Submitted:
26 February 2024
Posted:
01 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Sampling Locations, Collections and DNA Extraction
2.2. HT-qPCR
2.3. Bacterial 16S rRNA Sequencing of 64 Samples
2.4. Statistical Analysis
3. Results and Discussions
3.1. Microbial Profile
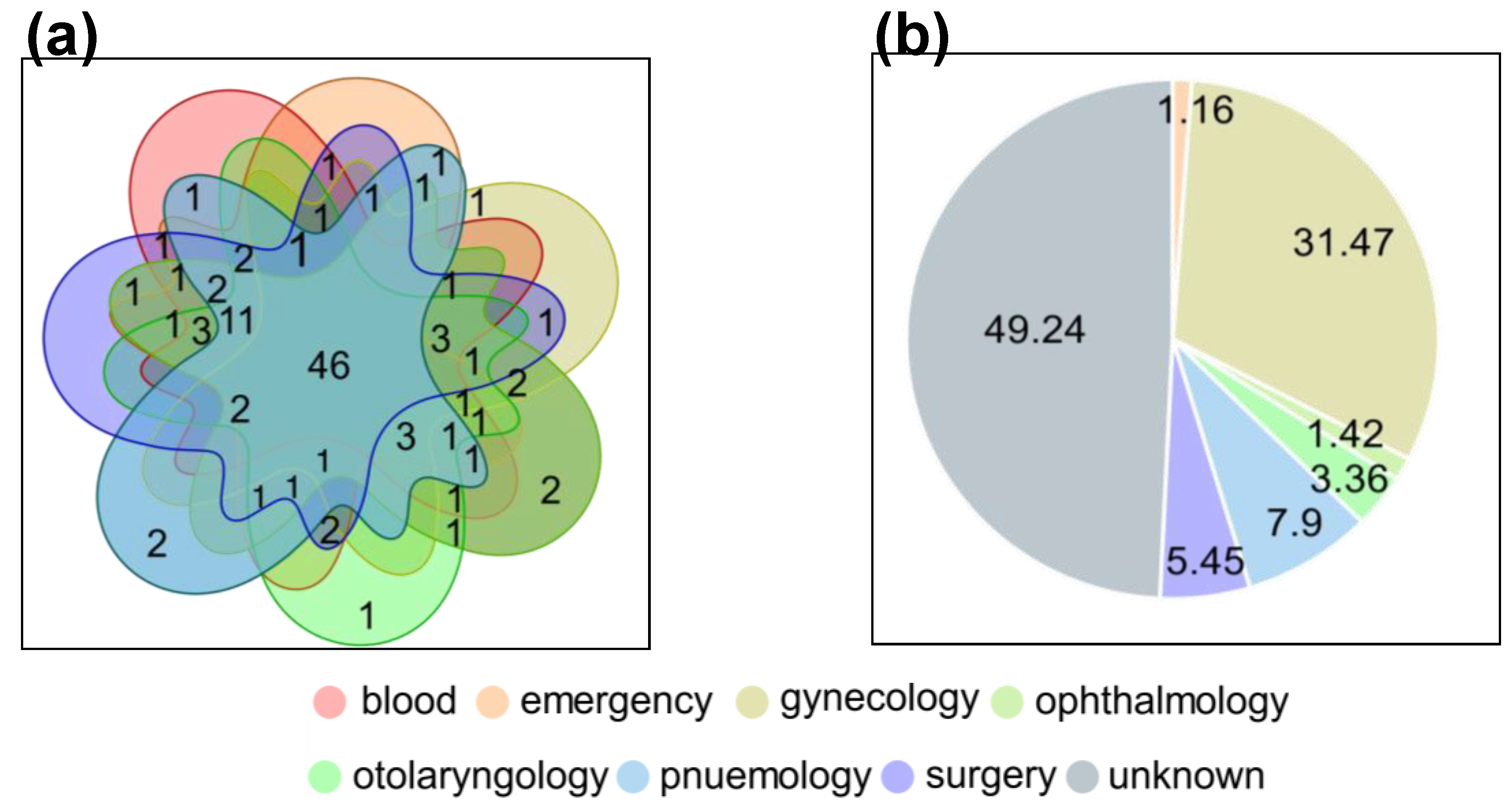
3.2. Broad-Spectrum Profile of ARGs and MGEs
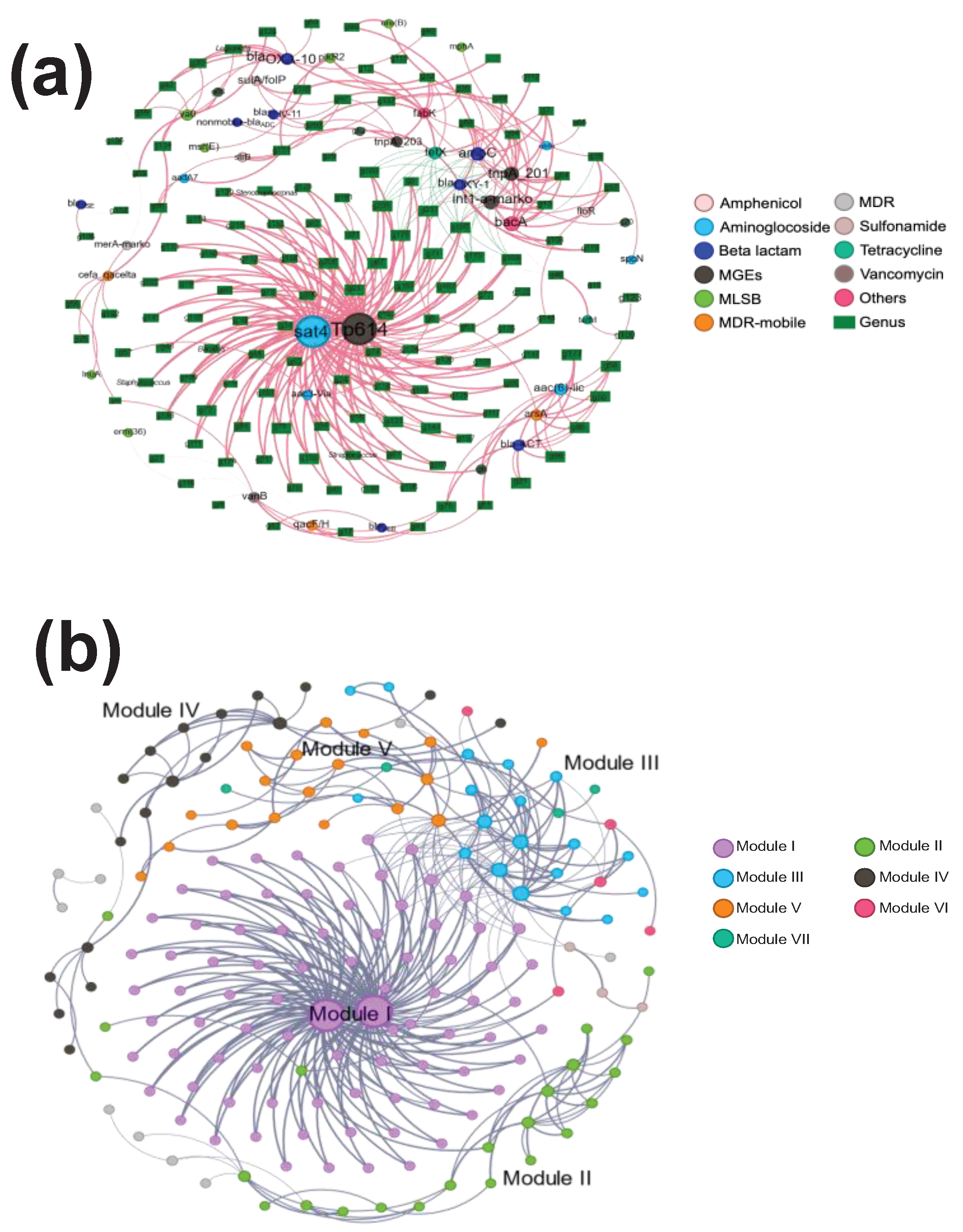
3.3. Co-Occurrence between Bacterial Communities and ARGs/MGEs in the Gynecology Department
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Alessandro, D.; Fara, G.M. Hospital Environments and Epidemiology of Healthcare-Associated Infections. In Indoor Air Quality in Healthcare Facilities; Capolongo, S., Settimo, G., Gola, M., Eds.; Springer International Publishing: Cham, 2017; pp. 41–52 ISBN 9783319491608.
