Submitted:
22 April 2024
Posted:
23 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Mass Spectrometry Analysis
2.2. Cell Culture and Transfections
2.3. Antibodies
2.4. Plasmids
2.5. Fusion Protein Purification and Binding Assays
2.6. Infectivity Assays
Pseudovirions Quantification
2.7. Synthetic HPV Particle Production and Labeling
2.8. Cell Viability Assay
3. Results
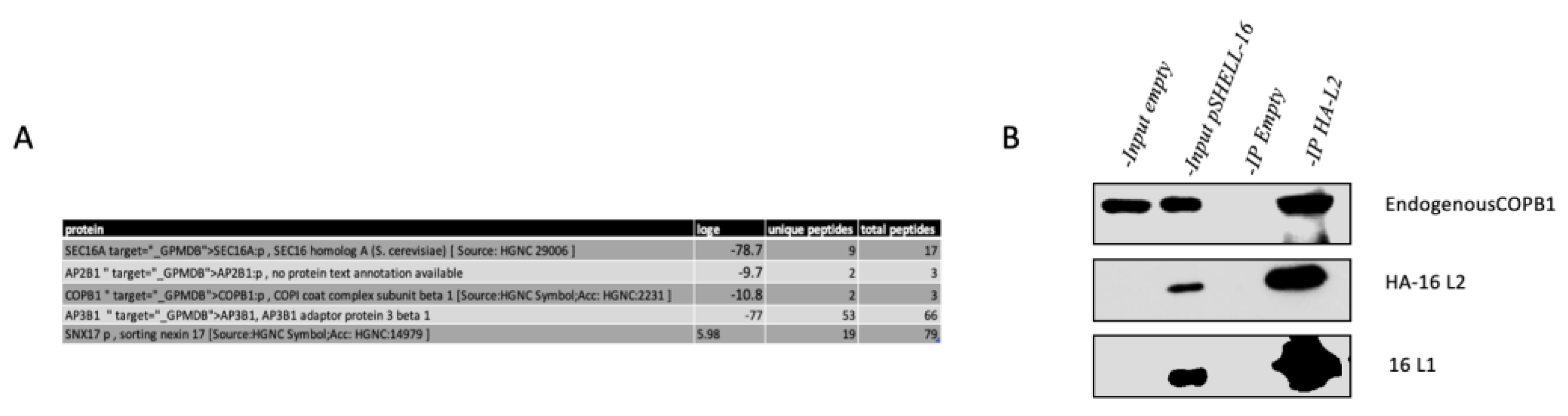
3.1. Mass Spectrometry Assay Identifies COPB1 Cargo Protein as an HPV-16 Interacting Protein
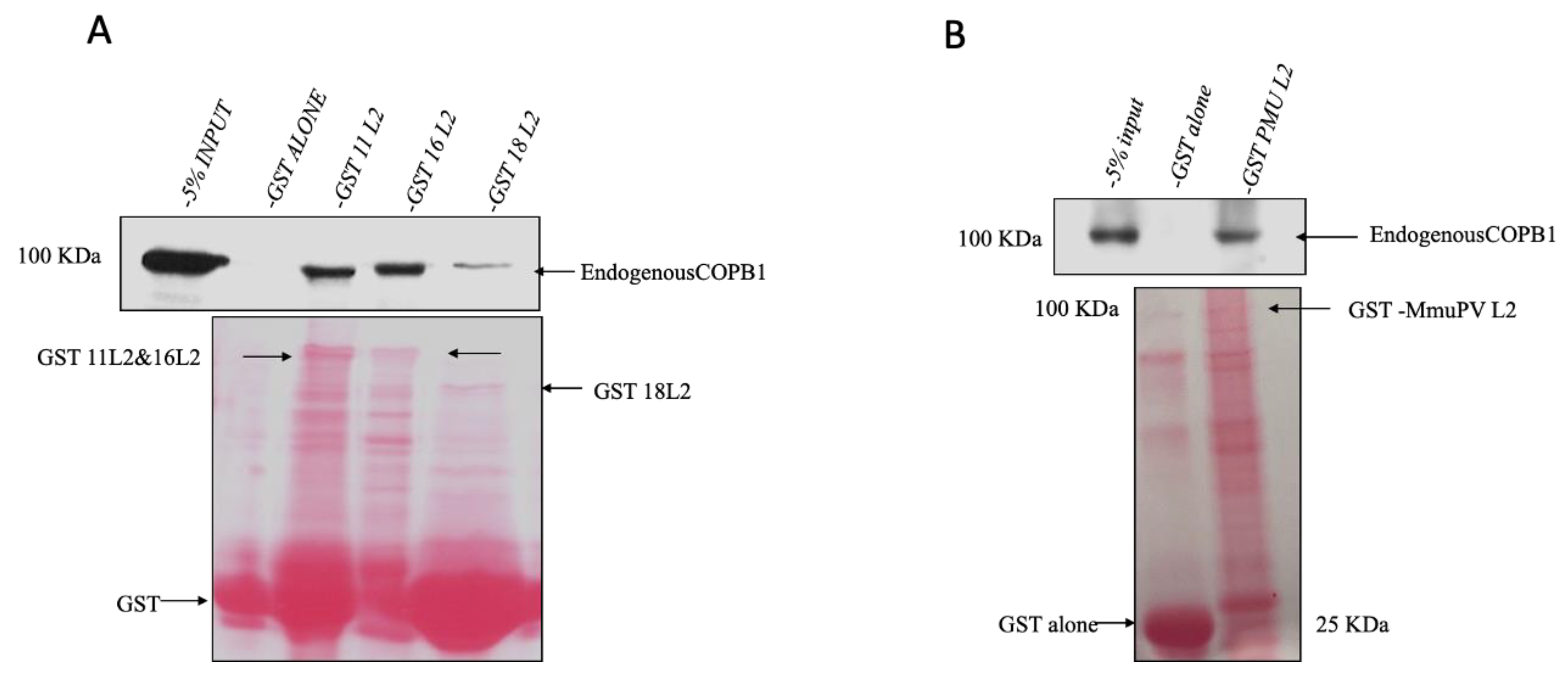
3.2. GST-L2 Fusion Protein Binds COPB1 In Vitro
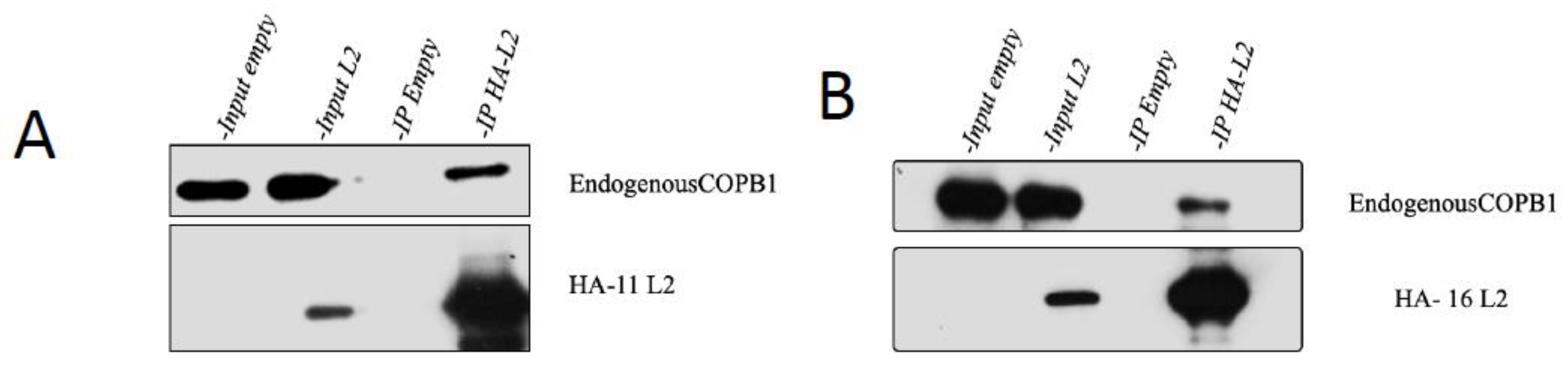
3.3. HPV-16 and HPV-11 L2 Interact with COPB1 In Vivo
