a Chrysoeriol and Luteolin Released from Alfalfa Seeds Induce nod Genes in Rhizobium meliloti. Plant Physiol. 1990, 92, 116–22. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, U. a; Joseph, C.M.; Phillips, D. a Flavonoids Released Naturally from Alfalfa Seeds Enhance Growth Rate of Rhizobium meliloti. Plant Physiol. 1991, 95, 797–803. [Google Scholar] [CrossRef]
- Hungria, M.; Phillips, D. Effects of a seed colour mutation on rhizobial nod-gene-inducing flavonoids and nodulation in common bean. Mol. Plant-Microbe Interact. 1993, 6, 418–422. [Google Scholar] [CrossRef]
- Cooper, J.E. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007, 103, 1355–1365. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Zhang, Y.; Zeng, X.; Li, P.; Wang, X.; Benedito, V.A.; Zhao, J. Isoflavone malonyl-CoA acyltransferase GmMaT2 is involved in nodulation of soybean by modifying synthesis and secretion of isoflavones. J. Exp. Bot. 2021, 72, 1349–1369. [Google Scholar] [CrossRef] [PubMed]
- Hungria, M.; Joseph, C.M.; Phillips, D.A. Anthocyanidins and Flavonols, Major nod Gene Inducers from Seeds of a Black-Seeded Common Bean. 1991, 751–758. [Google Scholar]
- Subramanian, S.; Stacey, G.; Yu, O. Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 2007, 12, 282–5. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.P.; Ankita Aski, M.S.; Tontang, M.T.; Choudhary, P.; Tripathi, K.; Singh, A.; Kumar, R.R.; Thimmegowda, V.; Stobdan, T.; et al. Morphological, molecular, and biochemical characterization of a unique lentil (Lens culinaris medik.) genotype showing seed-coat color anomalies due to altered anthocyanin pathway. Plants 2022, 11, 1815. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Gillaspie, A.G.; Morris, J.B.; Pittman, R.N.; Davis, J.; Pederson, G.A. Flavonoid content in different legume germplasm seeds quantified by HPLC. Plant Genet. Resour. 2008, 6, 62–69. [Google Scholar] [CrossRef]
- dos Santos Sousa, W.; Teixeira, I.R.; Souza Campos, T.; da Silva, G.C.; da Silva, M.; Guimarães Moreira, S. Supplementary reinoculation in topdressing of Rhizobium tropici in common bean crop: effects on nodulation, morphology, and grain yield. J. Plant Nutr. 2022, 45, 3094–3108. [Google Scholar] [CrossRef]
- Shumilina, J.; Soboleva, A.; Abakumov, E.; Shtark, O.Y.; Zhukov, V.A.; Frolov, A. Signaling in Legume--Rhizobia Symbiosis. Int. J. Mol. Sci. 2023, 24, 17397. [Google Scholar] [CrossRef] [PubMed]
- Compton, K.K.; Scharf, B.E. Rhizobial chemoattractants, the taste and preferences of legume symbionts. Front. Plant Sci. 2021, 12, 686465. [Google Scholar] [CrossRef] [PubMed]
- Samac, D.A.; Graham, M.A. Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol. 2007, 144, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Sivasakthi, K.; Marques, E.; Kalungwana, N.; Carrasquilla-Garcia, N.; Chang, P.L.; Bergmann, E.M.; Bueno, E.; Cordeiro, M.; Sani, S.G.A.S.; Udupa, S.M.; et al. Functional dissection of the chickpea (Cicer arietinum L.) stay-green phenotype associated with molecular variation at an ortholog of Mendel’s I gene for cotyledon color: implications for crop production and carotenoid biofortification. Int. J. Mol. Sci. 2019, 20, 5562. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, J.H.; Zabala, G.; Varala, K.; Hudson, M.; Vodkin, L.O. Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 2009, 21, 3063–3077. [Google Scholar] [CrossRef] [PubMed]
- McClean, P.E.; Bett, K.E.; Stonehouse, R.; Lee, R.; Pflieger, S.; Moghaddam, S.M.; Geffroy, V.; Miklas, P.; Mamidi, S. White seed color in common bean (Phaseolus vulgaris) results from convergent evolution in the P (pigment) gene. New Phytol. 2018, 219, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Moreau, C.; Lin-Wang, K.; Schwinn, K.E.; Thomson, S.J.; Fiers, M.W.E.J.; Frew, T.J.; Murray, S.R.; Hofer, J.M.I.; Jacobs, J.M.E.; et al. Identification of Mendel’s white flower character. PLoS One 2010, 5, e13230. [Google Scholar] [CrossRef] [PubMed]
- Varma Penmetsa, R.; Carrasquilla-Garcia, N.; Bergmann, E.M.; Vance, L.; Castro, B.; Kassa, M.T.; Sarma, B.K.; Datta, S.; Farmer, A.D.; Baek, J.-M.; et al. Multiple post-domestication origins of kabuli chickpea through allelic variation in a diversification-associated transcription factor. New Phytol. 2016, 211, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Mirali, M.; Purves, R.W.; Stonehouse, R.; Song, R.; Bett, K.; Vandenberg, A. Genetics and biochemistry of zero-tannin lentils. PLoS One 2016, 11, e0164624. [Google Scholar] [CrossRef]
