Submitted:
01 March 2024
Posted:
04 March 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
| Pre-Nested | Cycles | Duration | HRV Assay | RSV Assay |
|---|---|---|---|---|
| Initial denaturation | 1 | 45 minutes | 48°C | |
| PCR activation | 1 | 2 minutes | 94°C | |
| Denaturation | 30 seconds | 94°C | ||
| Annealing | 45 | 90 seconds | 60°C | 55°C |
| Extension | 60 seconds | 72°C | ||
| Final extension | 1 | 10 minutes | 72°C | |
| Final hold | 1 | Infinite | 4°C | |
| Nested | ||||
| PCR activation | 1 | 2 minutes | 94°C | |
| Denaturation | 30 seconds | 94°C | ||
| Annealing | 40 | 60 seconds | 55°C | 50°C |
| Extension | 30 seconds | 72°C | ||
| Final extension | 1 | 10 minutes | 72°C | |
| Final hold | 1 | Infinite | 4°C |
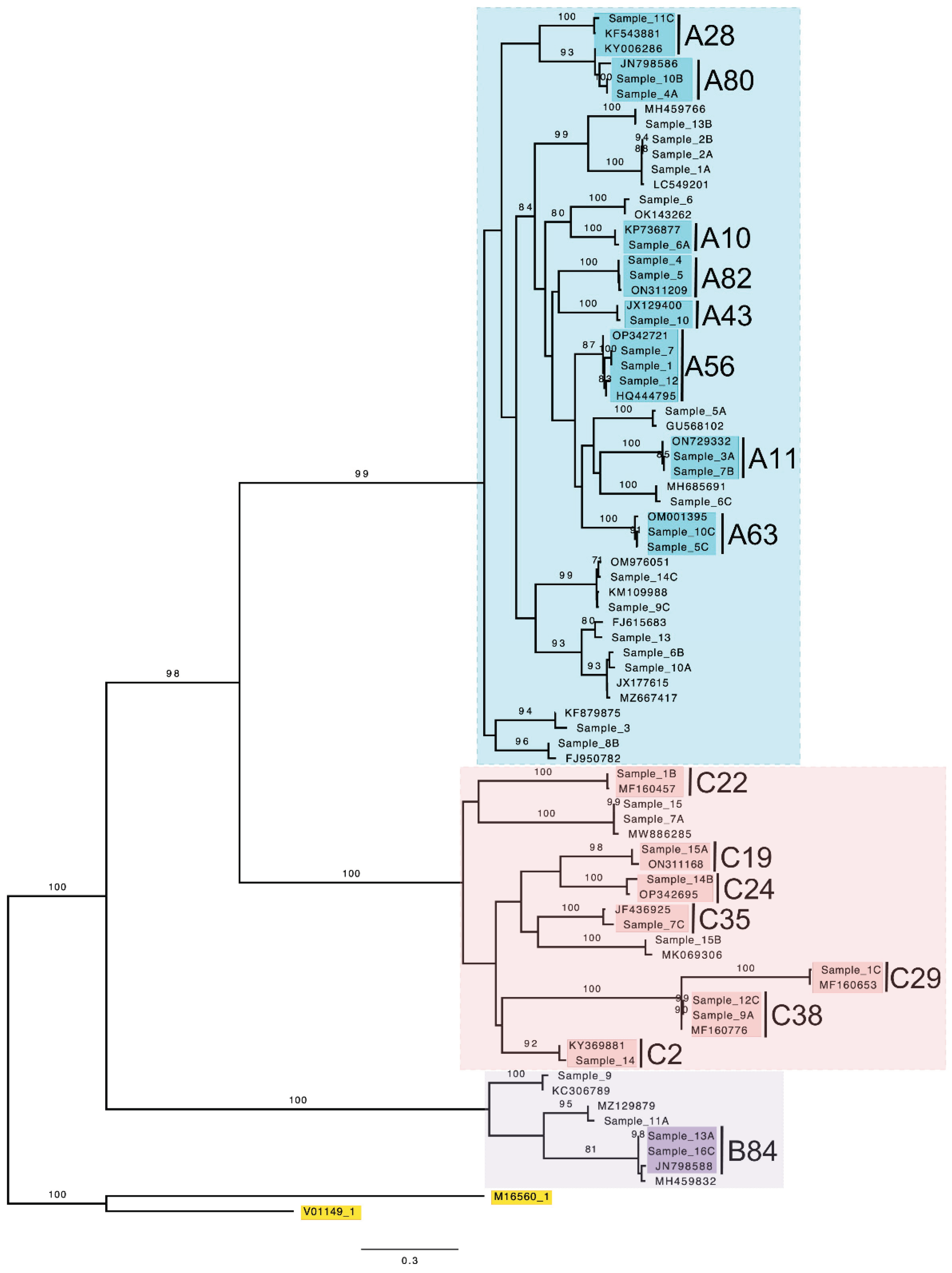
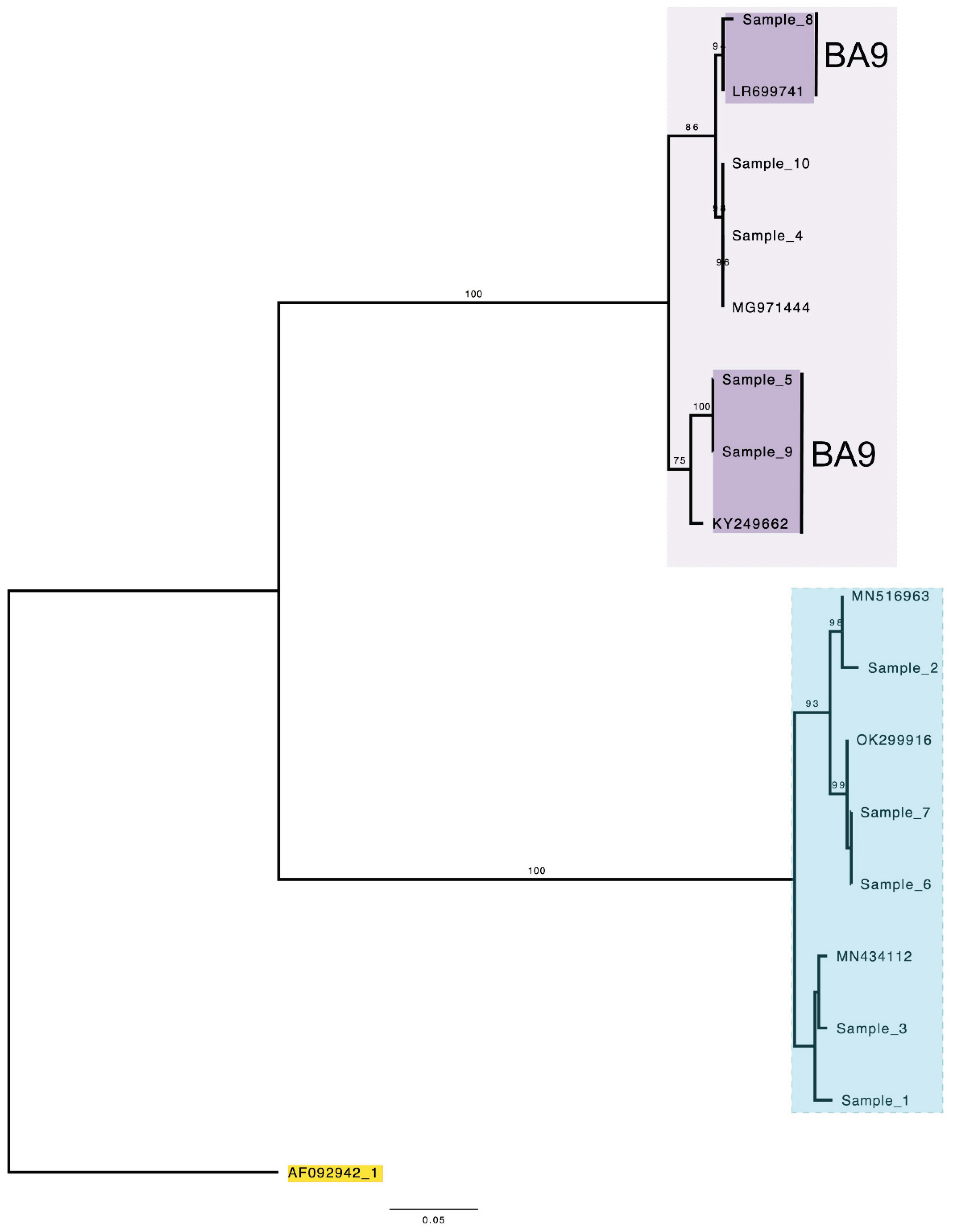
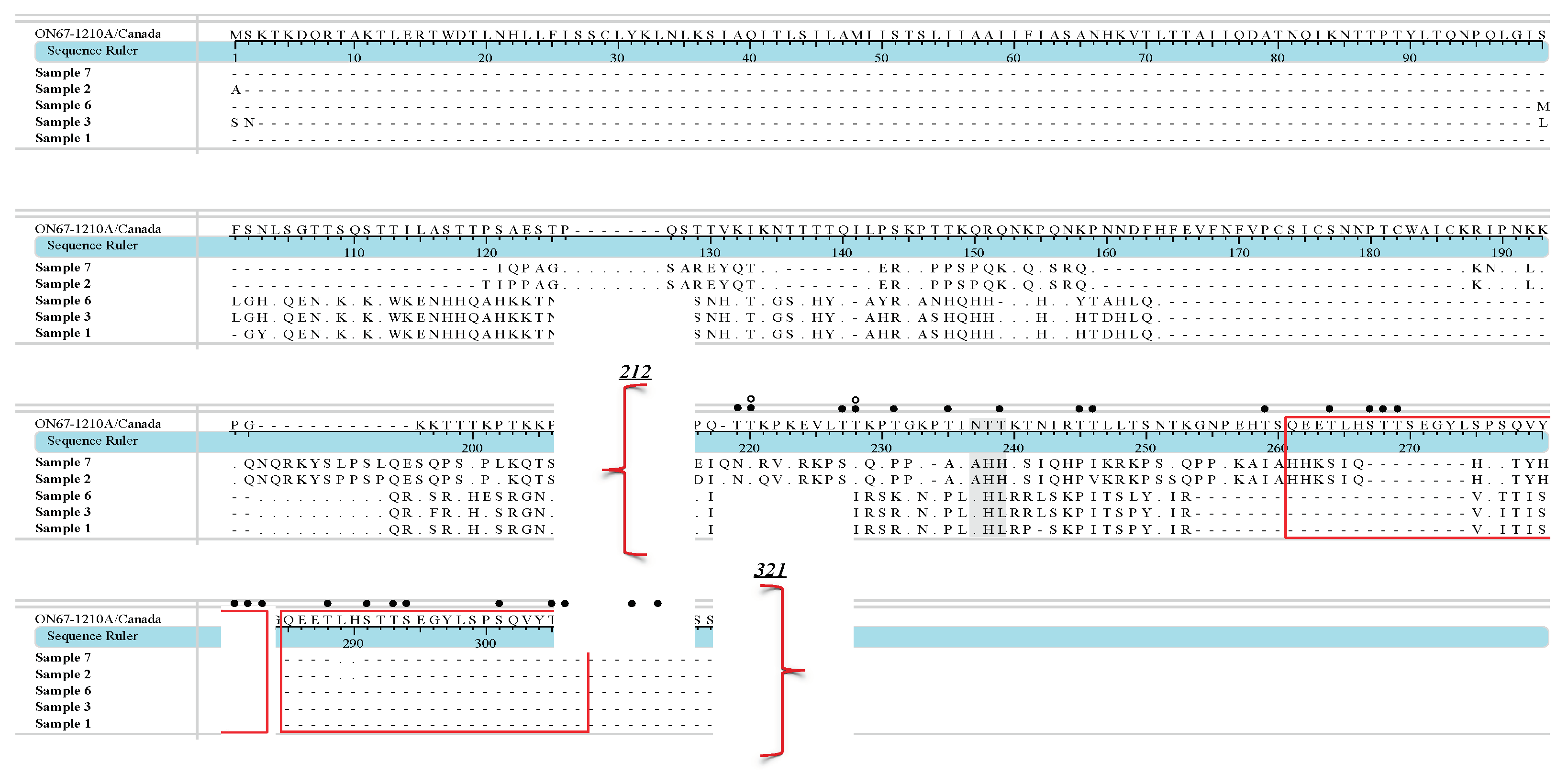
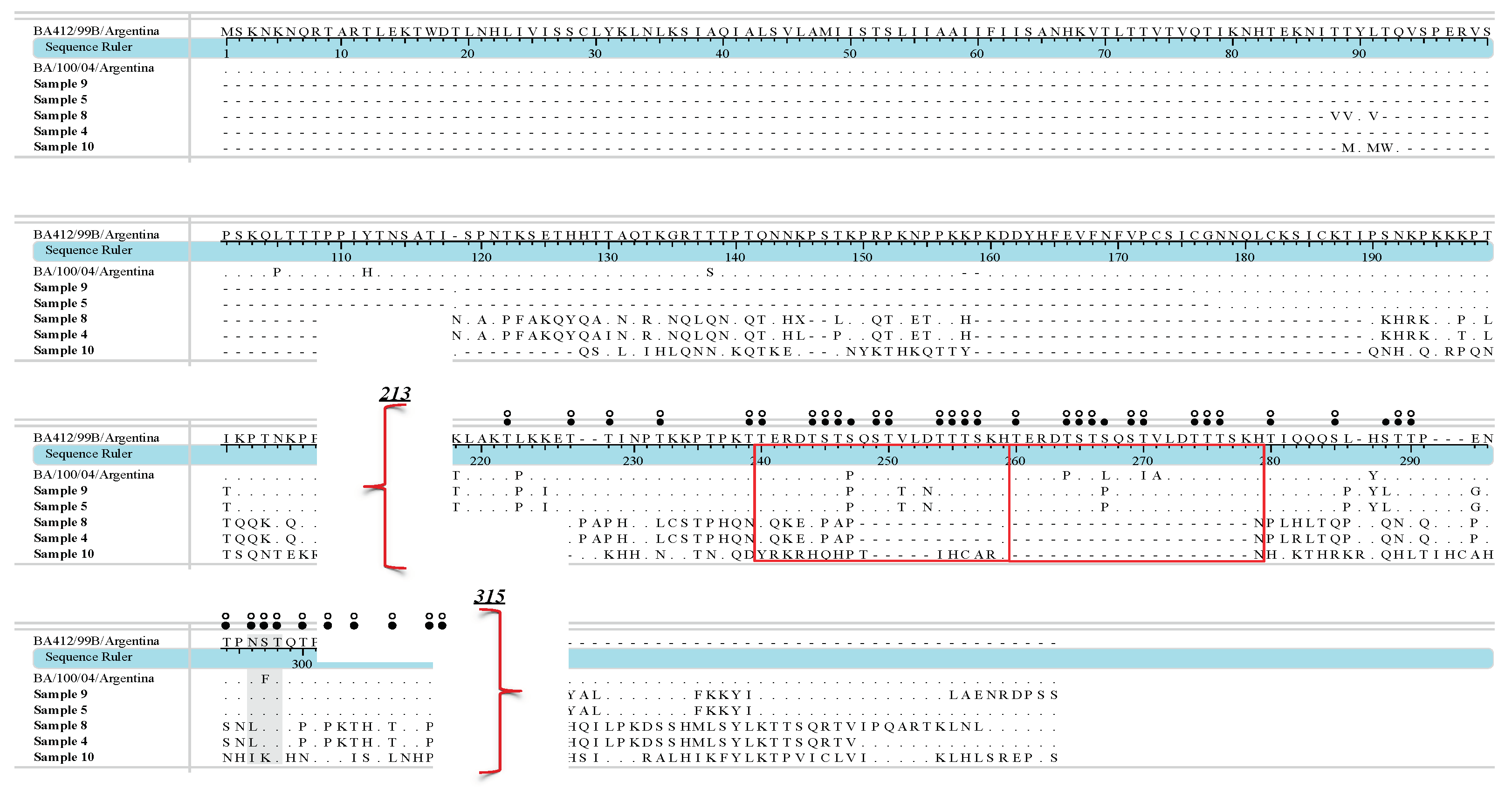
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geoghegan, S., Erviti, A., Caballero, M.T., Vallone, F., Zanone, S.M., Losada, J.V., Bianchi, A., Acosta, P.L., Talarico, L.B., Ferretti, A., Grimaldi, L.A., Sancilio, A., Dueñas, K., Sastre, G., Rodriguez, A., Ferrero, F., Barboza, E., Gago, G.F., Nocito, C., Flamenco, E., Perez, A.R., Rebec, B., Ferrola, F.M., Libser, R., Karron, R.A., Bergel, El., Polack, F.P. Mortality due to Respiratory Syncytial Virus. Burden and Risk Factors. Am J Respir Crit Care Med 2017, 195(1), 96–103. [CrossRef]
- Kotaniemi-Syrjänen, A., Vainionpää, R., Reijonen, T.M., Waris, M., Korhonen, K., Korppi, M. Rhinovirus-induced wheezing in infancy--the first sign of childhood asthma?. J Allergy Clin Immunol 2003, 111(1), 66–71. [CrossRef]
- Linsuwanon, P., Payungporn, S., Suwannakarn, K., Chieochansin, T., Theamboonlers, A., Poovorawan, Y. Complete coding sequence characterization and comparative analysis of the putative novel human rhinovirus (HRV) species C and B. Virology J 2003, 8, 5. [CrossRef]
- Smuts, H.E., Workman, L.J., Zar, H.J. Human rhinovirus infection in young African children with acute wheezing. BMC Infect Dis 2011, 11, 65. [CrossRef]
- Drysdale, S.B., Alcazar, M., Wilson, T., Smith, M., Zuckerman, M., Lauinger, I.L., Tong, C.Y., Broughton, S., Rafferty, G.F., Johnston, S.L., Greenough, A. Respiratory outcome of prematurely born infants following human rhinovirus A and C infections. Eur J Pediatr 2014, 173(7), 913–919. [CrossRef]
- Stone, C.A., Jr, Miller, E.K. Understanding the Association of Human Rhinovirus with Asthma. Clin Vaccine Immunol 2015, CVI, 23(1), 6–10. [CrossRef]
- Jartti, T., Gern, J.E. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clinical Immunol 2017, 140(4), 895–906. [CrossRef]