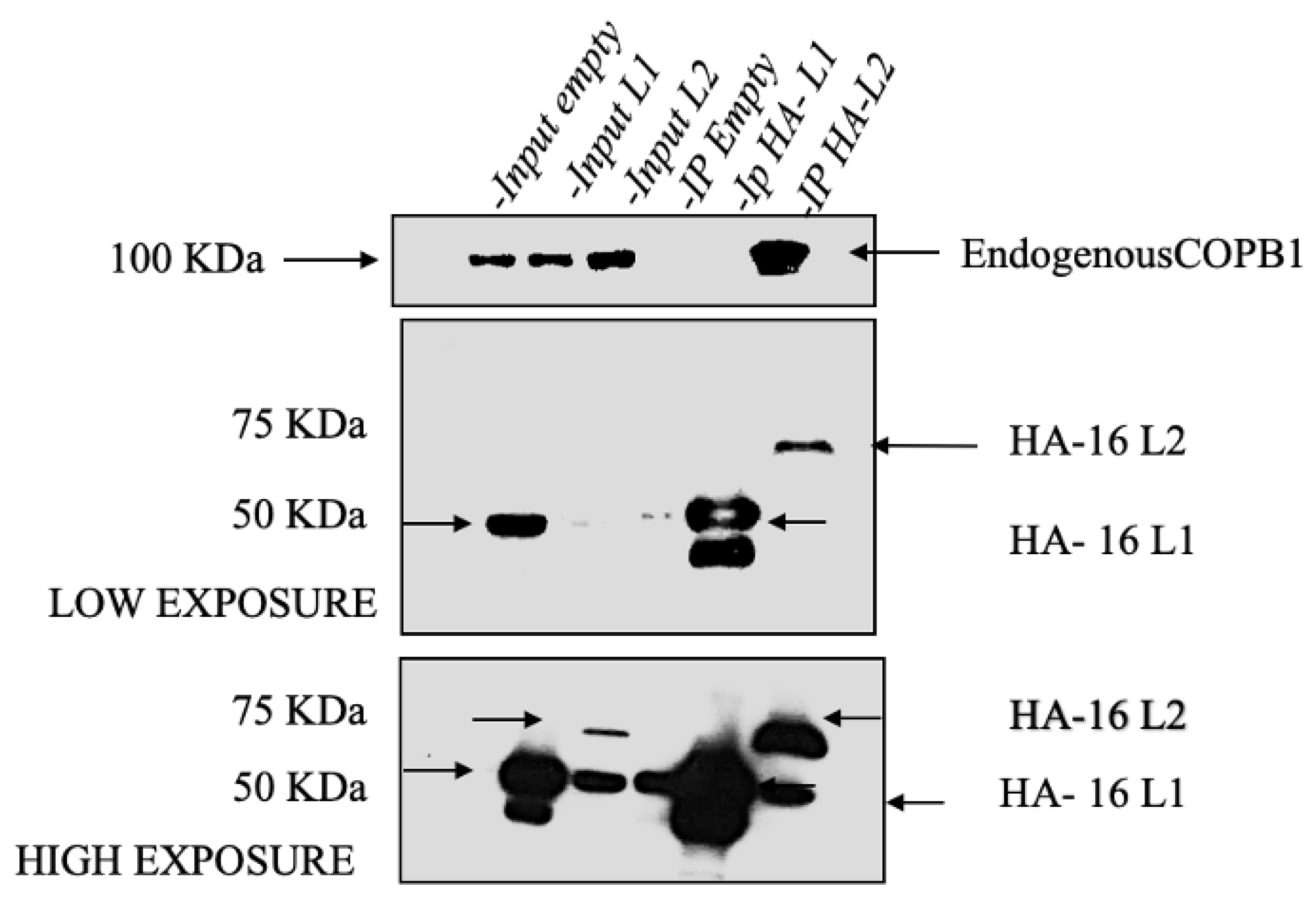
3.4. HPV-16 L2, but Not L1, Binds to COPB1
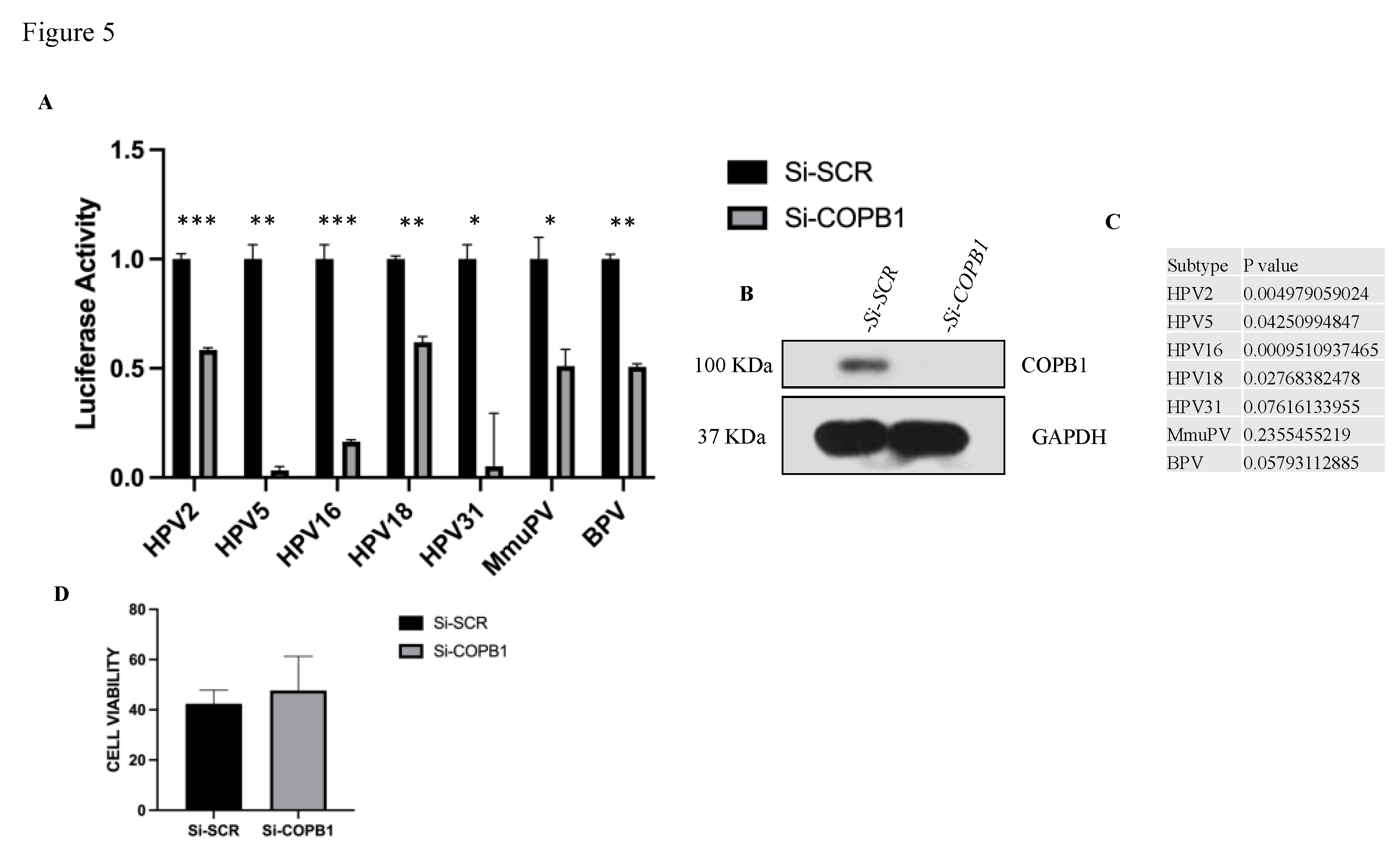
3.5. HPV Infection Requires COPB1
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schelhaas M, Shah B, Holzer M, Blattmann P, Kühling L, Day PM, Schiller JT, Helenius A. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8[4]:e1002657. Epub 2012 Apr 19. [CrossRef] [PubMed]
- Marušič MB, Mencin N, Ličen M, Banks L, Grm HŠ. 2010. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J Virol 84:11585–11589.
- Szymonowicz KA, Chen J. Biological and clinical aspects of HPV-related cancers. Cancer Biol Med. 2020 Nov 15;17[4]:864-878. Epub 2020 Dec 15. [CrossRef] [PubMed]