- Gutierrez, N.; Torres, A.M. Characterization and diagnostic marker for TTG1 regulating tannin and anthocyanin biosynthesis in faba bean. Sci. Rep. 2019, 9, 16174. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, N.; Avila, C.M.; Torres, A.M. The bHLH transcription factor VfTT8 underlies zt2, the locus determining zero tannin content in faba bean (Vicia faba L.). Sci. Rep. 2020, 10, 14299. [Google Scholar] [CrossRef] [PubMed]
- Garc\’\ia-Fernández, C.; Campa, A.; Ferreira, J.J. Dissecting the genetic control of seed coat color in a RIL population of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2021, 134, 3687–3698. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Rodr\’\iguez Madrera, R.; Jurado, M.; Garc\’\ia-Fernández, C.; Suárez Valles, B.; Ferreira, J.J. Genome-wide association study for the extractable phenolic profile and coat color of common bean seeds (Phaseolus vulgaris L.). BMC Plant Biol. 2023, 23, 158. [Google Scholar] [CrossRef] [PubMed]
- Herniter, I.A.; Munoz-Amatriain, M.; Lo, S.; Guo, Y.-N.; Lonardi, S.; Close, T.J. Identification of candidate genes controlling red seed coat color in cowpea (Vigna unguiculata [L.] Walp). Horticulturae 2024, 10, 161. [Google Scholar] [CrossRef]
- Belane, A.K.; Dakora, F.D. Measurement of N2 fixation in 30 cowpea (Vigna unguiculata L. Walp.) genotypes under field conditions in Ghana, using the15N natural abundance technique. Symbiosis 2009, 48, 47–56. [Google Scholar] [CrossRef]
- Belane, A.K.; Dakora, F.D. Symbiotic N2 fixation in 30 field-grown cowpea (Vigna unguiculata L. Walp.) genotypes in the Upper West Region of Ghana measured using 15 N natural abundance. Biol. Fertil. Soils 2010, 46, 191–198. [Google Scholar] [CrossRef]
- Pule-Meulenberg, F.; Belane, A.K.; Krasova-Wade, T.; Dakora, F.D. Symbiotic functioning and bradyrhizobial biodiversity of cowpea (Vigna unguiculata L. Walp.) in Africa. BMC Microbiol. 2010, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Herridge, D.; Rose, I. Breeding for enhanced nitrogen ® xation in crop legumes. 2000, 65. [Google Scholar]
- Egbadzor, K.F.; Yeboah, M.; Gamedoagbao, D.K.; Offei, S.K.; Danquah, E.Y.; Ofori, K. Inheritance of seed coat colour in cowpea (Vigna unguiculata (L.) Walp). 2014. [Google Scholar] [CrossRef]
- Mohammed, H.; Jaiswal, S.K.; Mohammed, M.; Mbah, G.C.; Dakora, F.D. Insights into nitrogen fixing traits and population structure analyses in cowpea (Vigna unguiculata L. Walp) accessions grown in Ghana. Physiol. Mol. Biol. Plants 2020, 26, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Jaiswal, S.K.; Sowley, E.N.K.; Ahiabor, B.D.K. Symbiotic N2 fixation and grain yield of endangered Kersting’s groundnut landraces in response to soil and plant associated Bradyrhizobium inoculation to promote ecological resource-use efficiency. Front. Microbiol. 2018, 9, 2105. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, M.J.; Staples, L.B. Origin, evaluation and use of Macrotyloma as forage. Trop Grassl 1993, 27, 16–29. [Google Scholar]
- Hassen, A.I.; Bopape, F.L.; van Vuuren, A.; Gerrano, A.S.; Morey, L. Symbiotic interaction of bambara groundnut (Vigna subterranea) landraces with rhizobia spp. from other legume hosts reveals promiscuous nodulation. South African J. Bot. 2023, 160, 493–503. [Google Scholar] [CrossRef]
- Wilker, J.; Navabi, A.; Rajcan, I.; Marsolais, F.; Hill, B.; Torkamaneh, D.; Pauls, K.P. Agronomic performance and nitrogen fixation of heirloom and conventional dry bean varieties under low-nitrogen field conditions. Front. Plant Sci. 2019, 10, 952. [Google Scholar] [CrossRef] [PubMed]
- Tsamo, A.T.; Ndibewu, P.P.; Dakora, F.D. Phytochemical profile of seeds from 21 Bambara groundnut landraces via UPLC-qTOF-MS. Food Res. Int. 2018, 112, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Tsamo, A.T.; Mohammed, M.; Ndibewu, P.P.; Dakora, F.D. Identification and quantification of anthocyanins in seeds of Kersting�s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet] landraces of varying seed coat pigmentation. J. Food Meas. Charact.
