Submitted:
29 February 2024
Posted:
29 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Neuronal Differentiation: Induced Pluripotent Stem Cell Culture
2.2. Neuronal Differentiation
2.3. Lentivirus Production
2.4. Neuronal Characterization by Immunocytochemistry
2.5. Amyloid-β Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Western Blotting for Tau: Protein Extraction
2.7. Detergent Compatible (DC) Protein Assay
2.8. Western Blotting
2.9. Flexstation to Measure Calcium Responses to Glutamate, NMDA, AMPA and Kainate
2.10. RT-qPCR for AMPAR Subunits: RNA Extraction and Purification
2.11. cDNA Synthesis
2.12. qPCR
2.13. Data Analysis
3. Results
3.1. Generation of iPSC-Derived Neurons from FAD and Control Lines
3.1.1. iPSC-Derived Neurons Express Neuronal Marker MAP2
3.2. FAD NGN2 Derived iPSC Neurons Did Not Show Evidence of Aβ Pathology at Day 35 of Maturation
3.3. FAD NGN2 Derived iPSC Neurons Did Not Show Tau Pathology at Day 35 of Maturation
3.4. FAD Neurons Demonstrated Increased AMPAR Ca2+ Signalling Compared to Isogenic Controls
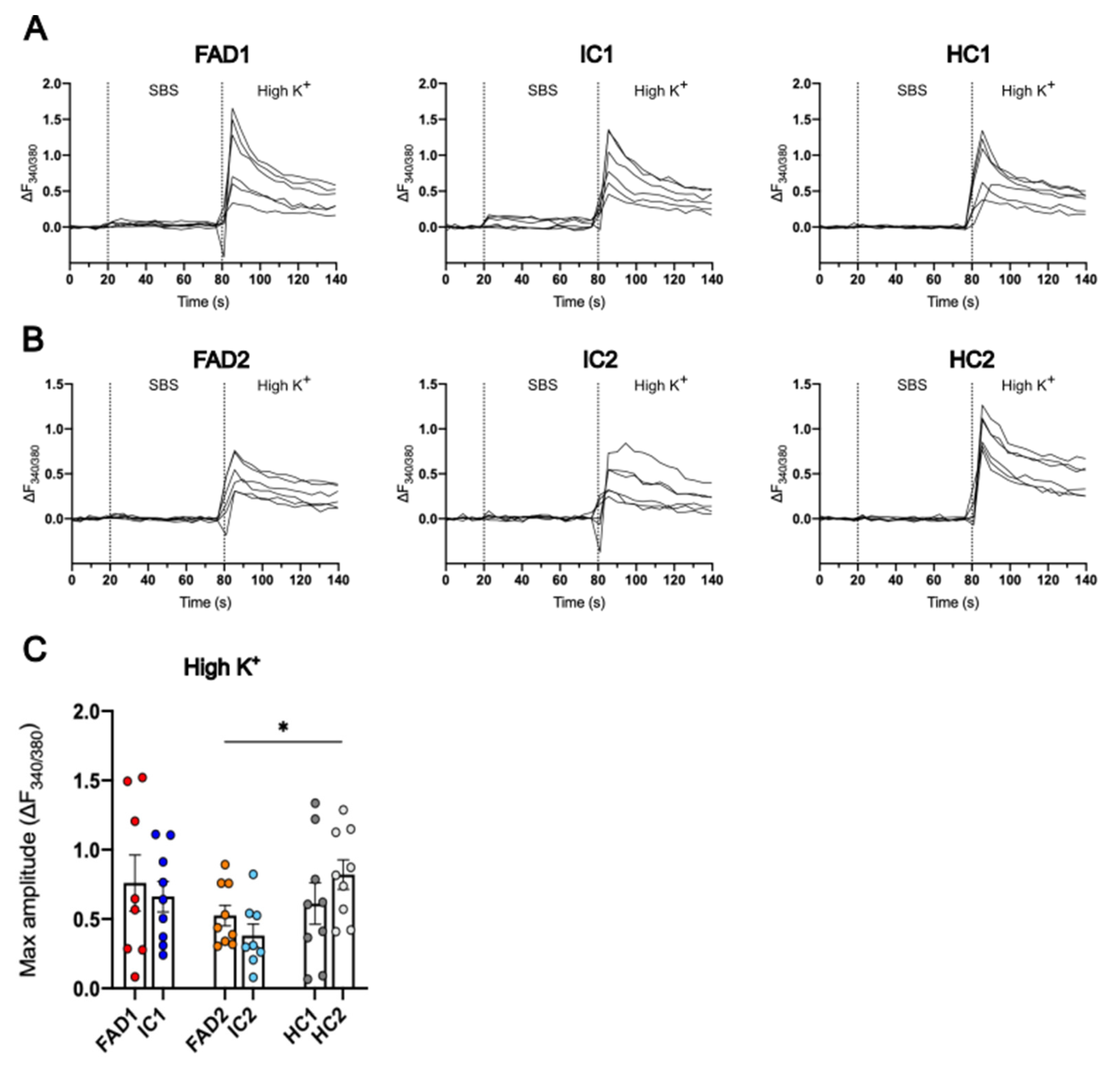
3.5. The iPSC-Derived Neurons Displayed Ca2+ Responses to High K+
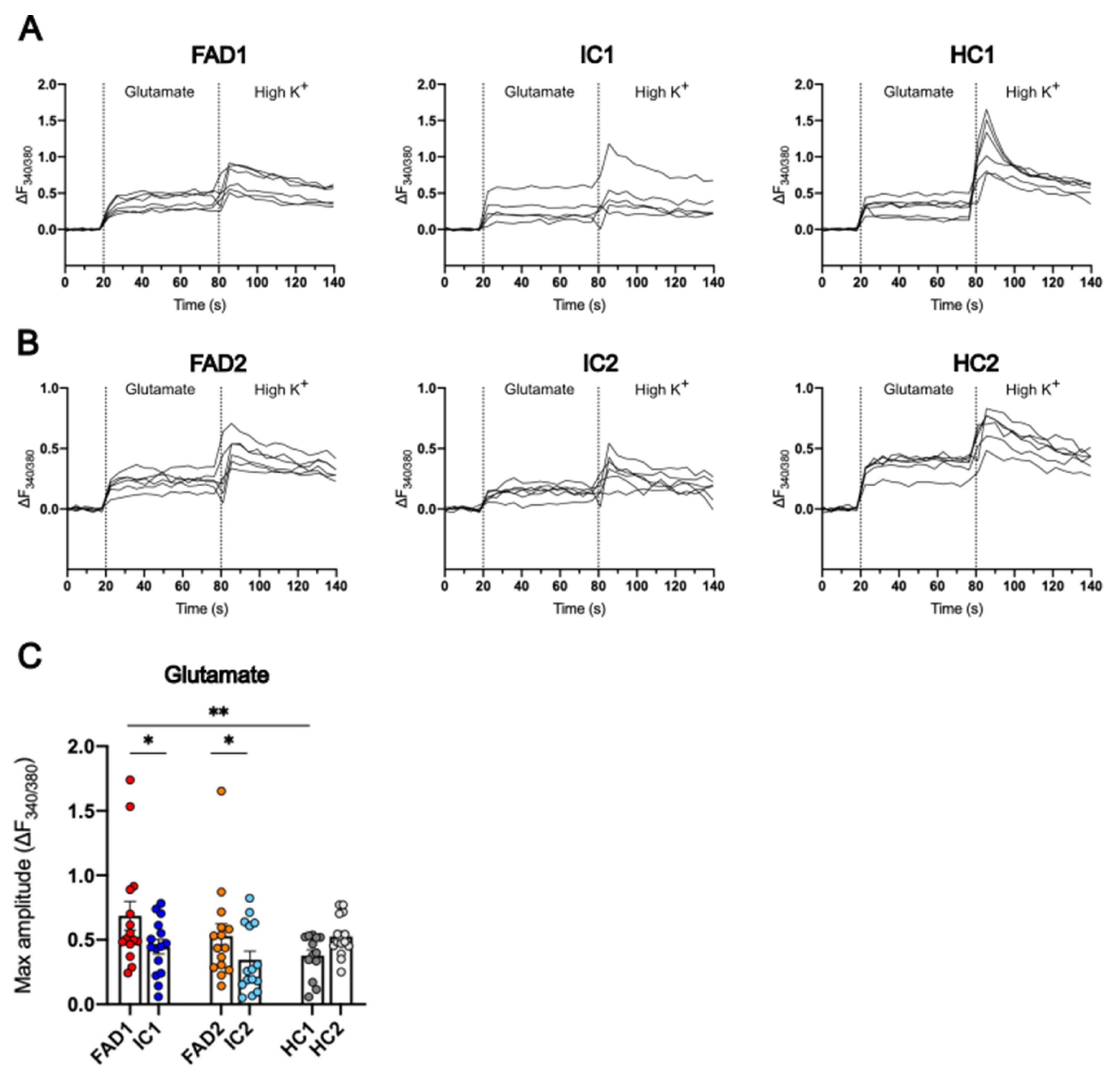
3.6. FAD Neurons Displayed Increased Ca2+ Responses to Glutamate
3.7. FAD Neurons Displayed Increased Ca2+ Responses to AMPA but Not NMDA or Kainate Compared to Their Isogenic Control Lines
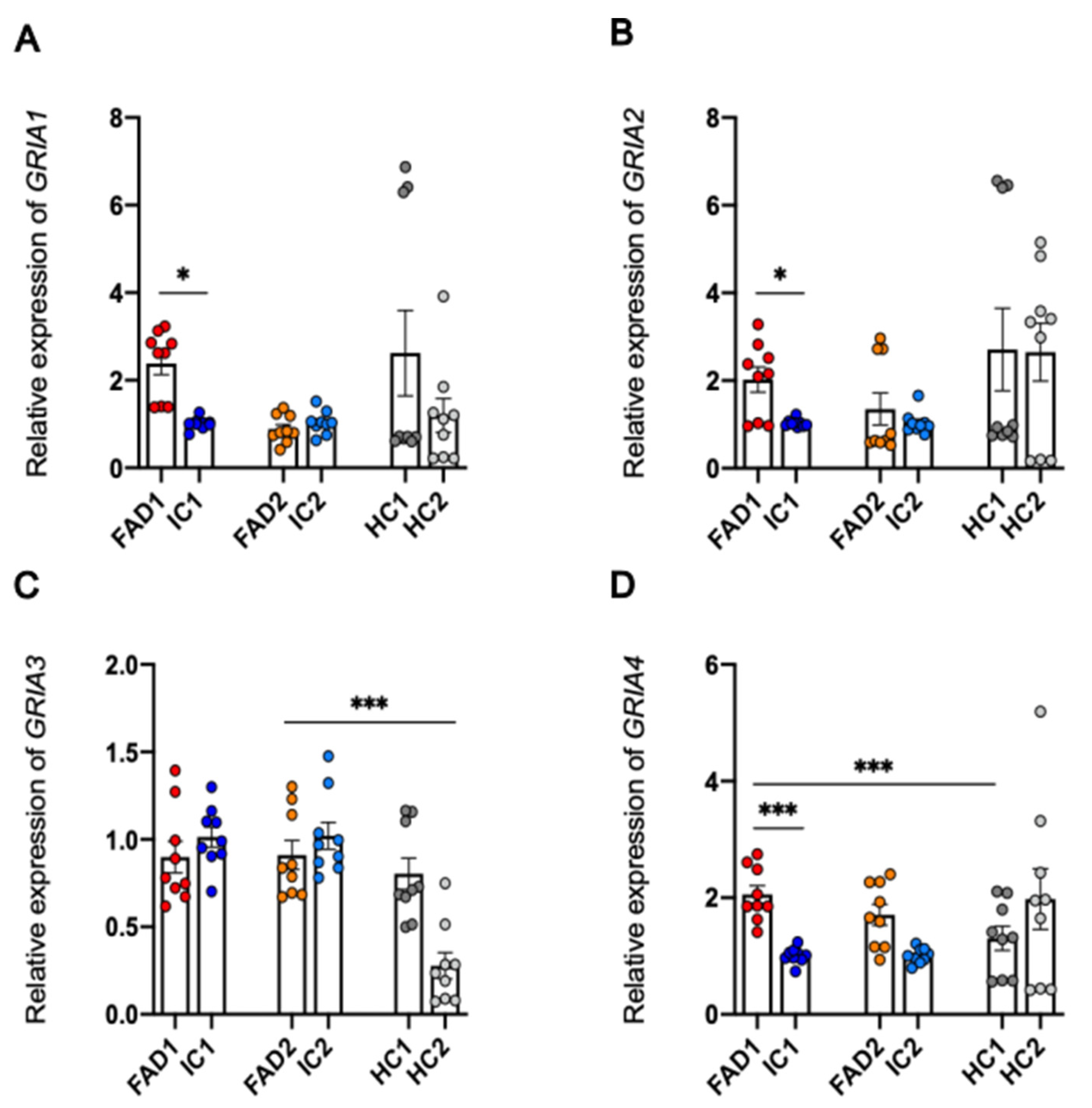
3.8. Regulation at the Level of mRNA or Protein of the AMPA Receptor Subunits Does Not Explain the Increased Calcium Responses to AMPA in FAD Neurons Compared to Isogenic Control Neurons
3.8.1. Protein Expression of GluA1 and GluA2 is not Significantly Different between FAD and Control Neurons
4. Discussion
4.1. FAD Neurons Lacking Aβ and Tau Pathology Show Elevated Ca2+ Responses to Glutamate and AMPA Compared to Isogenic Controls
4.2. Aberrant Ca2+ Signalling of FAD Neurons Occurs Independently of Changes in GluA1 and GluA2 Protein Expression
5. Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bird, T.D. Alzheimer Disease Overview. 1998.