- Shi, T., McAllister, D.A., O’Brien, K.L., Simoes, E.A.F., Madhi, S.A., Gessner, B.D., Polack, F.P., Balsells, E., Acacio, S., Aguayo, C., Alassani, I., Ali, A., Antonio, M., Awasthi, S., Awori, J.O., Azziz-Baumgartner, E., Baggett, H.C., Baillie, V.L., Balmaseda, A., Barahona, A., Basnet, S., Bassat, Q., Basualdo, W., Bigogo, G., Bont, L., Breiman, R.F., Brooks, W.A., Broor, S., Bruce, N., Bruden, D., Buchy, P., Campbell, S., Carosone-Link, P., Chadha, M., Chipeta, J., Chou, M., Clara, W., Cohen, C., de Cuellar, E., Dang, D.A., Dash-Yandag, B., Deloria-Knoll, M., Dherani, M., Eap, T., Ebruke, B.E., Echavarria, M., de Freitas Lázaro Emediato, C.C., Fasce, R.A., Feikin, D.R., Feng, L., Gentile, A., Gordon, A., Goswami, D., Goyet, S., Groome, M., Halasa, N., Hirve, S., Homaira, N., Howie, S.R.C., Jara, J., Jroundi, I., Kartasasmita, C.B., Khuri-Bulos, N., Kotloff, K.L., Krishnan, A., Libster, R., Lopez, O., Lucero, M.G., Lucion, F., Lupisan, S.P., Marcone, D.N., McCracken, J.P., Mejia, M., Moisi, J.C., Montgomery, J.M., Moore, D.P., Moraleda, C., Moyes, J., Munywoki, P., Mutyara, K., Nicol, M.P., Nokes, D.J., Nymadawa, P., da Costa Oliveira, M.T., Oshitani, H., Pandey, N., Paranhos-Baccalà, G., Phillips, L.N., Picot, V.S., Rahman, M., Rakoto-Andrianarivelo, M., Rasmussen, Z.A., Rath, B.A., Robinson, A., Romero, C., Russomando, G., Salimi, V., Sawatwong, P., Scheltema, N., Schweiger, B., Scott, J.A.G., Seidenberg, P., Shen, K., Singleton, R., Sotomayor, V., Strand, T.A., Sutanto, A., Sylla, M., Tapia, M.D., Thamthitiwat, S., Thomas, E.D., Tokarz, R., Turner, C., Venter, M., Waicharoen, S., Wang, J., Watthanaworawit, W., Yoshida, L.M., Yu, H., Zar, H.J., Campbell, H., Nair, H.; RSV Global Epidemiology Network. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet (London, England) 2017, 390(10098), 946–958. [CrossRef]
- Borchers, A.T., Chang, C., Gershwin, M.E., Gershwin, L.J. Respiratory syncytial virus--a comprehensive review. Clin Rev Allergy Immunol 2013, 45(3), 331–379. [CrossRef]
- Laudanno, S.L., Sánchez Yanotti, C.I., Polack, F.P. RSV Lower Respiratory Tract Illness in Infants of Low- and Middle-income Countries. Acta Med Acad 2020, 49(2), 191–197. [CrossRef]
- Zar, H.J., Nduru, P., Stadler, J.A.M., Gray, D., Barnett, W., Lesosky, M., Myer, L., Nicol, M.P. Early-life respiratory syncytial virus lower respiratory tract infection in a South African birth cohort: epidemiology and effect on lung health. Lancet Glob Health 2020, 8(10), e1316–e1325. [CrossRef]
- Jacobs, S.E., Lamson, D.M., St George, K., Walsh, T.J. Human rhinoviruses. Clin Microbiol Rev 2013, 26(1), 135–162. [CrossRef]
- Nakauchi, M., Nagata, N., Takayama, I., Saito, S., Kubo, H., Kaida, A., Oba, K., Odagiri, T., Kageyama, T. Propagation of Rhinovirus C in Differentiated Immortalized Human Airway HBEC3-KT Epithelial Cells. Viruses 2019, 11(3), 216. [CrossRef]
- Ashraf, S., Brockman-Schneider, R., Bochkov, Y.A., Pasic, T.R., Gern, J.E. Biological characteristics and propagation of human rhinovirus-C in differentiated sinus epithelial cells. Virology 2013, 436(1), 143–149. [CrossRef]
- Gomez-Perosanz, M., Sanchez-Trincado, J.L., Fernandez-Arquero, M., Sidney, J., Sette, A., Lafuente, E.M., Reche, P.A. Human rhinovirus-specific CD8 T cell responses target conserved and unusual epitopes. FASEB J 2021, 35(1), e21208. [CrossRef]
- Esneau, C., Duff, A.C., Bartlett, N.W. Understanding Rhinovirus Circulation and Impact on Illness. Viruses 2022, 14(1), 141. [CrossRef]