- Tsamo, A.T.; Mohammed, H.; Mohammed, M.; Papoh Ndibewu, P.; Dapare Dakora, F. Seed coat metabolite profiling of cowpea (Vigna unguiculata L. Walp.) accessions from Ghana using UPLC-PDA-QTOF-MS and chemometrics. Nat. Prod. Res. 2020, 34, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Mbah, G.C.; Dakora, F.D. Nitrate inhibition of N 2 fixation and its effect on micronutrient accumulation in shoots of soybean (Glycine max L. Merr.), Bambara groundnut (Vigna subterranea L. Vedc) and Kersting’s groundnut (Macrotyloma geocarpum Harms.). Symbiosis 2017, 1–12. [Google Scholar] [CrossRef]
- Zenabou, N.; Firmin, S.L.; Laurette, N.N.; Daniel, W.F.; Laurianne, T.N.; Martin, B.J. Nodulation Potential of Bambara Groundnut (Vigna subterranea L.) in Yaounde (Centre Region of Cameroon). Am. J. Food Nutr. 2022, 10, 34–39. [Google Scholar]
- Ouedraogo, M.; Kima, A.S.; Konaté, M.N.G.; Ouedraogo, H.M.; Ouoba, A.; Nikiema, B.; Kambou, D.J.; Zongo, K.F.; Nandkangre, H. Performance of bambara groundnut (Vigna subterranea [L.] Verdcourt) genotypes cropped on plinthite soil in the semi arid-zone, Burkina Faso. Int. J. Plant \& Soil Sci. 2022, 34, 1067–1075. [Google Scholar]
- Bitire, T.D.; Abberton, M.; Tella, E.O.; Edemodu, A.; Oyatomi, O.; Babalola, O.O. Impact of nitrogen-fixation bacteria on nitrogen-fixation efficiency of Bambara groundnut [Vigna subterranea (L) Verdc] genotypes. Front. Microbiol. 2023, 14, 1187250. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.K.; Mohammed, M.; Dakora, F.D. Microbial community structure in the rhizosphere of the orphan legume Kersting’s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet]. Mol. Biol. Rep. 2019, 46. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Jaiswal, S.K.; Dakora, F.D. Insights into the phylogeny, nodule function, and biogeographic distribution of microsymbionts nodulating the orphan Kersting’s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet] in African soils. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. African origin of Bradyrhizobium populations nodulating Bambara groundnut (Vigna subterranea L. Verdc) in Ghanaian and South African soils. PLoS One 2017, 12, e0184943. [Google Scholar] [CrossRef] [PubMed]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. Phylogeny and distribution of Bradyrhizobium symbionts nodulating cowpea (Vigna unguiculata L. Walp)and their association with the physicochemical properties of acidic African soils. Syst. Appl. Microbiol. 2019, 42. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, S.T.; Jaiswal, S.K.; Mohammed, M.; Dakora, F.D. Studies of Phylogeny, Symbiotic Functioning and Ecological Traits of Indigenous Microsymbionts Nodulating Bambara Groundnut (Vigna subterranea L. Verdc) in Eswatini. Microb. Ecol. 2021, 1–16. [Google Scholar] [CrossRef]