- Blacker, D.; Tanzi, R.E. The Genetics of Alzheimer Disease: Current Status and Future Prospects. Arch Neurol. 1998, 55, 294–296. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Brion, J.P.; Couck, A.M.; Passareiro, E.; Flament-Durand, J. Neurofibrillary tangles of Alzheimer’s disease: An immunohistochemical study. J Submicrosc Cytol. 1985, 17, 89–96. [Google Scholar] [PubMed]
- Ge, M.; Zhang, J.; Chen, S.; Huang, Y.; Chen, W.; He, L.; et al. Role of Calcium Homeostasis in Alzheimer’s Disease. Neuropsychiatr Dis Treat. 2022, 18, 487–498. [Google Scholar] [CrossRef]
- Bezprozvanny, I.; Mattson, M.P. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008, 31, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, S.; Herreman, A.; De Strooper, B.; Bezprozvanny, I. Role of presenilins in neuronal calcium homeostasis. J Neurosci. 2010, 30, 8566–8580. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.; Nelson, O.; Bezprozvanny, A.; Wang, Z.; Lee, S.F.; Hao, Y.H.; et al. Presenilins Form ER Ca2+ Leak Channels, a Function Disrupted by Familial Alzheimer’s Disease-Linked Mutations. Cell. 2006, 126, 981–993. [Google Scholar] [CrossRef]
- Green, K.N.; Demuro, A.; Akbari, Y.; Hitt, B.D.; Smith, I.F.; Parker, I.; et al. SERCA pump activity is physiologically regulated by presenilin and regulates amyloid β production. J Cell Biol. 2008, 181, 1107–1116. [Google Scholar] [CrossRef]
- Cheung, K-H; Shineman, D. ; Müller, M.; Cárdenas, C.; Mei, L.; Yang, J.; et al. supplemental data - Mechanism of Ca2+ disruption in Alzheimer’s disease by presenilin regulation of InsP3 receptor channel gating. Neuron. 2008, 58, 871–883. [Google Scholar] [CrossRef]
- Lee, J.H.; McBrayer, M.K.; Wolfe, D.M.; Haslett, L.J.; Kumar, A.; Sato, Y.; et al. Presenilin 1 maintains lysosomal Ca2+ homeostasis by regulating vATPase-mediated lysosome acidification. Cell Rep. 2015, 12, 1430. [Google Scholar] [CrossRef]
- Wisden, W.; Seeburg, P.H. Mammalian ionotropic glutamate receptors. Curr Opin Neurobiol. 1993, 3, 291–298. [Google Scholar] [CrossRef]
- Schwenk, J.; Baehrens, D.; Haupt, A.; Bildl, W.; Boudkkazi, S.; Roeper, J.; et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014, 84, 41–54. [Google Scholar] [CrossRef]
- Hsieh, H.; Boehm, J.; Sato, C.; Iwatsubo, T.; Tomita, T.; Sisodia, S.; et al. AMPAR Removal Underlies Aβ-Induced Synaptic Depression and Dendritic Spine Loss. Neuron. 2006, 52, 831–843. [Google Scholar] [CrossRef]
- Whitcomb, D.J.; Hogg, E.L.; Regan, P.; Piers, T.; Narayan, P.; Whitehead, G.; et al. Intracellular oligomeric amyloid-beta rapidly regulates GluA1 subunit of AMPA receptor in the hippocampus. Sci Reports 2015 51. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; et al. Tau Mislocalization to Dendritic Spines Mediates Synaptic Dysfunction Independently of Neurodegeneration. Neuron. 2010, 68, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci. 2011, 33, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-Y.; Tuo, Q.-Z.; Liuyang, Z.-Y.; Xie, A.-J.; Feng, X.-L.; Yan, X.; et al. Extrasynaptic NMDA receptor-induced tau overexpression mediates neuronal death through suppressing survival signaling ERK phosphorylation. Cell Death Dis. 2016, 7, e2449. [Google Scholar] [CrossRef] [PubMed]
- Lesné, S.; Ali, C.; Gabriel, C.; Croci, N.; MacKenzie, E.T.; Glabe, C.G.; et al. NMDA receptor activation inhibits α-secretase and promotes neuronal amyloid-β production. J Neurosci. 2005, 25, 9367–9377. [Google Scholar] [CrossRef]
- Bordji, K.; Becerril-Ortega, J.; Nicole, O.; Buisson, A. Activation of Extrasynaptic, But Not Synaptic, NMDA Receptors Modifies Amyloid Precursor Protein Expression Pattern and Increases Amyloid-β Production. J Neurosci. 2010, 30, 15927–15942. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hong, S.; Shepardson, N.E.; Walsh, D.M.; Shankar, G.M.; Selkoe, D. Soluble Oligomers of Amyloid β Protein Facilitate Hippocampal Long-Term Depression by Disrupting Neuronal Glutamate Uptake. Neuron. 2009, 62, 788–801. [Google Scholar] [CrossRef]