- Pereira da Fonseca, T.A.; Pessôa, R.; Felix, A.C.; Sanabani, S.S. Diversity of Bacterial Communities on Four Frequently Used Surfaces in a Large Brazilian Teaching Hospital. Int. J. Environ. Res. Public Health 2016, 13, 152. [CrossRef]
- Treakle, A.M.; Thom, K.A.; Furuno, J.P.; Strauss, S.M.; Harris, A.D.; Perencevich, E.N. Bacterial Contamination of Health Care Workers’ White Coats. Am. J. Infect. Control 2009, 37, 101–105. [CrossRef]
- Tang, P.H.P.; Worster, A.; Srigley, J.A.; Main, C.L. Examination of Staphylococcal Stethoscope Contamination in the Emergency Department (pilot) Study (EXSSCITED Pilot Study). CJEM 2011, 13, 239–244. [CrossRef]
- Li, X.; Wu, Z.; Dang, C.; Zhang, M.; Zhao, B.; Cheng, Z.; Chen, L.; Zhong, Z.; Ye, Y.; Xia, Y. A Metagenomic-Based Method to Study Hospital Air Dust Resistome. Chem. Eng. J. 2021, 406, 126854. [CrossRef]
- Franco, L.C.; Tanner, W.; Ganim, C.; Davy, T.; Edwards, J.; Donlan, R. A Microbiological Survey of Handwashing Sinks in the Hospital Built Environment Reveals Differences in Patient Room and Healthcare Personnel Sinks. Sci. Rep. 2020, 10, 8234. [CrossRef]
- Kotay, S.M.; Parikh, H.I.; Barry, K.; Gweon, H.S.; Guilford, W.; Carroll, J.; Mathers, A.J. Nutrients Influence the Dynamics of Klebsiella Pneumoniae Carbapenemase Producing Enterobacterales in Transplanted Hospital Sinks. Water Res. 2020, 176, 115707. [CrossRef]
- Kramer, A.; Schwebke, I.; Kampf, G. How Long Do Nosocomial Pathogens Persist on Inanimate Surfaces? A Systematic Review. BMC Infect. Dis. 2006, 6, 130.
- Boyce, J.M. Environmental Contamination Makes an Important Contribution to Hospital Infection. J. Hosp. Infect. 2007, 65 Suppl 2, 50–54. [CrossRef]
- Kelsey, M. Pseudomonas in Augmented Care: Should We Worry? J. Antimicrob. Chemother. 2013, 68, 2697–2700.
- Hota, B. Contamination, Disinfection, and Cross-Colonization: Are Hospital Surfaces Reservoirs for Nosocomial Infection? Clin. Infect. Dis. 2004, 39, 1182–1189.
- Moges, F.; Endris, M.; Belyhun, Y.; Worku, W. Isolation and Characterization of Multiple Drug Resistance Bacterial Pathogens from Waste Water in Hospital and Non-Hospital Environments, Northwest Ethiopia. BMC Res. Notes 2014, 7, 215. [CrossRef]
- Phoon, H.Y.P.; Hussin, H.; Hussain, B.M.; Lim, S.Y.; Woon, J.J.; Er, Y.X.; Thong, K.L. Distribution, Genetic Diversity and Antimicrobial Resistance of Clinically Important Bacteria from the Environment of a Tertiary Hospital in Malaysia. J Glob Antimicrob Resist 2018, 14, 132–140. [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the Wild: Antibiotic Resistance Genes in Natural Environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [CrossRef]
- Klassert, T.E.; Leistner, R.; Zubiria-Barrera, C.; Stock, M.; López, M.; Neubert, R.; Driesch, D.; Gastmeier, P.; Slevogt, H. Bacterial Colonization Dynamics and Antibiotic Resistance Gene Dissemination in the Hospital Environment after First Patient Occupancy: A Longitudinal Metagenetic Study. Microbiome 2021, 9, 169.