- Makoi, J.; Ndakidemi, P. Biological, ecological and agronomic significance of plant phenolic compounds in rhizosphere of the symbiotic legumes. African J. Biotechnol. 2007, 6. [Google Scholar]
- Ndakidemi, P.A.; Dakora, F.D. Review : Legume seed flavonoids and nitrogenous metabolites as signals and protectants in early seedling development. Funct. Plant Bilogy 2003, 30, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, J.M.M.; Ashby, A.M.; Richards, A.J.M.; Loake, G.J.; Watson, M.D.; Shaw, C.H. Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards Flavonoid Inducers of the Symbiotic Nodulation Genes. Microbiology 1988, 134, 2741–2746. [Google Scholar] [CrossRef]
- Begum, A.A. Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 2001, 52, 1537–1543. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–44. [Google Scholar] [CrossRef] [PubMed]
- Mabhaudhi, T.; Modi, A. Growth, phenological and yield responses of a bambara groundnut (Vigna subterranea L. Verdc) landrace to imposed water stress: II. Rain shelter conditions. Water SA 2013, 39, 191–198. [Google Scholar] [CrossRef]
- COLLINSON, S.T.; SIBUGA, K.P.; TARIMO, A.J.P.; AZAM-ALI, S.N. INFLUENCE OF SOWING DATE ON THE GROWTH AND YIELD OF BAMBARA GROUNDNUT LANDRACES IN TANZANIA. Exp. Agric. 2000, 36, 1–13. [Google Scholar] [CrossRef]
- Wamba, O.; Taffouo, V.; Youmbi, E.; Ngwene, B.; Amougou, A. Effects of organic and inorganic nutrient sources on growth, total chlorophyll and yield of three bambara groundnut landraces in the Coastal Region of Cameroom. J. Agron. 2012, 11, 31–42. [Google Scholar] [CrossRef]
- Nogués, S.; Allen, D.J.; Morison, J.I.L.; Baker, N.R. Ultraviolet-B radiation effects on water relations, leaf development, and photosynthesis in droughted pea plants. Plant Physiol. 1998, 117, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Del Bel, Z.; Andrade, A.; Lindström, L.; Alvarez, D.; Vigliocco, A.; Alemano, S. The role of the sunflower seed coat and endosperm in the control of seed dormancy and germination: phytohormone profile and their interaction with seed tissues. Plant Growth Regul. 2024, 102, 51–64. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Gao, P.; Yu, B.; Bing, D.; Datla, R.; Fobert, P.; Xiang, D. The seed coat’s impact on crop performance in pea (Pisum sativum L.). Plants 2022, 11, 2056. [Google Scholar] [CrossRef] [PubMed]
