Submitted:
29 February 2024
Posted:
04 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Clam Sampling and DNA Extraction
2.2. Intron Amplification and DNA Polymorphism Detection
2.3. Data Analysis
3. Results
3.1. Newly Sampled Populations
3.2. Analysis of the Pooled Set of Populations
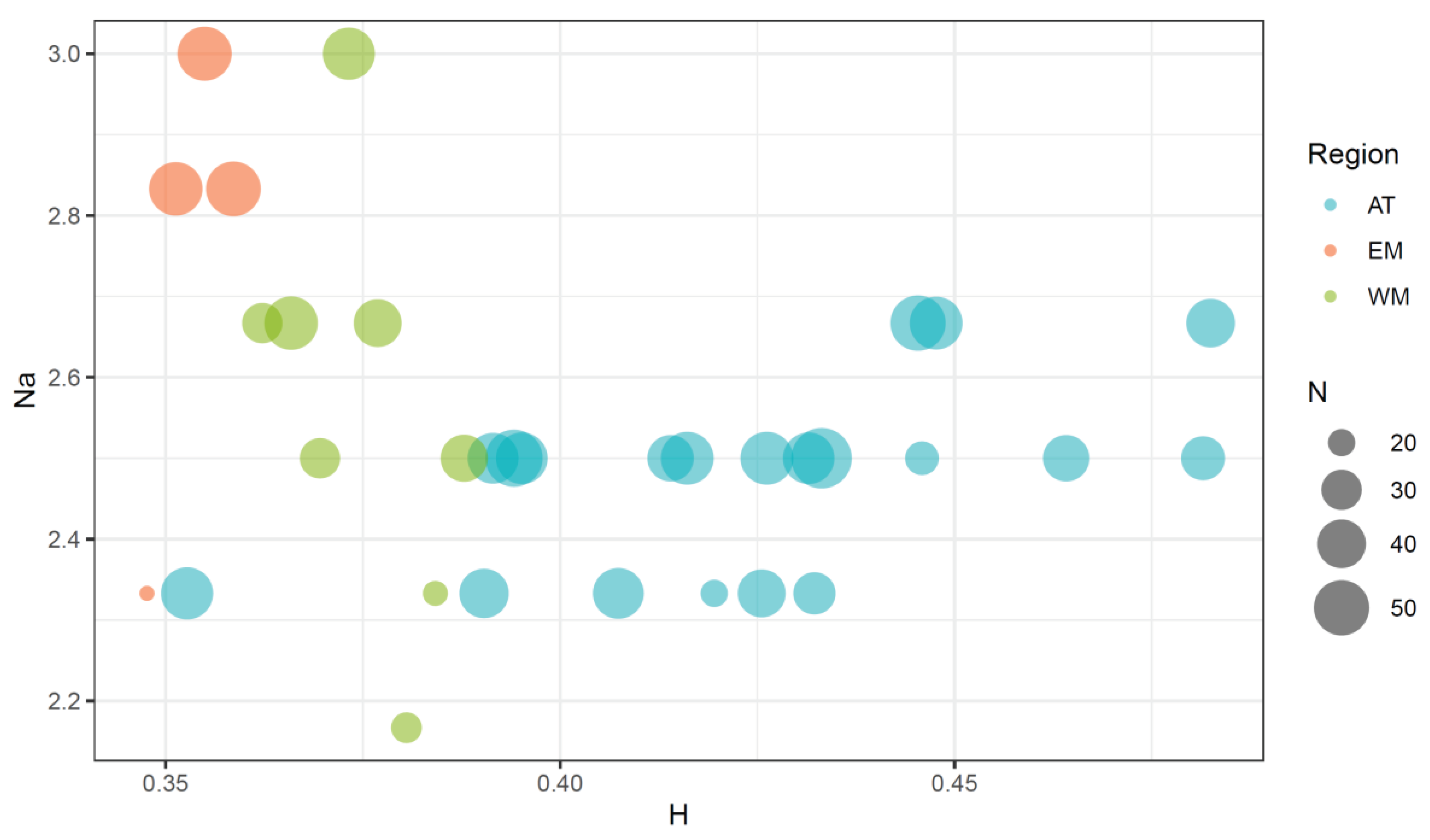
3.2.1. Genetic Variability
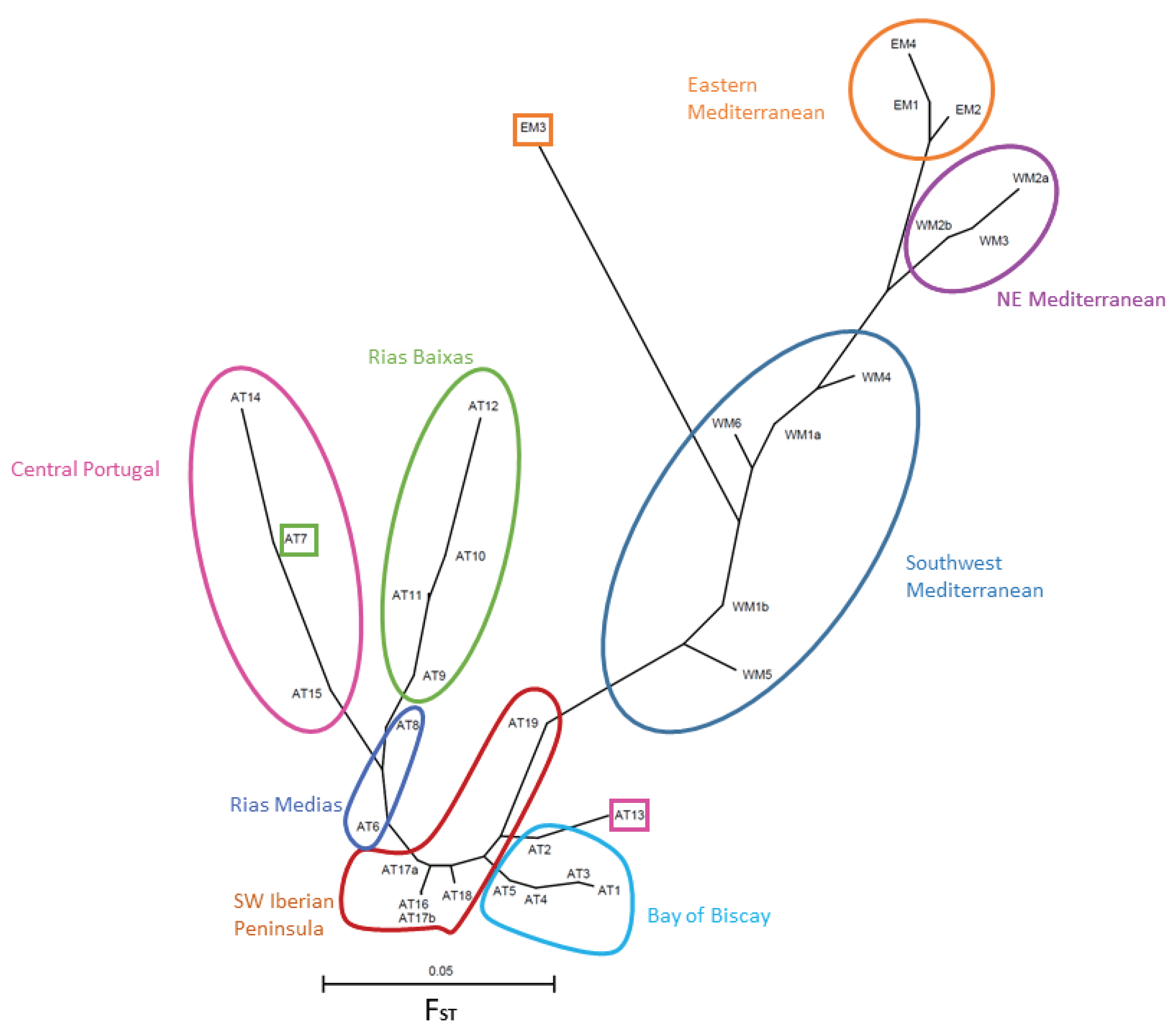
3.2.1. FST Statistics and Population Neighbor-Joining Tree
3.2.3. Hierarchical FST Analysis
3.2.4. Temporal Genetic Differentiation
3.2.5. Bayesian Analysis of Genetic Structure
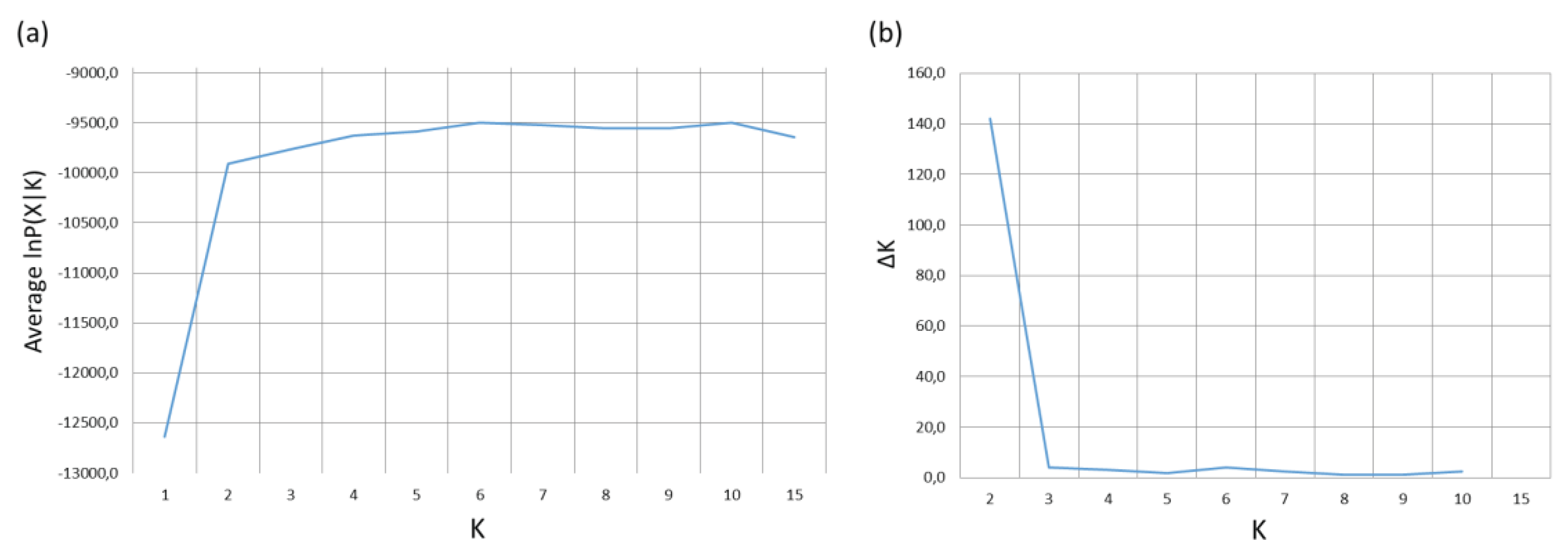
4. Discussion
4.1. Genetic Variability
4.2. Temporal Genetic Differentiation
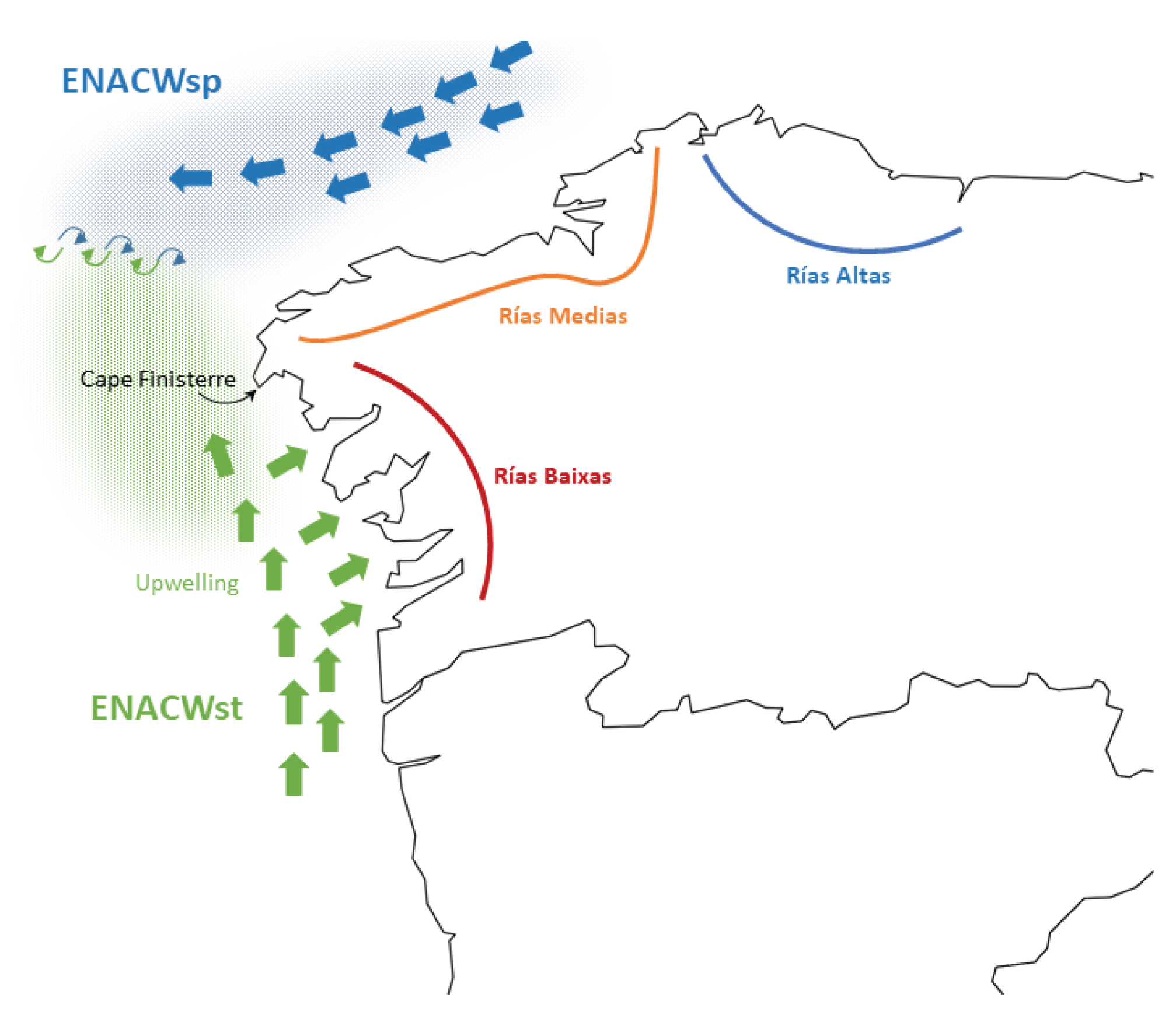
4.3. Geographic Genetic Differentiation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macho, G.; Woodin, S.A.; Wethey, D.S.; Vázquez, E. Impacts of Sublethal and Lethal High Temperatures on Clams Exploited in European Fisheries. Journal of Shellfish Research 2016, 35, 405–419. [Google Scholar] [CrossRef]
- Aranguren, R.; Gomez-León, J.; Balseiro, P.; Costa, M.M.; Novoa, B.; Figueras, A. Abnormal Mortalities of the Carpet Shell Clam Ruditapes Decussatus (Linnaeus 1756) in Natural Bed Populations: A Practical Approach. Aquaculture Research 2014, 45, 1303–1310. [Google Scholar] [CrossRef]
- Bidegain, G.; Bárcena, J.F.; García, A.; Juanes, J.A. Predicting Coexistence and Predominance Patterns between the Introduced Manila Clam (Ruditapes Philippinarum) and the European Native Clam (Ruditapes Decussatus). Estuarine, Coastal and Shelf Science 2015, 152, 162–172. [Google Scholar] [CrossRef]