- Sukhum, K.V.; Newcomer, E.P.; Cass, C.; Wallace, M.A.; Johnson, C.; Fine, J.; Sax, S.; Barlet, M.H.; Burnham, C.-A.D.; Dantas, G.; et al. Antibiotic-Resistant Organisms Establish Reservoirs in New Hospital Built Environments and Are Related to Patient Blood Infection Isolates. Commun. Med. 2022, 2, 62. [CrossRef]
- Cassone, M.; Mantey, J.; Gontjes, K.J.; Lansing, B.J.; Gibson, K.E.; Wang, J.; Mody, L. Seasonal Patterns in Incidence and Antimicrobial Resistance of Common Bacterial Pathogens in Nursing Home Patients and Their Rooms. Front Public Health 2021, 9, 671428. [CrossRef]
- Ramos, T.; Dedesko, S.; Siegel, J.A.; Gilbert, J.A.; Stephens, B. Spatial and Temporal Variations in Indoor Environmental Conditions, Human Occupancy, and Operational Characteristics in a New Hospital Building. PLoS One 2015, 10, e0118207. [CrossRef]
- ElRakaiby, M.T.; Gamal-Eldin, S.; Amin, M.A.; Aziz, R.K. Hospital Microbiome Variations As Analyzed by High-Throughput Sequencing. OMICS 2019, 23, 426–438. [CrossRef]
- Choe, Y.J.; Smit, M.A.; Mermel, L.A. Seasonality of Respiratory Viruses and Bacterial Pathogens. Antimicrob. Resist. Infect. Control 2019, 8, 125. [CrossRef]
- Ben Maamar, S.; Hu, J.; Hartmann, E.M. Implications of Indoor Microbial Ecology and Evolution on Antibiotic Resistance. J. Expo. Sci. Environ. Epidemiol. 2020, 30, 1–15.
- Bokulich, N.A.; Mills, D.A.; Underwood, M.A. Surface Microbes in the Neonatal Intensive Care Unit: Changes with Routine Cleaning and over Time. J. Clin. Microbiol. 2013, 51, 2617–2624. [CrossRef]
- Oberauner, L.; Zachow, C.; Lackner, S.; Högenauer, C.; Smolle, K.-H.; Berg, G. The Ignored Diversity: Complex Bacterial Communities in Intensive Care Units Revealed by 16S Pyrosequencing. Sci. Rep. 2013, 3, 1413. [CrossRef]
- Zhu, D.; Ding, J.; Yin, Y.; Ke, X.; O’Connor, P.; Zhu, Y.-G. Effects of Earthworms on the Microbiomes and Antibiotic Resistomes of Detritus Fauna and Phyllospheres. Environ. Sci. Technol. 2020, 54, 6000–6008. [CrossRef]
- Stedtfeld, R.D.; Guo, X.; Stedtfeld, T.M.; Sheng, H.; Williams, M.R.; Hauschild, K.; Gunturu, S.; Tift, L.; Wang, F.; Howe, A.; et al. Primer Set 2.0 for Highly Parallel qPCR Array Targeting Antibiotic Resistance Genes and Mobile Genetic Elements. FEMS Microbiol. Ecol. 2018, 94, doi:10.1093/femsec/fiy130. [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890.
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336.
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461.
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596.
- Oksanen, J.; Blanchet, F. G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, B.; Simpson, G.; Solymos, P.; Stevens, H.; Wagner, H. Vegan: Community ecology package. R Package Version 2.2−1 2015, 2, 1−2.
- Genuer, R.; Poggi, J.-M. Random Forests with R; Springer Nature, 2020; ISBN 9783030564858.
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinformatics 2011, 12, 35. [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. ICWSM 2009, 3, 361–362. [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763.
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer, 2016; ISBN 9783319242774.
- Bonadonna, L.; Briancesco, R.; Coccia, A.M. Analysis of Microorganisms in Hospital Environments and Potential Risks. In Indoor Air Quality in Healthcare Facilities; Capolongo, S., Settimo, G., Gola, M., Eds.; Springer International Publishing: Cham, 2017; pp. 53–62 ISBN 9783319491608.
- Petriglieri, F.; Singleton, C.; Peces, M.; Petersen, J.F.; Nierychlo, M.; Nielsen, P.H. “Candidatus Dechloromonas Phosphoritropha” and “Ca. D. Phosphorivorans”, Novel Polyphosphate Accumulating Organisms Abundant in Wastewater Treatment Systems. ISME J. 2021, 15, 3605–3614. [CrossRef]
- Bernardet, J.-F.; Segers, P.; Vancanneyt, M.; Berthe, F.; Kersters, K.; Vandamme, P. Cutting a Gordian Knot: Emended Classification and Description of the Genus Flavobacterium, Emended Description of the Family Flavobacteriaceae, and Proposal of Flavobacterium Hydatis Nom. Nov. (Basonym, Cytophaga Aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 1996, 46, 128–148.