- Clementi, N., Ghosh, S., De Santis, M., Castelli, M., Criscuolo, E., Zanoni, I., Clementi, M., Mancini, N. Viral Respiratory Pathogens and Lung Injury. Clin Microbiol Rev 2021, 34(3), e00103-20. [CrossRef]
- Han, M., Rajput, C., Hershenson, M.B. Rhinovirus Attributes that Contribute to Asthma Development. Immunol Allergy Clin North Am 2019, 39(3), 345–359. [CrossRef]
- Makris, S., Johnston, S. Recent advances in understanding rhinovirus immunity. F1000Res 2018, 7, F1000 Faculty Rev-1537. [CrossRef]
- Santti, J., Hyypiä, T., Kinnunen, L., Salminen, M. Evidence of recombination among enteroviruses. J Virol 1999, 73(10), 8741–8749. [CrossRef]
- Savolainen, C., Laine, P., Mulders, M.N., Hovi, T. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J Gen Virol 2004, 85(Pt 8), 2271–2277. [CrossRef]
- Arden, K.E., McErlean, P., Nissen, M.D., Sloots, T.P., Mackay, I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol 2006, 78(9), 1232–1240. [CrossRef]
- Ren, L., Yang, D., Ren, X., Li, M., Mu, X., Wang, Q., Cao, J., Hu, K., Yan, C., Fan, H., Li, X., Chen, Y., Wang, R., An, F., An, S., Luo, M., Wang, Y., Xiao, Y., Xiang, Z., Xiao, Y., Li, L., Huang, F., Gao, Z., Wang, J. Genotyping of human rhinovirus in adult patients with acute respiratory infections identified predominant infections of genotype A21. Sci Rep 2017, 7, 41601. [CrossRef]
- Zhao, Y., Shen, J., Wu, B., Liu, G., Lu, R., Tan, W. Genotypic Diversity and Epidemiology of Human Rhinovirus Among Children With Severe Acute Respiratory Tract Infection in Shanghai, 2013-2015. Front Microbiol 2018, 9, 1836. [CrossRef]
- Baillie, V.L., Moore, D.P., Mathunjwa, A., Morailane, P., Simões, E.A.F., Madhi, S.A. Molecular Subtyping of Human Rhinovirus in Children from Three Sub-Saharan African Countries. J Clin Microbiol 2019, 57(9), e00723-19. [CrossRef]
- Luka, M.M., Kamau, E., Adema, I., Munywoki, P.K., Otieno, G.P., Gicheru, E., Gichuki, A., Kibinge, N., Agoti, C.N., Nokes, D.J. Molecular Epidemiology of Human Rhinovirus From 1-Year Surveillance Within a School Setting in Rural Coastal Kenya. Open Forum Infect Dis 2020, 7(10), ofaa385. [CrossRef]
- da Costa Souza, L., Bello, E.J.M., Dos Santos, E.M., Nagata, T. Molecular and clinical characteristics related to rhinovirus infection in Brasília, Brazil. Braz J Microbiol 2021, 52(1), 289–298. [CrossRef]
- Pandya, M.C., Callahan, S.M., Savchenko, K.G., Stobart, C.C. A Contemporary View of Respiratory Syncytial Virus (RSV) Biology and Strain-Specific Differences. Pathogens 2019, 8(2), 67. [CrossRef]
- Garcia-Garcia, M.L., Calvo Rey, C., Del Rosal Rabes, T. Pediatric Asthma and Viral Infection. Asma y virus en el niño. Arch Bronconeumol 2016, 52(5), 269–273. [CrossRef]
- Karron, R.A., Zar, H.J. Determining the outcomes of interventions to prevent respiratory syncytial virus disease in children: what to measure?. Lancet Respir Med 2018, 6(1), 65–74. [CrossRef]
- Battles, M.B., McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat Rev Microbiol 2019, 17(4), 233–245. [CrossRef]