- Arias, C.; Arrieta, I.; Tapia, R. β-Amyloid peptide fragment 25–35 potentiates the calcium-dependent release of excitatory amino acids from depolarized hippocampal slices. J Neurosci Res. 1995, 41, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Parpura-Gill, A.; Beitz, D.; Uemura, E. The inhibitory effects of β-amyloid on glutamate and glucose uptakes by cultured astrocytes. Brain Res. 1997, 754, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.M.; Nong, Y.; Almeida, C.G.; Paul, S.; Moran, T.; Choi, E.Y.; et al. Regulation of NMDA receptor trafficking by amyloid-β. Nat Neurosci 2005 88. 2005, 8, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Winslow, B.T.; Onysko, M.K.; Stob, C.M.; Hazlewood, K.A. Treatment of Alzheimer disease. Am Fam Physician. 2011, 83, 1403–1412. [Google Scholar] [PubMed]
- Falcón-Moya, R.; Sihra, T.S.; Rodríguez-Moreno, A. Kainate receptors: Role in epilepsy. Front Mol Neurosci. 2018, 11, 1–7. [Google Scholar] [CrossRef]
- Barthet, G.; Moreira-De-Sá, A.; Zhang, P.; Deforges, S.; Castanheira, J.; Gorlewicz, A.; et al. Presenilin and APP Regulate Synaptic Kainate Receptors. J Neurosci. 2022, 42, 9253–9262. [Google Scholar] [CrossRef]
- Malenka, R.C.; Bear, M.F. LTP and LTD: An embarrassment of riches. Neuron. 2004, 44, 5–21. [Google Scholar] [CrossRef]
- Askenazi, M.; Kavanagh, T.; Pires, G.; Ueberheide, B.; Wisniewski, T.; Drummond, E. Compilation of all known protein changes in the human Alzheimer’s disease brain. BioRxiv 2023, 2023.04.13.536828. [Google Scholar]
- Choi, D.W. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988, 1, 623–634. [Google Scholar] [CrossRef]
- Cox, M.F.; Hascup, E.R.; Bartke, A.; Hascup, K.N. Friend or Foe? Defining the Role of Glutamate in Aging and Alzheimer’s Disease. Front Aging. 2022, 3, 65. [Google Scholar] [CrossRef]
- Targa Dias Anastacio, H.; Matosin, N.; Ooi, L. Neuronal hyperexcitability in Alzheimer’s disease: what are the drivers behind this aberrant phenotype? Transl Psychiatry. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Celone, K.A.; Calhoun, V.D.; Dickerson, B.C.; Atri, A.; Chua, E.F.; Miller, S.L.; et al. Alterations in memory networks in mild cognitive impairment and Alzheimer’s disease: An independent component analysis. J Neurosci. 2006, 26, 10222–10231. [Google Scholar] [CrossRef]
- Dickerson, B.C.; Salat, D.H.; Greve, D.N.; Chua, E.F.; Rand-Giovannetti, E.; Rentz, D.M.; et al. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 2005, 65, 404–411. [Google Scholar] [CrossRef]
- Balez, R.; Ooi, L. Getting to NO Alzheimer’s disease: Neuroprotection versus neurotoxicity mediated by nitric oxide. Oxid Med Cell Longev. 2016, 2016. [Google Scholar] [CrossRef]
- Ghatak, S.; Dolatabadi, N.; Trudler, D.; Zhang, X.; Wu, Y.; Mohata, M.; et al. Mechanisms of hyperexcitability in alzheimer’s disease hiPSC-derived neurons and cerebral organoids vs. Isogenic control. Elife. 2019, 8, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Šišková, Z.; Justus, D.; Kaneko, H.; Friedrichs, D.; Henneberg, N.; Beutel, T.; et al. Dendritic structural degeneration is functionally linked to cellular hyperexcitability in a mouse model of alzheimer’s disease. Neuron. 2014, 84, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Lerdkrai, C.; Asavapanumas, N.; Brawek, B.; Kovalchuk, Y.; Mojtahedi, N.; Del Moral, M.O.; et al. Intracellular Ca2+ stores control in vivo neuronal hyperactivity in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2018, 115, E1279–E1288. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Balez, R.; Muñoz, S.S.; Cabral-da-Silva, M.C.; Stevens, C.H.; Bax, M.; et al. Viral-free generation and characterization of a human induced pluripotent stem cell line from dermal fibroblasts. Stem Cell Res. 2018, 32, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, S.S.; Balez, R.; Castro, Cabral-da-Silva M e, Berg, T.; Engel, M.; Bax, M.; et al. Generation and characterization of human induced pluripotent stem cell lines from a familial Alzheimer’s disease PSEN1 A246E patient and a non-demented family member bearing wild-type PSEN1. Stem Cell Res. 2018, 31, 227–230. [CrossRef]