- Borsa, P.; Jarne, P.; Belkhir, K.; Bonhomme, F. Genetic Structure of the Palourde Ruditapes Decussatus L. the Mediterranean. In Genetics and evolution of aquatic organisms; Chapman and Hall: London, 1994; pp. 103–113. [Google Scholar]
- Gharbi, A.; Zitari-Chatti, R.; Van Wormhoudt, A.; Dhraief, M.N.; Denis, F.; Said, K.; Chatti, N. Allozyme Variation and Population Genetic Structure in the Carpet Shell Clam Ruditapes Decussatus across the Siculo-Tunisian Strait. Biochemical Genetics 2011, 49, 788–805. [Google Scholar] [CrossRef] [PubMed]
- Cordero, D.; Pe??a, J.B.; Saavedra, C. Phylogeographic Analysis of Introns and Mitochondrial DNA in the Clam Ruditapes Decussatus Uncovers the Effects of Pleistocene Glaciations and Endogenous Barriers to Gene Flow. Molecular Phylogenetics and Evolution 2014, 71, 274–287. [Google Scholar] [CrossRef]
- Sanna, D.; Lai, T.; Cossu, P.; Scarpa, F.; Dedola, G.L.; Cristo, B.; Francalacci, P.; Curini-Galletti, M.; Mura, L.; Fois, N.; et al. Cytochrome c Oxidase Subunit I Variability in Ruditapes Decussatus (Veneridae) from the Western Mediterranean. The European Zoological Journal 2017, 84, 554–565. [Google Scholar] [CrossRef]