- Ruckwardt, T.J., Morabito, K.M., Graham, B.S. Immunological Lessons from Respiratory Syncytial Virus Vaccine Development. Immunity 2019, 51(3), 429–442. [CrossRef]
- Driscoll, A.J., Arshad, S.H., Bont, L., Brunwasser, S.M., Cherian, T., Englund, J.A., Fell, D.B., Hammitt, L.L., Hartert, T.V., Innis, B.L., Karron, R.A., Langley, G.E., Mulholland, E.K., Munywoki, P.K., Nair, H., Ortiz, J.R., Savitz, D.A., Scheltema, N.M., Simões, E.A.F., Smith, P.G., Were, F., Zar, H.J., Feikin, D.R. Does respiratory syncytial virus lower respiratory illness in early life cause recurrent wheeze of early childhood and asthma? Critical review of the evidence and guidance for future studies from a World Health Organization-sponsored meeting. Vaccine 2020, 38(11), 2435–2448. [CrossRef]
- Bergeron, H.C., Tripp, R.A. Immunopathology of RSV: An Updated Review. Viruses 2021, 13(12), 2478. [CrossRef]
- Malinczak, C.A., Lukacs, N.W., Fonseca, W. Early-Life Respiratory Syncytial Virus Infection, Trained Immunity and Subsequent Pulmonary Diseases. Viruses 2020, 12(5), 505. [CrossRef]
- Nam, H.H., Ison, M.G. Respiratory syncytial virus infection in adults. Br Med J 2019, 366, l5021. [CrossRef]
- Liu, H., Lu, B., Tabor, D.E., Tovchigrechko, A., Wilkins, D., Jin, H., Madhi, S.A., Soofie, N., Esser, M.T., Nunes, M.C. Characterization of human respiratory syncytial virus (RSV) isolated from HIV-exposed-uninfected and HIV-unexposed infants in South Africa during 2015-2017. Influenza Other Respir Viruses 2020, 14(4), 403–411. [CrossRef]
- Bin Lu, Liu, H., Tabor, D.E., Tovchigrechko, A., Qi, Y., Ruzin, A., Esser, M.T., Jin, H. Emergence of new antigenic epitopes in the glycoproteins of human respiratory syncytial virus collected from a US surveillance study, 2015-17. Sci Rep 2019, 9(1), 3898. [CrossRef]
- Yun, K.W., Choi, E.H., Lee, H.J. Molecular epidemiology of respiratory syncytial virus for 28 consecutive seasons (1990-2018) and genetic variability of the duplication region in the G gene of genotypes ON1 and BA in South Korea. Arch Virol 2020, 165(5), 1069–1077. [CrossRef]
- Lau, S.K., Yip, C.C., Lin, A.W., Lee, R.A., So, L.Y., Lau, Y.L., Chan, K.H., Woo, P.C., Yuen, K.Y. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis 2009, 200(7), 1096–1103. [CrossRef]
- Miller, E.K., Khuri-Bulos, N., Williams, J.V., Shehabi, A.A., Faouri, S., Al Jundi, I., Chen, Q., Heil, L., Mohamed, Y., Morin, L.L., Ali, A., Halasa, N.B. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol 2009, 46(1), 85–89. [CrossRef]
- Pretorius, M.A., Tempia, S., Treurnicht, F.K., Walaza, S., Cohen, A.L., Moyes, J., Hellferscee, O., Variava, E., Dawood, H., Chhagan, M., Haffjee, S., Madhi, S.A., Cohen, C., Venter, M. Genetic diversity and molecular epidemiology of human rhinoviruses in South Africa. Influenza Other Respir Viruses 2014, 8(5), 567–573. [CrossRef]
- Hung, H.M., Yang, S.L., Chen, C.J., Chiu, C.H., Kuo, C.Y., Huang, K.A., Lin, T.Y., Hsieh, Y.C., Gong, Y. N., Tsao, K.C., Huang, Y.C. Molecular epidemiology and clinical features of rhinovirus infections among hospitalized patients in a medical center in Taiwan. J Microbiol Immunol Infect 2019, 52(2), 233–241. [CrossRef]