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the Mechanisms of Acinetobacter Baumannii Virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [CrossRef]
- Madebo, C.; Haile, A.; Eticha, T.; Solomon, F. Hospital-Based Air-Borne and Surface-Borne Bacterial Pathogens and Their Antimicrobial Profiles in Wolaita Sodo, Southern Ethiopia. Int. J. Microbiol. 2022, 2022, 5718341. [CrossRef]
- Perencevich, E.N.; McGregor, J.C.; Shardell, M.; Furuno, J.P.; Harris, A.D.; Morris, J.G., Jr; Fisman, D.N.; Johnson, J.A. Summer Peaks in the Incidences of Gram-Negative Bacterial Infection Among Hospitalized Patients. Infect. Control Hosp. Epidemiol. 2008, 29, 1124–1131. [CrossRef]
- Zhou, J.; Deng, Y.; Shen, L.; Wen, C.; Yan, Q.; Ning, D.; Qin, Y.; Xue, K.; Wu, L.; He, Z.; et al. Temperature Mediates Continental-Scale Diversity of Microbes in Forest Soils. Nat. Commun. 2016, 7, 12083. [CrossRef]
- Dé, I.; Rolston, K.V.I.; Han, X.Y. Clinical Significance of Roseomonas Species Isolated from Catheter and Blood Samples: Analysis of 36 Cases in Patients with Cancer. Clin. Infect. Dis. 2004, 38, 1579–1584. [CrossRef]
- Ryan, K.J.; Sherris, R.C.G. Medical Microbiology^ Available online: https://booksca.ca/wp-content/uploads/XPreview/Pharmacology/3/sherris-medical-microbiology-7th-edition-by-kenneth-j-ryan.pdf (accessed on 19 January 2024).
- Brown, J.H.; Gillooly, J.F.; Allen, A.P.; Savage, V.M.; West, G.B. Toward a Metabolic Theory of Ecology. Ecology 2004, 85, 1771–1789.
- Hospodsky, D.; Qian, J.; Nazaroff, W.W.; Yamamoto, N.; Bibby, K.; Rismani-Yazdi, H.; Peccia, J. Human Occupancy as a Source of Indoor Airborne Bacteria. PLoS One 2012, 7, e34867. [CrossRef]
- Leung, M.H.Y.; Tong, X.; Lee, P.K.H. Indoor Microbiome and Airborne Pathogens. Comprehensive Biotechnology 2019, 96.
- Li, Y.; Liao, H.; Yao, H. Prevalence of Antibiotic Resistance Genes in Air-Conditioning Systems in Hospitals, Farms, and Residences. Int. J. Environ. Res. Public Health 2019, 16, doi:10.3390/ijerph16050683. [CrossRef]
- Hartmann, E.M.; Hickey, R.; Hsu, T.; Betancourt Román, C.M.; Chen, J.; Schwager, R.; Kline, J.; Brown, G.Z.; Halden, R.U.; Huttenhower, C.; et al. Antimicrobial Chemicals Are Associated with Elevated Antibiotic Resistance Genes in the Indoor Dust Microbiome. Environ. Sci. Technol. 2016, 50, 9807–9815. [CrossRef]
- Prussin, A.J., 2nd; Marr, L.C. Sources of Airborne Microorganisms in the Built Environment. Microbiome 2015, 3, 78.