- Patarnello, T.; Volckaert, F.A.M.J.; Castilho, R. Pillars of Hercules: Is the Atlantic-Mediterranean Transition a Phylogeographical Break? Molecular Ecology 2007, 16, 4426–4444. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Rives, B.; Schunter, C.; MaCpherson, E. Impact of Life History Traits on Gene Flow: A Multispecies Systematic Review across Oceanographic Barriers in the Mediterranean Sea. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Bierne, N.; Welch, J.; Loire, E.; Bonhomme, F.; David, P. The Coupling Hypothesis: Why Genome Scans May Fail to Map Local Adaptation Genes. Molecular Ecology 2011, 20, 2044–2072. [Google Scholar] [CrossRef] [PubMed]
- El Ayari, T.; Trigui El Menif, N.; Hamer, B.; Cahill, A.E.; Bierne, N. The Hidden Side of a Major Marine Biogeographic Boundary: A Wide Mosaic Hybrid Zone at the Atlantic–Mediterranean Divide Reveals the Complex Interaction between Natural and Genetic Barriers in Mussels. Heredity 2019, 122, 770–784. [Google Scholar] [CrossRef]
- Arias-Pérez, A.; Cordero, D.; Borrell, Y.; Sánchez, J.A.; Blanco, G.; Freire, R.; Insua, A.; Saavedra, C. Assessing the Geographic Scale of Genetic Population Management with Microsatellites and Introns in the Clam Ruditapes Decussatus. Ecology and Evolution 2016, 6. [Google Scholar] [CrossRef]
- Gharbi, A.; Chatti, N.; Said, K.; Wormhoudt, A. Genetic Variation and Population Structure of the Carpet Shell Clam Ruditapes Decussatus along the Tunisian Coast Inferred from mtDNA and ITS1 Sequence Analysis. Biologia 2010, 65, 688–696. [Google Scholar] [CrossRef]