- Nehme, R.; Zuccaro, E.; Ghosh, S.D.; Li, C.; Sherwood, J.L.; Pietilainen, O.; et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep. 2018, 23, 2509–2523. [Google Scholar] [CrossRef]
- Hansson, O.; Lehmann, S.; Otto, M.; Zetterberg, H.; Lewczuk, P. Advantages and disadvantages of the use of the CSF Amyloid β (Aβ) 42/40 ratio in the diagnosis of Alzheimer’s Disease. Alzheimer’s Res Ther. 2019, 11, 1–15. [Google Scholar] [CrossRef]
- Schindler, S.E.; Bollinger, J.G.; Ovod, V.; Mawuenyega, K.G.; Li, Y.; Gordon, B.A.; et al. High-precision plasma β-amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019, 93, e1647. [Google Scholar] [CrossRef]
- Amft, M.; Ortner, M.; Eichenlaub, U.; Goldhardt, O.; Diehl-Schmid, J.; Hedderich, D.M.; et al. The cerebrospinal fluid biomarker ratio Aβ42/40 identifies amyloid positron emission tomography positivity better than Aβ42 alone in a heterogeneous memory clinic cohort. Alzheimer’s Res Ther. 2022, 14, 1–9. [Google Scholar] [CrossRef]
- Hansson, O.; Zetterberg, H.; Buchhave, P.; Andreasson, U.; Londos, E.; Minthon, L.; et al. Prediction of Alzheimer’s Disease Using the CSF Aβ42/Aβ40 Ratio in Patients with Mild Cognitive Impairment. Dement Geriatr Cogn Disord. 2007, 23, 316–320. [Google Scholar] [CrossRef]
- Balez, R.; Steiner, N.; Engel, M.; Muñoz, S.S.; Lum, J.S.; Wu, Y.; et al. Neuroprotective effects of apigenin against inflammation, neuronal excitability and apoptosis in an induced pluripotent stem cell model of Alzheimer’s disease. Sci Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef]
- Balez, R.; Stevens, C.H.; Lenk, K.; Sidhu, K.; Sutherland, G.; Ooi, L. Increased neuronal nitric oxide synthase in Alzheimer’s disease mediates spontaneous calcium signalling and divergent glutamatergic calcium responses. Antioxid Redox Signal. 2024. 1 February 2024. [CrossRef] [PubMed]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 2018, 98, 1141–1154e7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, H.; Ma, Y.; Shi, G.; Song, J.; Tang, Y.; et al. Early pathogenic event of Alzheimer’s disease documented in iPSCs from patients with PSEN1 mutations. Oncotarget. 2017, 8, 7900–7913. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Rodríguez, S.; Perry, G.; Luna-Muñoz, J.; Acevedo-Aquino, M.C.; Williams, S. Phosphorylation of tau protein at sites Ser396-404 is one of the earliest events in Alzheimer’s disease and Down syndrome. Neuropathol Appl Neurobiol. 2014, 40, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; et al. National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012, 8, 1. [Google Scholar] [CrossRef]
- Xia, Y.; Prokop, S.; Giasson, B.I. “Don’t Phos Over Tau”: recent developments in clinical biomarkers and therapies targeting tau phosphorylation in Alzheimer’s disease and other tauopathies. Mol Neurodegener 2021 161. 2021, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, H.; Mair, W.; Kumar, M.; Schlaffner, C.N.; Tang, S.; Beerepoot, P.; et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer’s Disease. Cell. 2020, 183, 1699–1713e13. [Google Scholar] [CrossRef] [PubMed]
- van der Kant, R.; Goldstein, L.S.B.; Ossenkoppele, R. Amyloid-β-independent regulators of tau pathology in Alzheimer disease. Nat Rev Neurosci. 2020, 21, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.S.; Washicosky, K.J.; Brand, E.; von Maydell, D.; Aronson, J.; Kim, S.; et al. Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer’s disease. Nat Commun 2020 111. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Guntupalli, S.; Widagdo, J.; Anggono, V. Amyloid- β -Induced Dysregulation of AMPA Receptor Trafficking. Neural Plast. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Jurado, S. AMPA receptor trafficking in natural and pathological aging. Front Mol Neurosci. 2018, 10, 446. [Google Scholar] [CrossRef] [PubMed]