- Venter, M., Madhi, S.A., Tiemessen, C.T., Schoub, B.D. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol 2001, 82(Pt 9), 2117–2124. [CrossRef]
- Robertson, S.E., Roca, A., Alonso, P., Simoes, E.A., Kartasasmita, C.B., Olaleye, D.O., Odaibo, G.N., Collinson, M., Venter, M., Zhu, Y., Wright, P.F. Respiratory syncytial virus infection: denominator-based studies in Indonesia, Mozambique, Nigeria and South Africa. Bull World Health Organ 2004, 82(12), 914–922.
- McMorrow, M.L., Tempia, S., Walaza, S., Treurnicht, F.K., Moyes, J., Cohen, A.L., Pretorius, M., Hellferscee, O., Wolter, N., von Gottberg, A., Nguweneza, A., McAnerney, J.M., Naby, F., Mekgoe, O., Venter, M., Madhi, S.A., Cohen, C. The Role of Human Immunodeficiency Virus in Influenza- and Respiratory Syncytial Virus-associated Hospitalizations in South African Children, 2011-2016. Clin Infect Dis 2019, 68(5), 773–780. [CrossRef]
- Xu, L., Gao, H., Zeng, J., Liu, J., Lu, C., Guan, X., Qian, S., Xie, Z. A fatal case associated with respiratory syncytial virus infection in a young child. BMC Infect Dis 2018, 18(1), 217. [CrossRef]
- Kubale, J., Kuan, G., Gresh, L., Ojeda, S., Azziz-Baumgartner, E., Sanchez, N., Lopez, R., Harris, E., Balmaseda, A., Gordon, A. Assessing the Incidence of Symptomatic Respiratory Syncytial Virus Illness Within a Prospective Birth Cohort in Managua, Nicaragua. Clin Infect Dis 2020, 70(10), 2029–2035. [CrossRef]
- Winterbach M, Hattingh C, Heathfield, LJ. Retrospective study of sudden unexpected death of infants in the Garden Route and Central Karoo districts of South Africa: Causes of death and epidemiological factors. S Afr J Child Health 2021, 15(2):74-82. [CrossRef]
- la Grange, H. Respiratory Pathogens in Cases of Sudden Unexpected Death in Infancy (SUDI) at Tygerberg Forensic Pathology Service Mortuary. Master of Science thesis, Stellenbosch University, Cape Town; 2013.
- Luoto, R., Jartti, T., Ruuskanen, O., Waris, M., Lehtonen, L., Heikkinen, T. Review of the clinical significance of respiratory virus infections in newborn infants. Acta Paediatr 2016, 105(10), 1132–1139. [CrossRef]
- Toizumi, M., Suzuki, M., Nguyen, H.A.T., Le, M.N., Ariyoshi, K., Moriuchi, H., Hashizume, M., Dang, D.A., Yoshida, L.M. Viral Acute Respiratory Illnesses in Young Infants Increase the Risk of Respiratory Readmission. Pediatr Infect Dis 2018, 37(12), 1217–1222. [CrossRef]
- Boonyaratanakornkit, J., Englund, J.A., Magaret, A.S., Bu, Y., Tielsch, J.M., Khatry, S.K., Katz, J., Kuypers, J., Shrestha, L., LeClerq, S.C., Steinhoff, M.C., Chu, H.Y. Primary and Repeated Respiratory Viral Infections Among Infants in Rural Nepal. J Pediatr Infect Dis Soc 2020, 9(1), 21–29. [CrossRef]
- Dempers, J.J., Burger, E.H., Toit-Prinsloo, L.D., Verster, J. A South African Perspective. In: SIDS Sudden Infant and Early Childhood Death: The Past, the Present and the Future. Duncan, J.R., Byard, R.W., Eds. University of Adelaide Press: Adelaide, Australia, 2018. Chapter 17. PMID: 30035954.