- Mo\"\ise, J.A.; Han, S.; Gudynait\ke-Savitch, L.; Johnson, D.A.; Miki, B.L.A. Seed coats: structure, development, composition, and biotechnology. Vitr. Cell. \& Dev. Biol. 2005, 41, 620–644. [Google Scholar]
- Shah, A.; Smith, D.L. Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Mohammed, M.; Ibny, F.Y.I.; Dakora, F.D. Rhizobia as a Source of Plant Growth-Promoting Molecules: Potential Applications and Possible Operational Mechanisms. Front. Sustain. Food Syst. 2021, 4, 1–14. [Google Scholar] [CrossRef]
- Makoi, J.H.J.R.; Belane, A.K.; Chimphango, S.B.M.; Dakora, F.D. Seed flavonoids and anthocyanins as markers of enhanced plant defence in nodulated cowpea (Vigna unguiculata L. Walp.). F. Crop. Res. 2010, 118, 21–27. [Google Scholar] [CrossRef]
- Herniter, I.A.; Jia, Z.; Kusi, F.; others. Market preferences for cowpea (Vigna unguiculata [L.] Walp) dry grain in Ghana. African J. Ag. Res 2019, 14, 928–934. [Google Scholar]
- Jiang, J.; Xiong, Y.L. Natural antioxidants as food and feed additives to promote health benefits and quality of meat products: A review. Meat Sci. 2016, 120, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: an overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
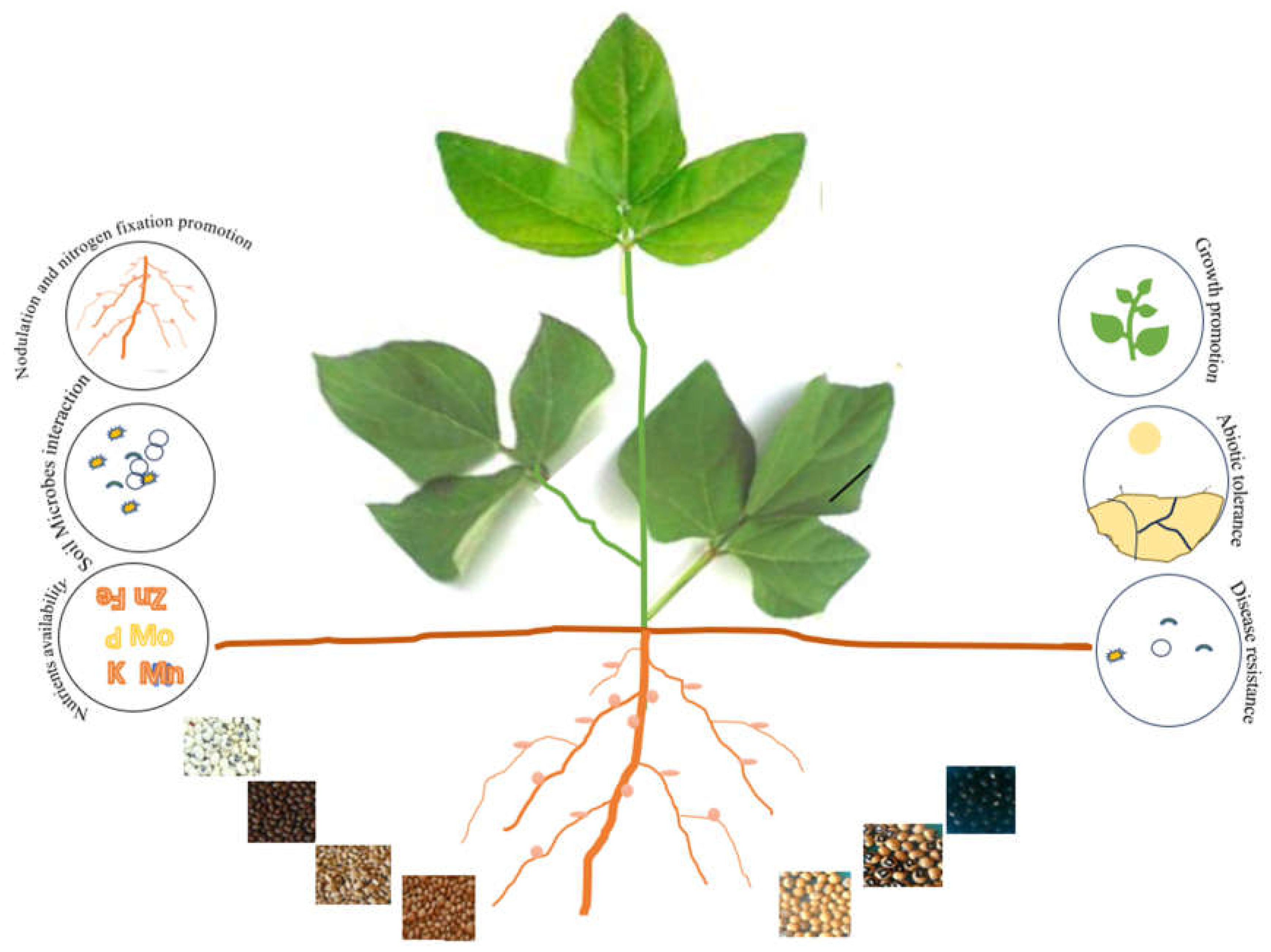
| Legume | Seedcoat color | Agronomical effect | References |
|---|---|---|---|
| Bambara Groundnut (Vigna Subterranea L. Verdc. | Black | Enhanced nodulation and nitrogen fixation | Puozaa et al. (2021) |
| Winged bean (Psophocarpus tetragonolobus) | Brown | Enhanced nodulation,and nitrogen fixation | Adegboyega et al. (2021) |
| Common bean (Phaseolus vulgaris L.) | light red | disease resistance and symbiotic nitrogen fixation | Wilker et al. (2020) |
| Soybean (Glycine max (L.) Merr.) | Black and Brown | Enhanced antioxidant activities and anthocyanins | Lim et al. (2021); Jung et al. (2022) |
| Soybean (Glycine max (L.) Merr.) | Yellow | Higher water absorption | Abati et al (2022) |
| Adzuki Bean (Vigna angularis L.) | Black | Higher accumulation of anthocyanins | Chu et al. (2021); Nagao et al. (2023) |
| Lentil (Lens culinaris Medik.) | Black | Higher nutraceutical values | Mishra et al. (2022) |
| Peanut (Arachis hypogaea L.) | Dark red | Higher polyphenol content | Nayak et al. 2020 |
| Kersting’s groundnut (Macrotyloma geocarpum Harms) | Black | Higher nitrogen fixation | Mohammed et al. (2018) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