- Marur, Shanthi, Gypsyamber D’Souza, William H Westra, and Arlene A Forastiere. ‘HPV-Associated Head and Neck Cancer: A Virus-Related Cancer Epidemic’. The Lancet Oncology 11, no. 8 [August 2010]: 781–89. [CrossRef]
- Aksoy P, Gottschalk EY, Meneses PI. HPV entry into cells. Mutat Res Rev Mutat Res. 2017 Apr-Jun;772:13-22. [CrossRef] [PubMed]
- Chan CK, Aimagambetova G, Ukybassova T, Kongrtay K, Azizan A. Human Papillomavirus Infection and Cervical Cancer: Epidemiology, Screening, and Vaccination-Review of Current Perspectives. J Oncol. 2019 Oct 10;2019:3257939. [CrossRef] [PubMed]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004 Mar 29;164[7]:1065-76. [CrossRef] [PubMed]
- Xie J, Zhang P, Crite M, Lindsay CV, DiMaio D. Retromer stabilizes transient membrane insertion of L2 capsid protein during retrograde entry of human papillomavirus. Sci Adv. 2021 Jun 30;7[27]:eabh4276. [CrossRef] [PubMed]
- Skoulakis, A., Fountas, S., Mantzana-Peteinelli, M. et al. Prevalence of human papillomavirus and subtype distribution in male partners of women with cervical intraepithelial neoplasia [CIN]: a systematic review. BMC Infect Dis 19, 192 [2019]. [CrossRef]
- Thomas M, Massimi P, Jenkins J, Banks L. 1995. HPV-18 E6 mediated inhibition of p53 DNA binding activity is independent of E6 induced degradation. Oncogene 10:261–268.