- Greenough, M.A.; Lane, D.J.R.; Balez, R.; Anastacio, H.T.D.; Zeng, Z.; Ganio, K.; et al. Selective ferroptosis vulnerability due to familial Alzheimer’s disease presenilin mutations. Cell Death Differ. 2022, 29, 2123–2136. [Google Scholar] [CrossRef]
- Barbour, A.; Gourmaud, S.; Li, X.; Stewart, D.; Irwin, D.; Talos, D.; et al. Seizures exacerbate excitatory: inhibitory imbalance in Alzheimer’s disease with attenuation after rapamycin treatment in 5XFAD mice. BioRxiv, 2023; 2023.03.02.530499. [Google Scholar]
- Guo, C.; Ma, Y.Y. Calcium Permeable-AMPA Receptors and Excitotoxicity in Neurological Disorders. Front Neural Circuits. 2021, 15, 82. [Google Scholar] [CrossRef]
- Kawahara, Y.; Ito, K.; Sun, H.; Kanazawa, I.; Kwak, S. Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur J Neurosci. 2003, 18, 23–33. [Google Scholar] [CrossRef]
- Wright, A.; Vissel, B. The essential role of AMPA receptor GluA2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci. 2012, 0, 34. [Google Scholar]
- Gaisler-Salomon, I.; Kravitz, E.; Feiler, Y.; Safran, M.; Biegon, A.; Amariglio, N.; et al. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol Aging. 2014, 35, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; et al. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA. 2016, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, S.; Smith, M.A.; Jones, E.G. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995, 699, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Konen, L.M.; Wright, A.L.; Royle, G.A.; Morris, G.P.; Lau, B.K.; Seow, P.W.; et al. A new mouse line with reduced GluA2 Q/R site RNA editing exhibits loss of dendritic spines, hippocampal CA1-neuron loss, learning and memory impairments and NMDA receptor-independent seizure vulnerability. Mol Brain. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Pachernegg, S.; Münster, Y.; Muth-Köhne, E.; Fuhrmann, G.; Hollmann, M. GluA2 is rapidly edited at the Q/R site during neural differentiation in vitro. Front Cell Neurosci. 2015, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Shi, Y.; Jackson, A.C.; Bjorgan, K.; During, M.J.; Sprengel, R.; et al. Subunit Composition of Synaptic AMPA Receptors Revealed by a Single-Cell Genetic Approach. Neuron. 2009, 62, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Wenthold, R.J.; Petralia, R.S.; Blahos, J.; Niedzielski, A.S. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci. 1996, 16, 1982–1989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, C.; Gearing, M.; Wang, P.G.; Chin, L.S.; Li, L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer’s disease. Acta Neuropathol Commun. 2018, 6, 19. [Google Scholar] [CrossRef]
- Mendonça, C.F.; Kuras, M.; Nogueira, F.C.S.; Plá, I.; Hortobágyi, T.; Csiba, L.; et al. Proteomic signatures of brain regions affected by tau pathology in early and late stages of Alzheimer’s disease. Neurobiol Dis. 2019, 130. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Yin, L.; Thambisetty, M.; Troncoso, J.C.; et al. Deep proteomic network analysis of Alzheimer’s disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Mol Neurodegener. 2018, 13. [Google Scholar] [CrossRef]
- Xu, J.; Patassini, S.; Rustogi, N.; Riba-Garcia, I.; Hale, B.D.; Phillips, A.M.; et al. Regional protein expression in human Alzheimer’s brain correlates with disease severity. Commun Biol. 2019, 2. [Google Scholar] [CrossRef]
- Yeung, J.H.Y.; Walby, J.L.; Palpagama, T.H.; Turner, C.; Waldvogel, H.J.; Faull, R.L.M.; et al. Glutamatergic receptor expression changes in the Alzheimer’s disease hippocampus and entorhinal cortex. Brain Pathol. 2021, 31, e13005. [Google Scholar] [CrossRef]