- Garstang, J., Watson, D., Pease, A., Ellis, C., Blair, P.S., Fleming, P. Improving engagement with services to prevent Sudden Unexpected Death in Infancy (SUDI) in families with children at risk of significant harm: A systematic review of evidence. Child Care Health Dev 2021, 47(5), 713–731. [CrossRef]
- Weber, M.A., Ashworth, M.T., Risdon, R.A., Hartley, J.C., Malone, M., Sebire, N.J. The role of post-mortem investigations in determining the cause of sudden unexpected death in infancy. Arch Dis Child 2008, 93(12), 1048–1053. [CrossRef]
- la Grange, H., Verster, J., Dempers, J.J., de Beer, C. Review of immunological and virological aspects as contributory factors in Sudden Unexpected Death in Infancy (SUDI). For Sci Int 2014, 245, 12–16. [CrossRef]
- Dempers, J.J., Coldrey, J., Burger, E.H., Thompson, V., Wadee, S.A., Odendaal, H.J., Sens, M.A., Randall, B.B., Folkerth, R.D., Kinney, H.C., PASS Network. The Institution of a Standardized Investigation Protocol for Sudden Infant Death in the Eastern Metropole, Cape Town, South Africa. J Forensic Sci 2016, 61(6), 1508–1514. [CrossRef]
- du Toit-Prinsloo, L., Dempers, J., Verster, J., Hattingh, C., Nel, H., Brandt, V.D., Jordaan, J., Saayman, G. Toward a standardized investigation protocol in sudden unexpected deaths in infancy in South Africa: a multicenter study of medico-legal investigation procedures and outcomes. Forensic Sci Med Pathol 2013, 9(3), 344–350. [CrossRef]
- Osei-Poku, G.K., Thomas, S., Mwananyanda, L., Lapidot, R., Elliott, P.A., Macleod, W.B., Somwe, S.W., Gill, C.J. A systematic review of the burden and risk factors of sudden infant death syndrome (SIDS) in Africa. J Glob Health 2021, 11, 04075. [CrossRef]
- Shipstone, R.A., Young, J., Kearney, L., Thompson, J.M.D. Applying a Social Exclusion Framework to Explore the Relationship Between Sudden Unexpected Deaths in Infancy (SUDI) and Social Vulnerability. Front Public Health 2020, 8, 563573. [CrossRef]
- Coiras, M.T., Aguilar, J.C., García, M.L., Casas, I., Pérez-Breña, P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol 2004, 72(3), 484–495. [CrossRef]
- Slovic, A., Ivancic-Jelecki, J., Ljubin-Sternak, S., Galinović, G.M., Forcic, D. A molecular epidemiological study of human respiratory syncytial virus in Croatia, 2011-2014. Infect Genet Evol 2016, 44, 76–84. [CrossRef]
- Venter, M., Collinson, M., Schoub, B.D. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: Comparison of viruses and genotypes responsible for different disease manifestations. J Med Virol 2002, 68(3), 452–461. [CrossRef]
- Fawkner-Corbett, D.W., Khoo, S.K., Duarte, C.M., Bezerra, P.G., Bochkov, Y.A., Gern, J.E., Le Souef, P.N., McNamara, P.S. Rhinovirus-C detection in children presenting with acute respiratory infection to hospital in Brazil. J Med Virol 2016, 88(1), 58–63. [CrossRef]
- Daleno, C., Piralla, A., Scala, A., Senatore, L., Principi, N., Esposito, S. Phylogenetic analysis of human rhinovirus isolates collected from otherwise healthy children with community-acquired pneumonia during five successive years. PloS One 2013, 8(11), e80614. [CrossRef]
- Oluwasemowo, O.O., Nejo, Y.T., Abokede, J.O., Lawson, M., Motayo, B.O. Genotypes of rhinovirus detected among children in two communities of South-West Nigeria. Virus genes 2021, 57(3), 276–279. [CrossRef]
- Haddad-Boubaker, S., Mefteh, K., Mejri, C., Bouaffsoun, A., El Moussi, A., Boutiba, I., Mnif, K., Slim, A., Kechrid, A., Smaoui, H. High genotypic diversity of Rhinoviruses obtained from Tunisian children with severe acute respiratory infection. J Infect Dev Ctries 2021, 15(5), 726–735. [CrossRef]
- Li, W., Yu, B., Zhou, J., Wang, Y., Xue, B., Pan, J., Ran, Y., Yang, X., Wang, X., Yang, F., Li, H. Genetic diversity and epidemiology of human rhinovirus among children with severe acute respiratory tract infection in Guangzhou, China. Virol J 2021, 18(1), 174. [CrossRef]
- Panda, S., Mohakud, N.K., Panda, S., Kumar, S. Epidemiology and phylogenetic analysis of human rhinovirus/Enterovirus in Odisha, Eastern India. Indian J Med Microbiol 2019, 37(4), 569–573. [CrossRef]
- Peltola, V., Waris, M., Osterback, R., Susi, P., Ruuskanen, O., Hyypiä, T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis 2008, 197(3), 382–389. [CrossRef]
- Jia, R., Lu, L., Li, S., Liu, P., Xu, M., Cao, L., Su, L., Xu, J. Human rhinoviruses prevailed among children in the setting of wearing face masks in Shanghai, 2020. BMC Infect Dis 2022, 22(1), 253. [CrossRef]
- Zlateva, K.T., de Vries, J.J., Coenjaerts, F.E., van Loon, A.M., Verheij, T., Little, P., Butler, C.C., Goossens, H., Ieven, M., Claas, E.C., GRACE Study Group. Prolonged shedding of rhinovirus and re-infection in adults with respiratory tract illness. Eur Respir J 2014, 44(1), 169–177. [CrossRef]
- Prasetyo, A.A., Desyardi, M.N., Tanamas, J., Suradi, Reviono, Harsini, Kageyama, S., Chikumi, H., Shimizu, E. Respiratory viruses and torque teno virus in adults with acute respiratory infections. Intervirology 2015, 58(1), 57–68. [CrossRef]
- Kamau, E., Onyango, C.O., Otieno, G.P., Kiyuka, P.K., Agoti, C.N., Medley, G.F., Cane, P.A., Nokes, D.J., Munywoki, P.K. An Intensive, Active Surveillance Reveals Continuous Invasion and High Diversity of Rhinovirus in Households. J Infect Dis 2019, 219(7), 1049–1057. [CrossRef]
- Heathfield, L., Martin, L., Ramesar, R. A 5-year retrospective analysis of infant death at Salt River Mortuary, Cape Town. S Afr J Child Health 2020, 14(3), 148-154. [CrossRef]
- Baker, T., Schandl, C., Presnell, S.E., Madory, J., Nolte, F.S., Batalis, N. Use of an Automated Nested Multiplex Respiratory Pathogen PCR Panel Postmortem in the Pediatric Forensic Setting. J For Sci 2017, 62(5), 1223–1228. [CrossRef]