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462.
- Keiffer TR, Soorya S, Sapp MJ. Recent Advances in Our Understanding of the Infectious Entry Pathway of Human Papillomavirus Type 16. Microorganisms. 2021 Oct 1;9(10):2076. [CrossRef] [PubMed]
- Bazan SB, de Alencar Muniz Chaves A, Aires KA, Cianciarullo AM, Garcea RL, Ho PL. Expression and characterization of HPV-16 L1 capsid protein in Pichia pastoris. Arch Virol. 2009;154(10):1609-17. Epub 2009 Sep 10. [CrossRef] [PubMed]
- Darshan MS, Lucchi J, Harding E, Moroianu J. The l2 minor capsid protein of human papillomavirus type 16 interacts with a network of nuclear import receptors. J Virol. 2004 Nov;78(22):12179-88. [CrossRef] [PubMed]
- Tsakogiannis D, Nikolaidis M, Zagouri F, Zografos E, Kottaridi C, Kyriakopoulou Z, Tzioga L, Markoulatos P, Amoutzias GD, Bletsa G. Mutation Profile of HPV16 L1 and L2 Genes in Different Geographic Areas. Viruses. 2022 Dec 31;15[1]:141. [CrossRef] [PubMed]
- Skoulakis, A., Fountas, S., Mantzana-Peteinelli, M. et al. Prevalence of human papillomavirus and subtype distribution in male partners of women with cervical intraepithelial neoplasia [CIN]: a systematic review. BMC Infect Dis 19, 192 [2019]. [CrossRef]
- Park SY, Guo X. Adaptor protein complexes and intracellular transport. Biosci Rep. 2014 Jul 29;34[4]:e00123. [CrossRef] [PubMed]
- Siddiqa A, Broniarczyk J, Banks L. ‘Papillomaviruses and Endocytic Trafficking’. International Journal of Molecular Sciences 19, no. 9 [September 2018]: 2619. [CrossRef]
- Morante AV, Baboolal DD, Simon X, Pan EC, Meneses PI. Human Papillomavirus Minor Capsid Protein L2 Mediates Intracellular Trafficking into and Passage beyond the Endoplasmic Reticulum. Microbiol Spectr. 2022 Jun 29;10[3]:e0150522. Epub 2022 May 24. [CrossRef] [PubMed]
- Calton CM, Bronnimann MP, Manson AR, Li S, Chapman JA, Suarez-Berumen M, Williamson TR, Molugu SK, Bernal RA, Campos SK. Translocation of the papillomavirus L2/vDNA complex across the limiting membrane requires the onset of mitosis. PLoS Pathog. 2017 May 2;13[5]: e1006200. [CrossRef] [PubMed]
- Broniarczyk J, Ring N, Massimi P, Giacca M, Banks L. HPV-16 virions can remain infectious for 2 weeks on senescent cells but require cell cycle re-activation to allow virus entry. Sci Rep. 2018 Jan 16;8[1]:811. [CrossRef] [PubMed]
- Graham FL, Van der Eb A. 1973. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology 52:456–467.