- Stepler, K.E.; Mahoney, E.R.; Kofler, J.; Hohman, T.J.; Lopez, O.L.; Robinson, R.A.S. Inclusion of African American/Black adults in a pilot brain proteomics study of Alzheimer’s disease. Neurobiol Dis. 2020, 146, 105129. [Google Scholar] [CrossRef] [PubMed]
- Sweet, R.A.; MacDonald, M.L.; Kirkwood, C.M.; Ding, Y.; Schempf, T.; Jones-Laughner, J.; et al. Apolipoprotein E∗4 (APOE∗4) genotype is associated with altered levels of glutamate signaling proteins and synaptic coexpression networks in the prefrontal cortex in mild to moderate Alzheimer disease. Mol Cell Proteomics. 2016, 15, 2252–2262. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Carter, E.K.; Dammer, E.B.; Duong, D.M.; Gerasimov, E.S.; Liu, Y.; et al. Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat Neurosci. 2022, 25, 213–225. [Google Scholar] [CrossRef]
- Lu, W.; Roche, K.W. Posttranslational regulation of AMPA receptor trafficking and function. Curr Opin Neurobiol. 2012, 22, 470–479. [Google Scholar] [CrossRef]
- Roche, K.W.; O’Brien, R.J.; Mammen, A.L.; Bernhardt, J.; Huganir, R.L. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996, 16, 1179–1188. [Google Scholar] [CrossRef]
- Derkach, V.; Barria, A.; Soderling, T.R. Ca2+/calmodulin-kinase II enhances channel conductance of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc Natl Acad Sci U S A. 1999, 96, 3269–3274. [Google Scholar] [CrossRef] [PubMed]
- Banke, T.G.; Bowie, D.; Lee, H.K.; Huganir, R.L.; Schousboe, A.; Traynelis, S.F. Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J Neurosci. 2000, 20, 89–102. [Google Scholar] [CrossRef]
- Fox, C.J.; Russell, K.; Titterness, A.K.; Yu, T.W.; Christie, B.R. Tyrosine phosphorylation of the GluR2 subunit is required for long-term depression of synaptic efficacy in young animals in vivo. Hippocampus. 2007, 17, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, G.; Ju, W.; Liu, L.; Wyszynski, M.; Lee, S.H.; Dunah, A.W.; et al. Tyrosine phosphorylation of GluR2 is required for insulin-stimulated AMPA receptor endocytosis and LTD. EMBO J. 2004, 23, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu Rev Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Chai, Y.J.; Ridder, M.C.; Chau, Y.Q.; Johnson, R.C.; Sah, P.; et al. Activity-Dependent ubiquitination of GluA1 and GluA2 regulates AMPA receptor intracellular sorting and degradation. Cell Rep. 2015, 10, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, L.A.; Hall, B.J.; Patrick, G.N. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010, 30, 16718–16729. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Guntupalli, S.; Jang, S.E.; Anggono, V. Regulation of AMPA receptor trafficking by protein ubiquitination. Front Mol Neurosci. 2017, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hou, Q.; Jarzylo, L.; Amato, S.; Gilbert, J.; Shang, F.; et al. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem. 2011, 119, 27–39. [Google Scholar] [CrossRef]
- Huo, Y.; Khatri, N.; Hou, Q.; Gilbert, J.; Wang, G.; Man, H.Y. The deubiquitinating enzyme USP46 regulates AMPA receptor ubiquitination and trafficking. J Neurochem. 2015, 134, 1067–1080. [Google Scholar] [CrossRef]
- Lussier, M.P.; Herring, B.E.; Nasu-Nishimura, Y.; Neutzner, A.; Karbowski, M.; Youle, R.J.; et al. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc Natl Acad Sci U S A. 2012, 109, 19426–19431. [Google Scholar] [CrossRef]
- Hayashi, T.; Rumbaugh, G.; Huganir, R.L. Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron. 2005, 47, 709–723. [Google Scholar] [CrossRef]
- Lin, D.T.; Makino, Y.; Sharma, K.; Hayashi, T.; Neve, R.; Takamiya, K.; et al. Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci. 2009, 12, 879–887. [Google Scholar] [CrossRef]
- Itoh, M.; Yamashita, M.; Kaneko, M.; Okuno, H.; Abe, M.; Yamazaki, M.; et al. Deficiency of AMPAR–palmitoylation aggravates seizure susceptibility. J Neurosci. 2018, 38, 10220–10235. [Google Scholar] [CrossRef]