- L’Huillier, A.G., Kaiser, L., Petty, T.J., Kilowoko, M., Kyungu, E., Hongoa, P., Vieille, G., Turin, L., Genton, B., D’Acremont, V., Tapparel, C. Molecular Epidemiology of Human Rhinoviruses and Enteroviruses Highlights Their Diversity in Sub-Saharan Africa. Viruses 2015, 7(12), 6412–6423. [CrossRef]
- Annamalay, A.A., Lanaspa, M., Khoo, S.K., Madrid, L., Acácio, S., Zhang, G., Laing, I.A., Gern, J., Goldblatt, J., Bizzintino, J., Lehmann, D., Le Souëf, P.N., Bassat, Q. Rhinovirus species and clinical features in children hospitalised with pneumonia from Mozambique. Trop Med Int Health 2016, 21(9), 1171–1180. [CrossRef]
- Milanoi, S., Ongus, J.R., Gachara, G., Coldren, R., Bulimo, W. Serotype and genetic diversity of human rhinovirus strains that circulated in Kenya in 2008. Influenza Other Respir Viruses 2016, 10(3), 185–191. [CrossRef]
- Morobe, J.M., Nyiro, J.U., Brand, S., Kamau, E., Gicheru, E., Eyase, F., Otieno, G.P., Munywoki, P.K., Agoti, C.N., Nokes, D.J. Human rhinovirus spatial-temporal epidemiology in rural coastal Kenya, 2015-2016, observed through outpatient surveillance. Wellcome Open Res 2019, 3, 128. [CrossRef]
- Basnet, S., Palmenberg, A.C., Gern, J.E. Rhinoviruses and Their Receptors. Chest 2019, 155(5), 1018–1025. [CrossRef]
- Lamborn, I.T., Su, H.C. Genetic determinants of host immunity against human rhinovirus infections. Hum Genet 2020, 139(6-7), 949–959. [CrossRef]
- Song, J., Wang, H., Shi, J., Cui, A., Huang, Y., Sun, L., Xiang, X., Ma, C., Yu, P., Yang, Z., Li, Q., Ng, T.I., Zhang, Y., Zhang, R., Xu, W. Emergence of BA9 genotype of human respiratory syncytial virus subgroup B in China from 2006 to 2014. Sci Rep 2017, 7(1), 16765. [CrossRef]
- A Al-Sharif, H., El-Kafrawy, S.A., Yousef, J.M., Kumosani, T.A., Kamal, M.A., Khathlan, N.A., Kaki, R.M., Alnajjar, A.A., Azhar, E.I. Dominance of the ON1 Genotype of RSV-A and BA9 Genotype of RSV-B in Respiratory Cases from Jeddah, Saudi Arabia. Genes 2020, 11(11), 1323. [CrossRef]
- Tabor, D.E., Fernandes, F., Langedijk, A.C., Wilkins, D., Lebbink, R.J., Tovchigrechko, A., Ruzin, A., Kragten-Tabatabaie, L., Jin, H., Esser, M.T., Bont, L.J., Abram, M.E., INFORM-RSV Study Group. Global Molecular Epidemiology of Respiratory Syncytial Virus from the 2017-2018 INFORM-RSV Study. J Clin Microbiol 2020, 59(1), e01828-20. [CrossRef]
- Madhi, S.A., Venter, M., Alexandra, R., Lewis, H., Kara, Y., Karshagen, W.F., Greef, M., Lassen, C. Respiratory syncytial virus associated illness in high-risk children and national characterisation of the circulating virus genotype in South Africa. J Clin Virol 2003, 27(2), 180–189. [CrossRef]
- van Niekerk, S., Venter, M. Replacement of previously circulating respiratory syncytial virus subtype B strains with the BA genotype in South Africa. J Virol 2011, 85(17), 8789–8797. [CrossRef]
- Ihling, C.M., Schnitzler, P., Heinrich, N., Mangu, C., Sudi, L., Souares, A., Gies, S., Sié, A., Coulibaly, B., Ouédraogo, A.T., Mordmüller, B., Held, J., Adegnika, A.A., Fernandes, J.F., Eckerle, I., May, J., Hogan, B., Eibach, D., Tabatabai, J. Molecular epidemiology of respiratory syncytial virus in children in sub-Saharan Africa. Trop Med Int Health 2021, 26(7), 810–822. [CrossRef]
- Pangesti, K.N.A., Abd El Ghany, M., Walsh, M.G., Kesson, A.M., Hill-Cawthorne, G.A. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 2018, 28(2), 10.1002/rmv.1968. [CrossRef]
- Kang, H.M., Park, K.C., Park, J., Kil, H.R., Yang, E.A. Circulating Respiratory Syncytial Virus Genotypes and Genetic Variability of the G Gene during 2017 and 2018/2019 Seasonal Epidemics Isolated from Children with Lower Respiratory Tract Infections in Daejeon, Korea. Journal of Korean medical science 2020, 35(49), e422. [CrossRef]
- Korsun, N., Angelova, S., Trifonova, I., Voleva, S., Grigorova, I., Tzotcheva, I., Mileva, S., Alexiev, I., Perenovska, P. Predominance of ON1 and BA9 genotypes of respiratory syncytial virus (RSV) in Bulgaria, 2016-2018. J Med Virol 2021, 93(6), 3401–3411. [CrossRef]
- Razanajatovo Rahombanjanahary, N.H., Rybkina, K., Randriambolamanantsoa, T.H., Razafimanjato, H., Heraud, J.M. Genetic diversity and molecular epidemiology of respiratory syncytial virus circulated in Antananarivo, Madagascar, from 2011 to 2017: Predominance of ON1 and BA9 genotypes. J Clin Virol 2020, 129, 104506. [CrossRef]
- Wang, B., Song, J., Song, J., Mao, N., Liang, J., Chen, Y., Qi, Y., Bai, L., Xie, Z., Zhang, Y. An Outbreak of Severe Neonatal Pneumonia Caused by Human Respiratory Syncytial Virus BA9 in a Postpartum Care Centre in Shenyang, China. Microbiol Spectr 2022, 10(4), e0097422. [CrossRef]
- Valley-Omar, Z., Muloiwa, R., Hu, N.C., Eley, B., Hsiao, N.Y. Novel respiratory syncytial virus subtype ON1 among children, Cape Town, South Africa, 2012. Emerg Infect Dis 2013, 19(4), 668–670. [CrossRef]
- Tabatabai, J., Prifert, C., Pfeil, J., Grulich-Henn, J., Schnitzler, P. Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during winter season 2012-13. PloS One 2014, 9(10), e109191. [CrossRef]
- Tavakoli, F., Izadi, A., Yavarian, J., Sharifi-Zarchi, A., Salimi, V., Mokhtari-Azad, T. Determination of genetic characterization and circulation pattern of Respiratory Syncytial Virus (RSV) in children with a respiratory infection, Tehran, Iran, during 2018-2019. Virus Res 2021, 305, 198564. [CrossRef]
- Kenmoe, S., Vernet, M.A., Miszczak, F., Dina, J., Schoenhals, M., Beng, V.P., Vabret, A., Njouom, R. Genetic diversity of human respiratory syncytial virus isolated among children with acute respiratory infections in Southern Cameroon during three consecutive epidemic seasons, 2011-2013. Trop Med Health 2018, 46, 7. [CrossRef]