- Cruz, A.; da Costa, F.; Fernández-Pérez, J.; Nantón, A.; Fernández-Boo, S.; Insua, A.; Méndez, J. Genetic Variability in Ruditapes Decussatus Clam Combined with Perkinsus Infection Level to Support Founder Population Selection for a Breeding Program. PeerJ 2020, 8, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, C.; Milan, M.; Leite, R.B.; Cordero, D.; Patarnello, T.; Cancela, M.L.; Bargelloni, L. Transcriptional Profiling of Populations in the Clam Ruditapes Decussatus Suggests Genetically Determined Differentiation in Gene Expression along Parallel Temperature Gradients and between Races of the Atlantic Ocean and West Mediterranean Sea. Fishes 2023, 8, 203. [Google Scholar] [CrossRef]
- Nikula, R.; V�in�l�, R. Phylogeography of Cerastoderma Glaucum (Bivalvia: Cardiidae) across Europe: A Major Break in the Eastern Mediterranean. Marine Biology 2003, 143, 339–350. [Google Scholar] [CrossRef]
- Bahri-Sfar, L.; Lemaire, C.; Hassine, O.K.B.; Bonhomme, F. Fragmentation of Sea Bass Populations in the Western and Eastern Mediterranean as Revealed by Microsatellite Polymorphism. Proceedings of the Royal Society of London Series B 2000, 267, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. GENEPOP’007: A Complete Re-Implementation of the GENEPOP Software for Windows and Linux. Molecular Ecology Resources 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.W.; Thompson, E.A. Performing the Exact Test of Hardy-Weinberg Proportion for Multiple Alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Molecular Ecology Resources 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L. Maximum Likelihood Estimation of the Frequency of Null Alleles at Microsatellite Loci. Conserv Genet 2006, 7, 991–995. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Molecular Biology and Evolution 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Chybicki, I.J.; Burczyk, J. Simultaneous Estimation of Null Alleles and Inbreeding Coefficients. Journal of Heredity 2009, 100, 106–113. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- N, S.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees’. Molecular Biology and Evolution 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Molecular Ecology 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Leys, M.; Petit, E.J.; El-Bahloul, Y.; Liso, C.; Fournet, S.; Arnaud, J. Spatial Genetic Structure in Beta Vulgaris Subsp. Maritima and Beta Macrocarpa Reveals the Effect of Contrasting Mating System, Influence of Marine Currents, and Footprints of Postglacial Recolonization Routes. Ecology and Evolution 2014, 4, 1828–1852. [Google Scholar] [CrossRef]
- Piñeira, J.; Quesada, H.; Rolán-Alvarez, E.; Caballero, A. Genetic Discontinuity Associated with an Environmentally Induced Barrier to Gene Exchange in the Marine Snail Littorina Saxatilis. Mar. Ecol. Prog. Ser. 2008, 357, 175–184. [Google Scholar] [CrossRef]
- Alberto, F.; Santos, R.; Leitão, J. Assessing Patterns of Geographic Dispersal of Gelidium Sesquipedale (Rhodophyta) through RAPD Differentiation of Populations. Mar. Ecol. Prog. Ser. 1999, 191, 101–108. [Google Scholar] [CrossRef]
- Fraga, F.; Mouriño, C.; Manríquez, M. Res.Exp.Cient. 1982, pp. 51–77.