- Baudry, M.; Massicotte, G.; Hauge, S. Phosphatidylserine increases the affinity of the AMPA/quisqualate receptor in rat brain membranes. Behav Neural Biol. 1991, 55, 137–140. [Google Scholar] [CrossRef]
- Frank, C.; Rufini, S.; Tancredi, V.; Forcina, R.; Grossi, D.; D’Arcangelo, G. Cholesterol depletion inhibits synaptic transmission and synaptic plasticity in rat hippocampus. Exp Neurol. 2008, 212, 407–414. [Google Scholar] [CrossRef]
- Korinek, M.; Gonzalez-Gonzalez, I.M.; Smejkalova, T.; Hajdukovic, D.; Skrenkova, K.; Krusek, J.; et al. Cholesterol modulates presynaptic and postsynaptic properties of excitatory synaptic transmission. Sci Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Korinek, M.; Vyklicky, V.; Borovska, J.; Lichnerova, K.; Kaniakova, M.; Krausova, B.; et al. Cholesterol modulates open probability and desensitization of NMDA receptors. J Physiol. 2015, 593, 2279–2293. [Google Scholar] [CrossRef] [PubMed]
- Antonini, A.; Caioli, S.; Saba, L.; Vindigni, G.; Biocca, S.; Canu, N.; et al. Membrane cholesterol depletion in cortical neurons highlights altered NMDA receptor functionality in a mouse model of amyotrophic lateral sclerosis. Biochim Biophys Acta - Mol Basis Dis. 2018, 1864, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mejia, R.O.; Newman, J.W.; Toh, S.; Yu, G.-Q; Zhou, Y.; Halabisky, B.; et al. . Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer’s disease. Nat Neurosci. 2008, 11, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J.; Chabot, C.; Gagné, J.; Baudry, M.; Massicotte, G. Melittin increases AMPA receptor affinity in rat brain synaptoneurosomes. Brain Res. 1995, 671, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Ménard, C.; Patenaude, C.; Massicotte, G. Phosphorylation of AMPA receptor subunits is differentially regulated by phospholipase A2 inhibitors. Neurosci Lett. 2005, 389, 51–56. [Google Scholar] [CrossRef]
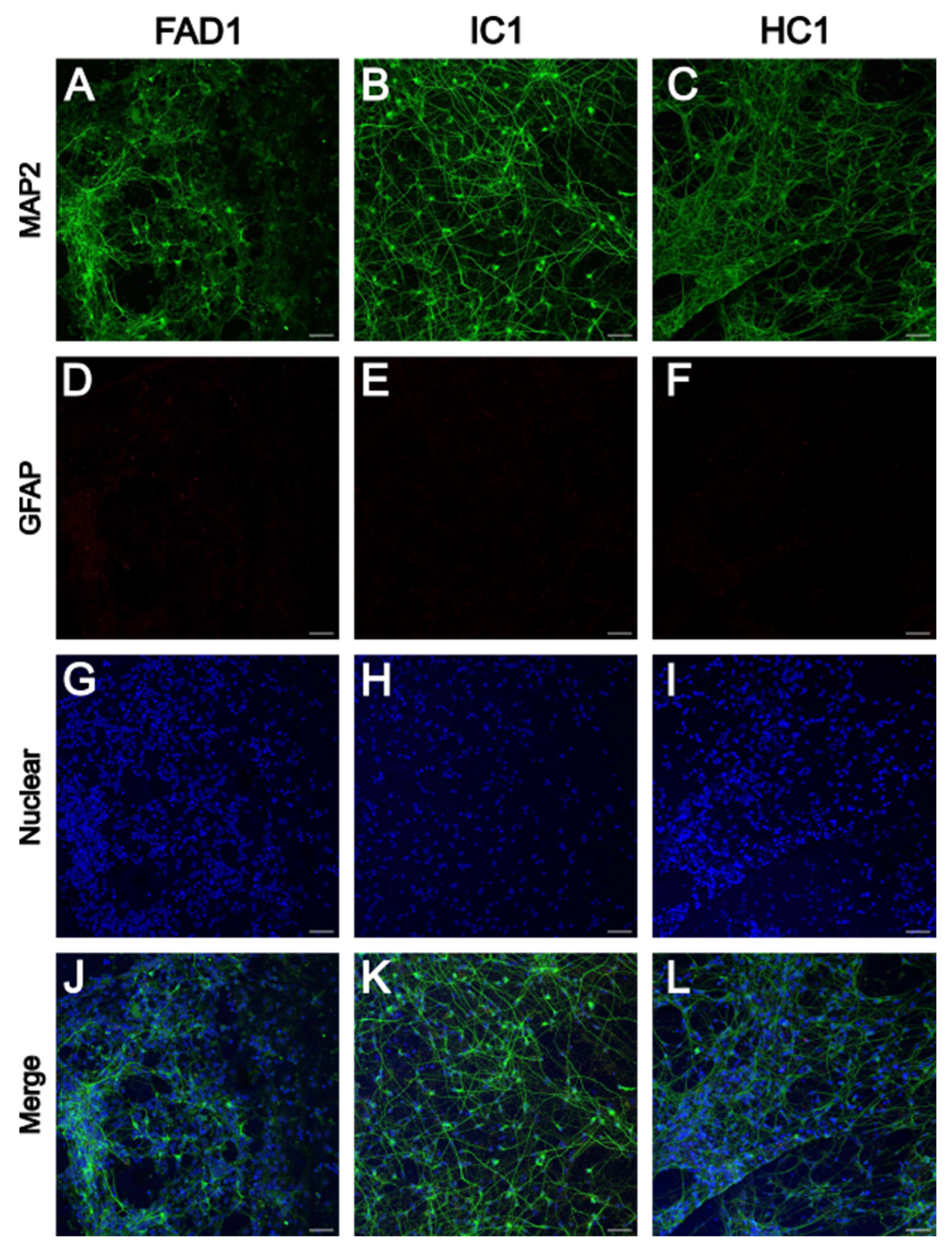
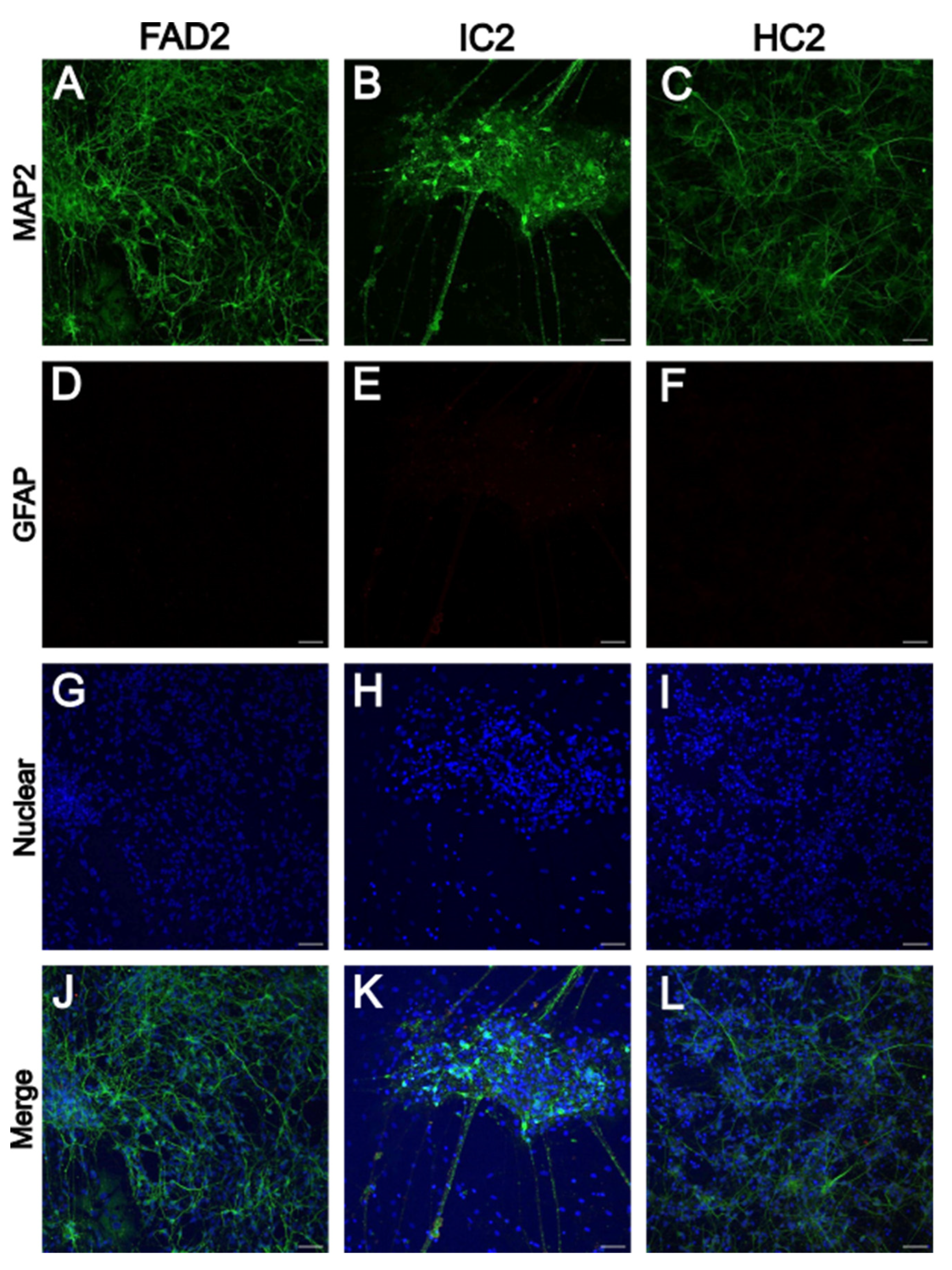
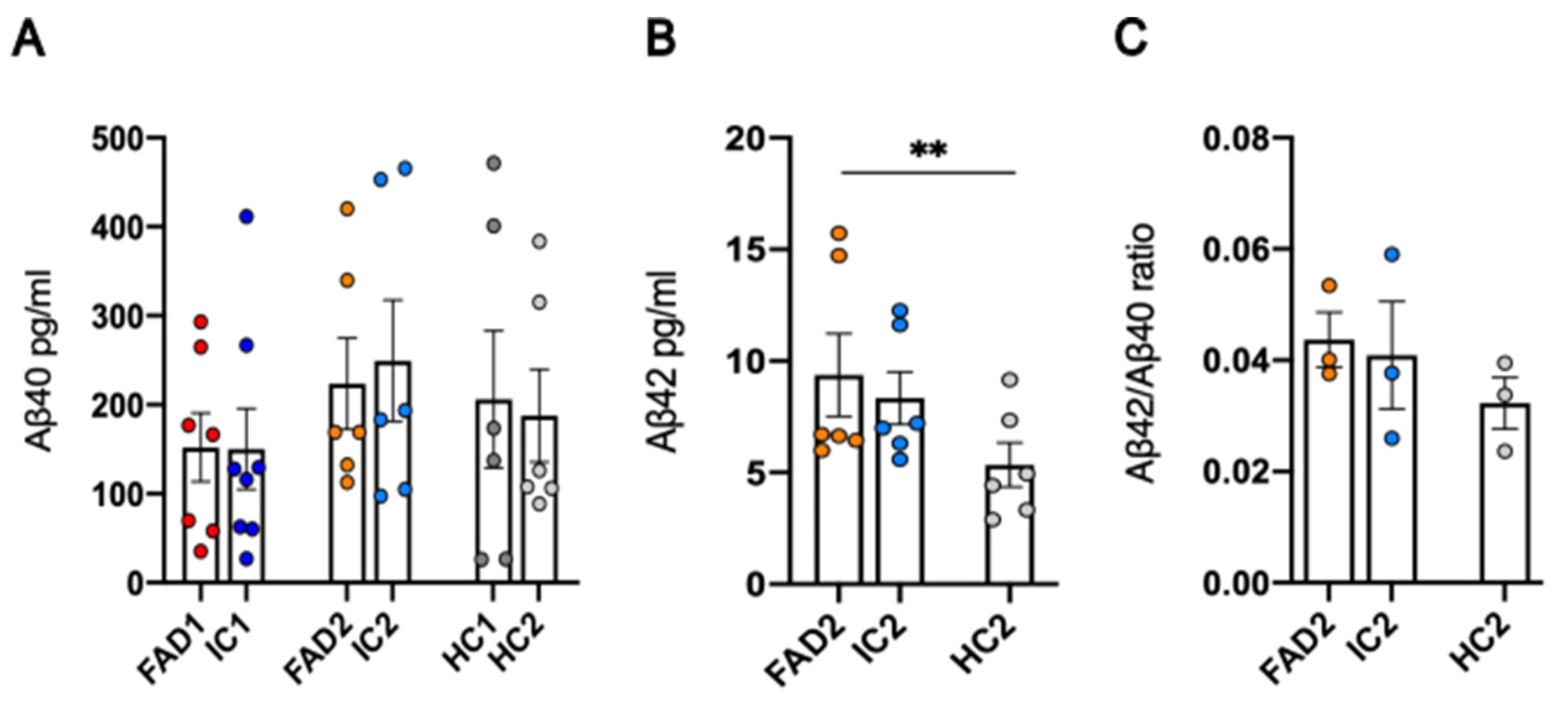
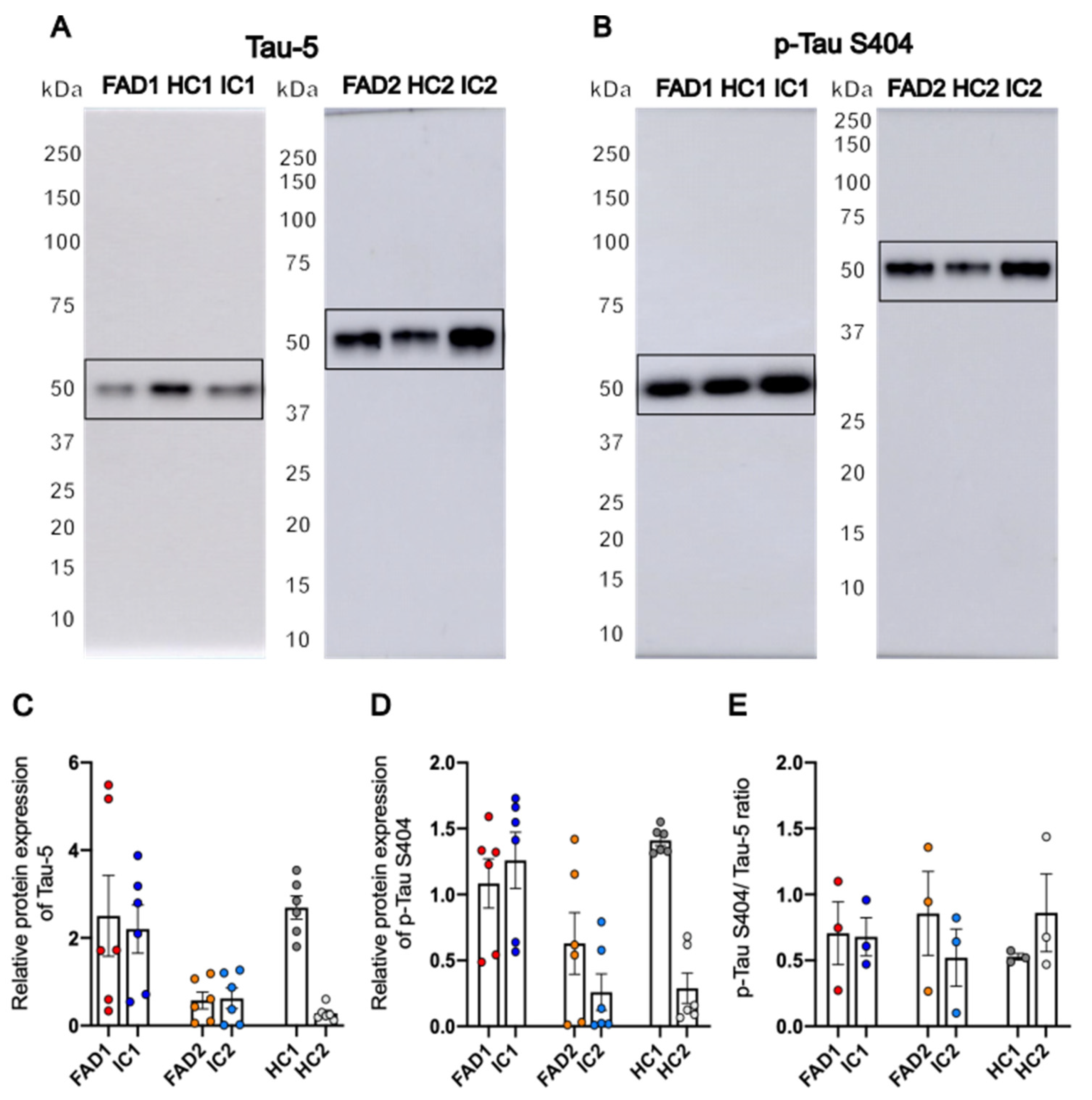


| iPSC name | Disease status | WT/mutation | APOE genotype | Age at collection | Sex | iPSC line characterisation |
|---|---|---|---|---|---|---|
| FAD1 | Familial AD | PSEN1S290C | Ɛ3/3 | 47 | M | |
| IC1 | Isogenic control | PSEN1WT | Ɛ3/3 | 47 | M | |
| HC1 | Healthy control | PSEN1WT | Ɛ2/4 | 57 | F | (Engel et al., 2018) |
| FAD2 | Familial AD | PSEN1A246E | Ɛ3/4 | 56 | F | (Muñoz et al., 2018) |
| IC2 | Isogenic control | PSEN1WT | Ɛ3/4 | 56 | F | |
| HC2 | Healthy control | PSEN1WT | Ɛ2/3 | 75 | F | (Muñoz et al., 2018) |
| Target | Sequence | Tm |
|---|---|---|
| B2M | F: AAGGACTGGTCTTTCTATCTC R: GATCCCACTTAACTATCTTGG |
55 °C |
| GAPDH | F: GAGCACAAGAGGAAGAGAGAGACCC R: GTTGAGCACAGGGTACTTTATTGATGGTACATG |
58 °C |
| GRIA1 | F: CTAGAAGATCCTTATGTGATGC R: CTCCGTATTTTCCATCACTG |
58 °C |
| GRIA2 | F: GGAATCTCCGTATGTTATGATG R: TTGTACTTGAACCCACAATG |
55 °C |
| GRIA3 | F: TATTGTATCTGGGGCGTTAC R: TTGAGAACTCAAGAAGGGAG |
55 °C |
| GRIA4 | F: GGTACGATAAAGGTGAATGTG R: AAAAGGTCAGCTTCATTCTC |
58 °C |
| GRIN1 | F: CGACCCTTACTTTTGAGAAC R: AGGCAATCTCGATGAAAATC |
55 °C |
| GRIN2A | F: AACAATTCAACCAATGAGGG R: CAGATAGAGGTCGTAAGTAAAC |
58 °C |
| GRIN2B | F: AGTTCCAGAGACCTAATGAC R: GAGCAATGCATCATCTACAC |
55 °C |
| GRIK1 | F: AGGTCTAATTCGTCTACAAGAG R: TATCACATAGAACTCCTTGCC |
58 °C |
| GRIK2 | F: GAAAAGAGAGCCAAGACTAAG R: AAGATGGTGATGATGACAAC |
58 °C |
| GRIK3 | F: CAGATACAAGMATGAGCCC R: TTTTTACTCCAGGGCAAATC |
63 °C |
| GRIK4 | F: GGAAGATCACAGAGCTAAAG R: CTGAGAGTCCATAAAAACTCC |
58 °C |
| GRIK5 | F: ATATCTGTGGAGAGAAGGAG R: GTTGAAGGACTTGAGGATTC |
58 °C |
| HPRT1 |
F: TGACACTGGCAAAACAATGCA R: GGTCCTTTTCACCAGCAAGCT |
58 °C |
| POU5F1 | F: GATCACCCTGGGATATACAC R: GCTTTGCATATCTCCTGAAG |
58 °C |
| NANOG |
F: CCAGAACCAGAGAATGAAATC R: TGGTGGTAGGAAGAGTAAAG |
58 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