- Jiao, X.; Guo, W.; Li, X.; Yao, F.; Zeng, M.; Yuan, Y.; Guo, X.; Wang, M.; Xie, Q.D.; Cai, L.; et al. New Insight into the Microbiome, Resistome, and Mobilome on the Dental Waste Water in the Context of Heavy Metal Environment. Front. Microbiol. 2023, 14, 1106157. [CrossRef]
- Markkanen, M.A.; Haukka, K.; Pärnänen, K.M.M.; Dougnon, V.T.; Bonkoungou, I.J.O.; Garba, Z.; Tinto, H.; Sarekoski, A.; Karkman, A.; Kantele, A.; et al. Metagenomic Analysis of the Abundance and Composition of Antibiotic Resistance Genes in Hospital Wastewater in Benin, Burkina Faso, and Finland. mSphere 2023, 8, e0053822. [CrossRef]
- Wu, D.; Jin, L.; Xie, J.; Liu, H.; Zhao, J.; Ye, D.; Li, X.-D. Inhalable Antibiotic Resistomes Emitted from Hospitals: Metagenomic Insights into Bacterial Hosts, Clinical Relevance, and Environmental Risks. Microbiome 2022, 10, 19. [CrossRef]
- Leigh, R.J.; McKenna, C.; McWade, R.; Lynch, B.; Walsh, F. Comparative Genomics and Pangenomics of Vancomycin-Resistant and Susceptible Enterococcus Faecium from Irish Hospitals. J. Med. Microbiol. 2022, 71, doi:10.1099/jmm.0.001590. [CrossRef]
- Liassine, N.; Assouvie, L.; Descombes, M.-C.; Tendon, V.D.; Kieffer, N.; Poirel, L.; Nordmann, P. Very Low Prevalence of MCR-1/MCR-2 Plasmid-Mediated Colistin Resistance in Urinary Tract Enterobacteriaceae in Switzerland. Int. J. Infect. Dis. 2016, 51, 4–5.
- Saadatian Farivar, A.; Nowroozi, J.; Eslami, G.; Sabokbar, A. RAPD PCR Profile, Antibiotic Resistance, Prevalence of armA Gene, and Detection of KPC Enzyme in Klebsiella Pneumoniae Isolates. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 6183162.
- Martinez, J.L.; Fajardo, A.; Garmendia, L.; Hernandez, A.; Linares, J.F.; Martínez-Solano, L.; Sánchez, M.B. A Global View of Antibiotic Resistance. FEMS Microbiol. Rev. 2009, 33, 44–65. [CrossRef]
- Whittle, G.; Whitehead, T.R.; Hamburger, N.; Shoemaker, N.B.; Cotta, M.A.; Salyers, A.A. Identification of a New Ribosomal Protection Type of Tetracycline Resistance Gene, tet(36), from Swine Manure Pits. Appl. Environ. Microbiol. 2003, 69, 4151–4158. [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and Network Analysis Reveal Wide Distribution and Co-Occurrence of Environmental Antibiotic Resistance Genes. ISME J. 2015, 9, 2490–2502. [CrossRef]
- Pappa, O.; Chochlakis, D.; Sandalakis, V.; Dioli, C.; Psaroulaki, A.; Mavridou, A. Antibiotic Resistance of Legionella Pneumophila in Clinical and Water Isolates—A Systematic Review. Int. J. Environ. Res. Public Health 2020, 17, 5809. [CrossRef]
- Sharaby, Y.; Nitzan, O.; Brettar, I.; Höfle, M.G.; Peretz, A.; Halpern, M. Antimicrobial Agent Susceptibilities of Legionella Pneumophila MLVA-8 Genotypes. Sci. Rep. 2019, 9, 6138. [CrossRef]
- Nimmo, G.R.; Bull, J.Z. Comparative Susceptibility of Legionella Pneumophila and Legionella Longbeachae to 12 Antimicrobial Agents. J. Antimicrob. Chemother. 1995, 36, 219–223. [CrossRef]
- Golshani, Z.; Sharifzadeh, A. Prevalence of blaOxa10 Type Beta-Lactamase Gene in Carbapenemase Producing Pseudomonas Aeruginosa Strains Isolated from Patients in Isfahan. Jundishapur Journal of Microbiology 2013.
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377.
- Almahmoud, I.; Kay, E.; Schneider, D.; Maurin, M. Mutational Paths towards Increased Fluoroquinolone Resistance in Legionella Pneumophila. J. Antimicrob. Chemother. 2009, 64, 284–293. [CrossRef]
- Enany, S.; Alexander, L.C. The Rise of Virulence and Antibiotic Resistance in Staphylococcus Aureus; BoD – Books on Demand, 2017; ISBN 9789535129837.
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus Spp. Reveals Distinct Species and Antimicrobial Resistance Diversity across a One-Health Continuum. Sci. Rep. 2020, 10, 3937.



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