- Alvarez, I.; Gomez-Gesteira, M.; deCastro, M.; Lorenzo, M.N.; Crespo, A.J.C.; Dias, J.M. Comparative Analysis of Upwelling Influence between the Western and Northern Coast of the Iberian Peninsula. Continental Shelf Research 2011, 31, 388–399. [Google Scholar] [CrossRef]
- Rodríguez-Moscoso, E.; Arnaiz, R. Gametogenesis and Energy Storage in a Population of the Grooved Carpet-Shell Clam, Tapes Decussatus (Linne, 1787), in Northwest Spain. Aquaculture 1998, 162, 125–139. [Google Scholar] [CrossRef]
- Ojea, J.; Martínez, D.; Novoa, S.; Cerviño-Otero, A. Ciclo Gametogénico de Una Población de Almeja Japonesa Ruditapes Philippinarum (Adams & Reeve, 1850) En La Ría de Camariñas (Noroeste de España) y Relación Con La Composición Bioquímica Mayoritaria. Boletin - Instituto Espanol de Oceanografia 2005, 21, 337–342. [Google Scholar]
- Peres, P.A.; Bracken-Grissom, H.; Timm, L.E.; Mantelatto, F.L. Genomic Analyses Implicate the Amazon–Orinoco Plume as the Driver of Cryptic Speciation in a Swimming Crab. Genes 2022, 13, 2263. [Google Scholar] [CrossRef] [PubMed]
- Narváez-Barandica, J.C.; Quintero-Galvis, J.F.; Aguirre-Pabón, J.C.; Castro, L.R.; Betancur, R.; Acero Pizarro, A. A Comparative Phylogeography of Three Marine Species with Different PLD Modes Reveals Two Genetic Breaks across the Southern Caribbean Sea. Animals 2023, 13, 2528. [Google Scholar] [CrossRef] [PubMed]