- Luo, H.J., Huang, X.B., Zhong, H.L., Ye, C.X., Tan, X., Zhou, K., Yuan, L., Zhang, S.F., Zhu, X., Lin, C.J., Wang, W.J., Xu, L., Cao, K.Y. Epidemiological characteristics and phylogenic analysis of human respiratory syncytial virus in patients with respiratory infections during 2011-2016 in southern China. Int J Infect Dis 2020, 90, 5–17. [CrossRef]
- Lee, C.Y., Fang, Y.P., Wang, L.C., Chou, T.Y., Liu, H.F. Genetic Diversity and Molecular Epidemiology of Circulating Respiratory Syncytial Virus in Central Taiwan, 2008-2017. Viruses 2021, 14(1), 32. [CrossRef]
- Auksornkitti, V., Kamprasert, N., Thongkomplew, S., Suwannakarn, K., Theamboonlers, A., Samransamruajkij, R., Poovorawan, Y. Molecular characterization of human respiratory syncytial virus, 2010-2011: identification of genotype ON1 and a new subgroup B genotype in Thailand. Arch Virol 2014, 159(3), 499–507. [CrossRef]
- Etemadi, M.R., Sekawi, Z., Othman, N., Lye, M.S., Moghaddam, F.Y. Circulation of human respiratory syncytial virus strains among hospitalized children with acute lower respiratory infection in malaysia. Evol Bioinform Online 2013, 9, 151–161. [CrossRef]
- Ábrego, L.E., Delfraro, A., Franco, D., Castillo, J., Castillo, M., Moreno, B., López-Vergès, S., Pascale, J. M., Arbiza, J. Genetic variability of human respiratory syncytial virus group B in Panama reveals a novel genotype BA14. J Med Virol 2017, 89(10), 1734–1742. [CrossRef]
- Komoyo, G.F., Yambiyo, B.M., Manirakiza, A., Gody, J.C., Muller, C.P., Hübschen, J.M., Nakoune, E., Snoeck, C.J. Epidemiology and genetic characterization of respiratory syncytial virus in children with acute respiratory infections: Findings from the influenza sentinel surveillance network in Central African Republic, 2015 to 2018. Health Sci Rep 2021, 4(2), e298. [CrossRef]
- Raghuram S,V., Khan, W.H., Deeba, F., Sullender, W., Broor, S., Parveen, S. Retrospective phylogenetic analysis of circulating BA genotype of human respiratory syncytial virus with 60 bp duplication from New Delhi, India during 2007-2010. Virusdisease 2015, 26(4), 276–281. [CrossRef]
- Bandla, S.S., Devadiga, S., Bhatt, R., Dsa, O.C., Govindakarnavar, A. Molecular epidemiology of respiratory syncytial virus among children and adults in India 2016 to 2018. Virus Genes 2021, 57(6), 489–501. [CrossRef]
- Hindupur, A., Menon, T., Dhandapani, P. Genetic diversity of human respiratory syncytial virus in children with acute respiratory infections in Chennai, South India. Ind J Med Microbiol 2019, 37(2), 248–254. [CrossRef]
- Do, L.A.H., Wilm, A., van Doorn, H.R., Lam, H.M., Sim, S., Sukumaran, R., Tran, A.T., Nguyen, B.H., Tran, T.T.L., Tran, Q.H., Vo, Q.B., Dac, N.A.T., Trinh, H.N., Nguyen, T.T.H., Binh, B.T.L., Le, K., Nguyen, M.T., Thai, Q.T., Vo, T.V., Ngo, N.Q.M., Dang, T.K.H., Cao, N.H., Tran, T.V., Ho, L.V., Farrar, J., de Jong, M., Chen, S., Nagarajan, N., Bryant, J.E., Hibberd, M.L. Direct whole-genome deep-sequencing of human respiratory syncytial virus A and B from Vietnamese children identifies distinct patterns of inter- and intra-host evolution. J Gen Virol 2015, 96(12), 3470–3483. [CrossRef]
- Thongpan, I., Mauleekoonphairoj, J., Vichiwattana, P., Korkong, S., Wasitthankasem, R., Vongpunsawad, S., Poovorawan, Y. Respiratory syncytial virus genotypes NA1, ON1, and BA9 are prevalent in Thailand, 2012-2015. PeerJ 2017, 5, e3970. [CrossRef]
- Jia, R., Lu, L., Su, L., Lin, Z., Gao, D., Lv, H., Xu, M., Liu, P., Cao, L., Xu, J. Resurgence of Respiratory Syncytial Virus Infection During COVID-19 Pandemic Among Children in Shanghai, China. Front Microbiol 2022, 13, 938372. [CrossRef]
| Pre-Nested Master Mix | Final concentration | Volume (μl) |
|---|---|---|
| Nuclease Free Water | - | 26 |
| AMV/Tfl 5X Reaction Buffer | 1X | 10 |
| dNTP Mix | 0.2 mM of each | 1 |
| Forward Primer | 0.2 pmol/μl | 1 |
| Reverse Primer | 0.2 pmol/μl | 1 |
| MgSO4 | 1 mM | 4 |
| AMV Reverse Transcriptase | 0.1 u/μl | 1 |
| Tfl DNA Polymerase | 0.1 u/μl | 1 |
| Template RNA | - | 5 |
| Nested Master Mix | ||
| Nuclease Free Water | - | 30.5 |
| 5X GoTaq® Flexi Buffer | 1X | 10 |
| PCR nucleotide mix | 0.2 mM of each | 1 |
| Forward Primer | 0.2 pmol/μl | 1 |
| Reverse Primer | 0.2 pmol/μl | 1 |
| MgCl2 | 2 mM | 4 |
| Taq DNA Polymerase | 1.25 u | 0.5 |
| Pre-Nested PCR product | - | 2 |
| PCR Stage | Name | Sequence 5′ → 3′ |
|---|---|---|
| Pre-Nested | RV-Forward 1 | CTC CGG CCC CTG AAT RYG GCT AA |
| RV-Reverse 1 | TCI GGI ARY TTC CAS YAC CAI CC | |
| Nested | HRV 01.3 | TAC TTT GGG TGT CCG TGT TTC |
| HRV 02.3 | GGC AAC TTC CAC CAC CC |
| Pre-Nested Master Mix | Final concentration | Volume (μl) |
|---|---|---|
| Nuclease Free Water | - | 20.6 |
| AMV/Tfl 5X Reaction Buffer | 1X | 10 |
| dNTP Mix | 0.2 mM of each | 1 |
| Forward Primer | 0.2 pmol/μl | 1 |
| Reverse Primer | 0.2 pmol/μl | 1 |
| MgSO4 | 1 mM | 4 |
| AMV Reverse Transcriptase | 0.1 u/μl | 1 |
| Tfl DNA Polymerase | 0.1 u/μl | 1 |
| Dithiothreitol | 0.4 | |
| Template RNA | - | 10 |
| Nested Master Mix | ||
| Nuclease Free Water | - | 30.5 |
| 5X GoTaq® Flexi Buffer | 1X | 10 |
| PCR nucleotide mix | 0.2 mM of each | 1 |
| Forward Primer | 0.2 pmol/μl | 1 |
| Reverse Primer | 0.2 pmol/μl | 1 |
| MgCl2 | 2 mM | 4 |
| Taq DNA Polymerase | 1.25u | 0.5 |
| Pre-Nested PCR product | - | 2 |
| PCR Stage | Name | Sequence 5′ → 3′ |
|---|---|---|
| Pre-Nested | SH1 | CAC AGT KAC TGA CAA YAA AGG AGC |
| F164 | GTT ATG ACA CTG GTA TAC CAA CC | |
| Nested | ABG490 | ATG ATT WYC AYT TTG AAG TGT TC |
| F9AB | CAA CTC CAT KRT TAT TTG CC |
| Study | Virus | Expected | Detected | Selected for sequencing | PCR products obtained |
|---|---|---|---|---|---|
| 2015/2016 | HRV | 68 | 49 | 49 | 45 |
| RSV | 11 | 10 | 7 | 7 | |
| 2018/2019 | HRV | 38 | 15 | 15 | 15 |
| RSV | 45 | 5 | 3 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