- Prego, R.; Barciela, M.D.C.; Varela, M. Nutrient Dynamics in the Galician Coastal Area (Northwestern Iberian Peninsula): Do the Rias Bajas Receive More Nutrient Salts than the Rias Altas? Continental Shelf Research 1999, 19, 317–334. [Google Scholar] [CrossRef]
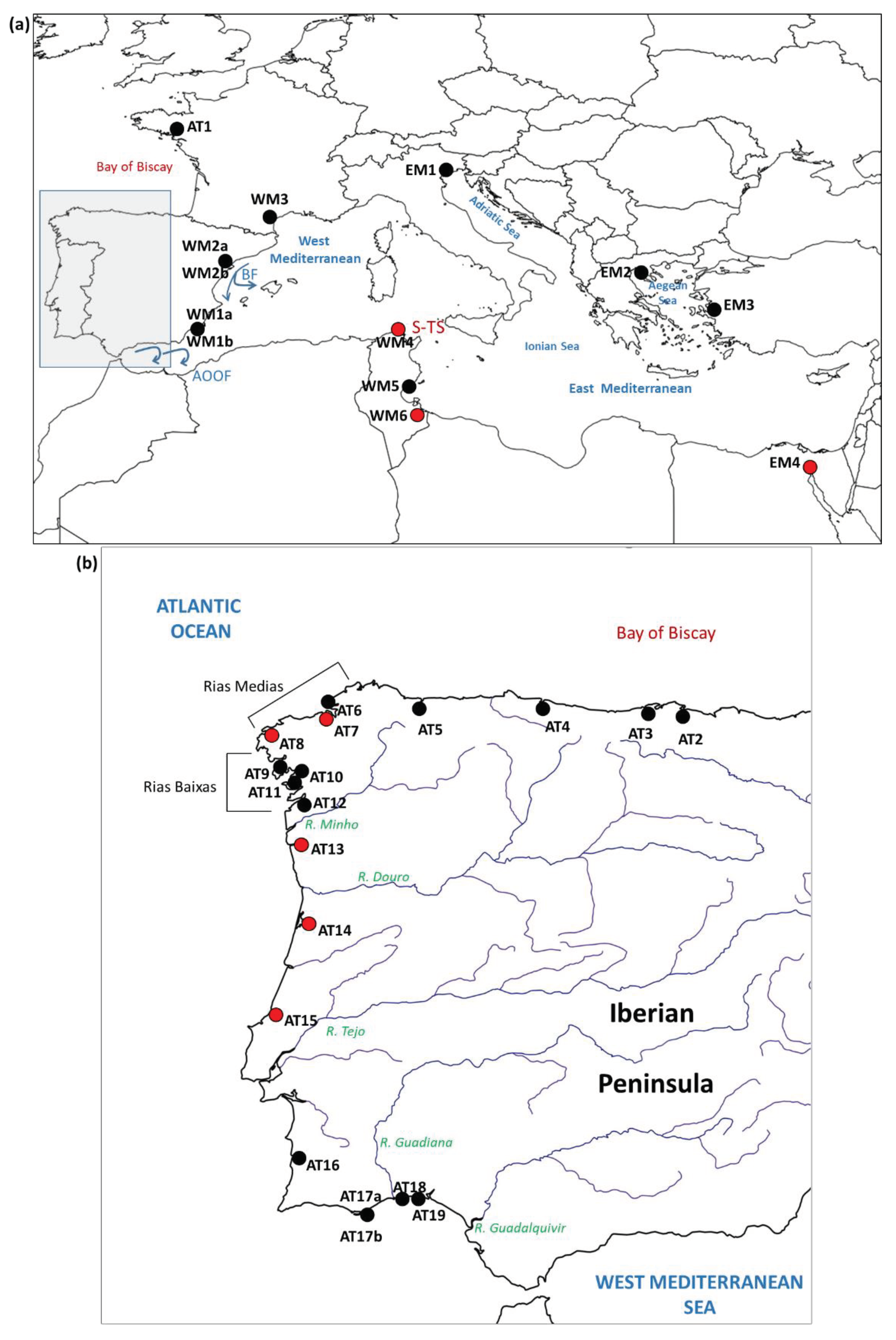
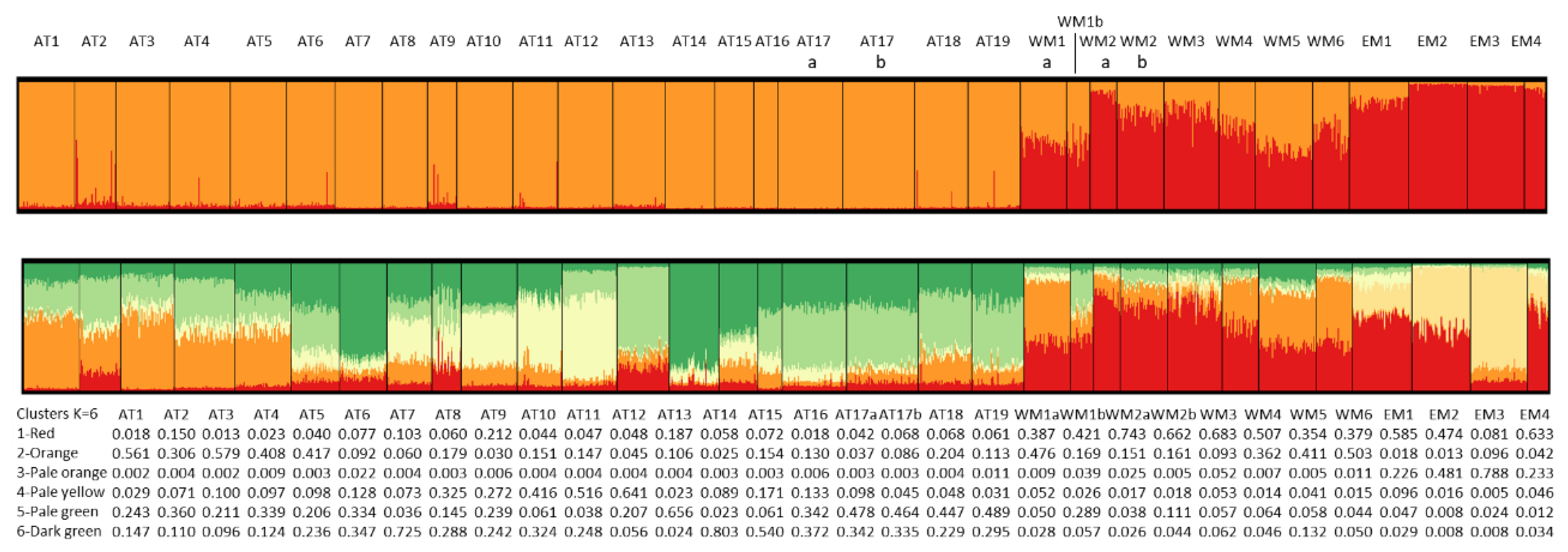
| Population code | Locality | Sampling year | Reference | Population code in reference |
|---|---|---|---|---|
| AT1 | Golfe du Morbihan (France) | 2005 | [6] | GMO |
| AT2 | Carasa (Spain) | 2010 | [12] | Car |
| AT3 | Pontejos (Spain) | 2010 | [12] | Pon |
| AT4 | Villaviciosa (Spain) | 2009 | [12] | Vil |
| AT5 | Ría del Eo (Spain) | 2009 | [12] | Eo |
| AT6 | Mugardos (Spain) | 2004 | [6] | MUG |
| AT7 | A Coruña (Spain) | 2005 | This study | - |
| AT8 | Camariñas (Spain) | 2019 | This study | - |
| AT9 | Ría de Noia (Spain) | 2011 | [15] | AN |
| AT10 | Lombos do Ulla (Spain) | 2008 | [6] | LUL |
| AT11 | Cambados (Spain) | 2009 | [12] | Cam |
| AT12 | Redondela (Spain) | 2009 | [12] | Red |
| AT13 | Viana do Castelo (Portugal) | 2021 | This study | - |
| AT14 | Aveiro (Portugal) | 2018 | This study | - |
| AT15 | Lagoa de Óbidos (Portugal) | 2018 | This study | - |
| AT16 | Milfontes (Portugal) | 2008 | [6] | MLF |
| AT17a | Ria Formosa (Portugal) | 2004 | [6] | FOR |
| AT17b | Ria Formosa (Portugal) | 2011 | [15] | AS |
| AT18 | Isla Cristina (Spain) | 2008 | [12] | Isl |
| AT19 | Río Piedras (Spain) | 2008 | [12] | Rio |
| WM1a | Mar Menor (Spain) | 2004 | [6] | MME |
| WM1b | Mar Menor (Spain) | 2011 | [15] | MS |
| WM2a | Ebro delta (Spain) | 2005 | [6] | EBR |
| WM2b | Ebro delta (Spain) | 2009 | [12] | Del |
| WM3 | Thau lagoon (France) | 2011 | [15] | MN |
| WM4 | Bizerte (Tunisia) | 2010 | This study | - |
| WM5 | Sfax (Tunisia) | 2008 | [6] | SFX |
| WM6 | Biben (Tunisia) | 2010 | This study | - |
| EM1 | Venice (Italy) | 2005 | [6] | VEN |
| EM2 | Halkidiki (Greece) | 2008 | [6] | HAL |
| EM3 | Izmir (Turkey) | 2007 | [6] | IZM |
| EM4 | Suez Canal (Egypt) | 2010 | This study | - |
|
Model |
F-statistics |
||
|---|---|---|---|
| FST | FSC | FCT | |
| Models without subdivision | |||
| 1 - All populations | 0,132*** | - | - |
| 2 - AT populations | 0,065*** | - | - |
| 3 - WM + EM populations | 0,080*** | - | - |
| 4 - WM populations | 0,036*** | - | - |
| 5 - EM populations | 0,101*** | - | - |
| Models with geographical subdivision | |||
| 6 - AT/WM/EM | 0,186*** | 0,063*** | 0,131*** |
| 7 - WM/EM | 0,100*** | 0,060*** | 0,043** |
| 8 - Bay of Biscay / NW Spain / Central Portugal + AT13 / SW Iberian Peninsula | 0,073*** | 0,037*** | 0,038*** |
| 9 - Bay of Biscay / NW Spain + AT13 / Central Portugal / SW Iberian Peninsula | 0,075*** | 0,033*** | 0,043*** |
| 10 - Bay of Biscay+AT13 / NW Spain / Central Portugal / SW Iberian Peninsula | 0,076*** | 0,027*** | 0,050*** |
| Model with temporal subdivision | |||
| 11 – a vs. b | 0,129*** | 0,185*** | -0,068 |
| * P < 0.05; ** P < 0.01; *** P < 0.001 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