- Siddiqa A, Broniarczyk J, Banks L. Papillomaviruses and Endocytic Trafficking. Int J Mol Sci. 2018 Sep 4;19[9]:2619. [CrossRef] [PubMed]
- DiGiuseppe S, Bienkowska-Haba M, Guion LGM, Keiffer TR, Sapp M. Human Papillomavirus Major Capsid Protein L1 Remains Associated with the Incoming Viral Genome throughout the Entry Process. J Virol. 2017 Jul 27;91[16]:e00537-17. [CrossRef] [PubMed]
- Broniarczyk J, Massimi P, Pim D, Bergant Marušič M, Myers MP, Garcea RL, Banks L. Phosphorylation of Human Papillomavirus Type 16 L2 Contributes to Efficient Virus Infectious Entry. J Virol. 2019 Jun 14;93[13]:e00128-19. [CrossRef] [PubMed]
- DiGiuseppe S, Keiffer TR, Bienkowska-Haba M, Luszczek W, Guion LG, Müller M, Sapp M. Topography of the Human Papillomavirus Minor Capsid Protein L2 during Vesicular Trafficking of Infectious Entry. J Virol. 2015 Oct;89[20]:10442-52. Epub 2015 Aug 5. [CrossRef] [PubMed]
- Broniarczyk J, Massimi P, Bergant M, Banks L. Human Papillomavirus Infectious Entry and Trafficking Is a Rapid Process. J Virol. 2015 Sep;89[17]:8727-32. Epub 2015 Jun 10. [CrossRef] [PubMed]
- Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol. 2013 Apr;87[7]:3862-70. Epub 2013 Jan 23. [CrossRef] [PubMed]
- Siddiqa A, Massimi P, Pim D, Broniarczyk J, Banks L. 2018. Human papillomavirus 16 infection induces VAP-dependent endosomal tubulation. J Virol 92:1–14.
- Day PM, Weisberg AS, Thompson CD, Hughes MM, Pang YY, Lowy DR, Schiller JT. Human Papillomavirus 16 Capsids Mediate Nuclear Entry during Infection. J Virol. 2019 Jul 17;93[15]:e00454-19. [CrossRef] [PubMed]
- Bergant M, Banks L. SNX17 facilitates infection with diverse papillomavirus types. J Virol. 2013 Jan;87[2]:1270-3. Epub 2012 Oct 31. [CrossRef] [PubMed]
- Bugnon Valdano M, Massimi P, Broniarczyk J, Pim D, Myers M, Gardiol D, Banks L. Human Papillomavirus infection requires the CCT Chaperonin Complex. J Virol. 2021 ;95[11]:e01943-20. Epub 2021 Mar 17. [CrossRef] [PubMed]
- Pim D, Broniarczyk J, Siddiqa A, Massimi P, Banks L. Human Papillomavirus 16 L2 Recruits both Retromer and Retriever Complexes during Retrograde Trafficking of the Viral Genome to the Cell Nucleus. J Virol. 2021 Jan 13;95[3]:e02068-20. [CrossRef] [PubMed]
- Duden, R. ER-to-Golgi transport: COP I and COP II function [Review]. Mol Membr Biol. 2003 Jul-Sep;20[3]:197-207. [CrossRef] [PubMed]
- McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004 Aug;16[4]:379-91. [CrossRef] [PubMed]
- Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009 Sep 3;583[17]:2701-9. Epub 2009 Jul 22. Erratum in: FEBS Lett. 2009 Nov 3;583[21]:3541. Ravet, M [corrected to Rawet, M]. [CrossRef] [PubMed]
- Matsuoka, K., Morimitsu, Y., Uchida, K., & Schekman, R. [1998]. Coat Assembly Directs v-SNARE Concentration into Synthetic COPII Vesicles. Molecular Cell, 2[5], 703-708. [CrossRef]
- Szul T, Sztul E. ‘COPII and COPI Traffic at the ER-Golgi Interface’. Physiology 26, no. 5 [October 2011]: 348–64. [CrossRef]
- Schoppe J, Schubert E, Apelbaum A, Yavavli E, Birkholz O, Stephanowitz H, Han Y, Perz A, Hofnagel O, Liu F, Piehler J, Raunser S, Ungermann C. Flexible open conformation of the AP-3 complex explains its role in cargo recruitment at the Golgi. J Biol Chem. 2021 Nov;297[5]:101334. Epub 2021 Oct 22. [CrossRef] [PubMed]
- 40. Golgi Apparatus. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26941/.
- Peotter J, Kasberg W, Pustova I, Audhya A. COPII-mediated trafficking at the ER/ERGIC interface. Traffic. 2019 Jul;20[7]:491-503. Epub 2019 May 30. [CrossRef] [PubMed]
- Gomez-Navarro N, and Miller EA. ‘COP-Coated Vesicles’. Current Biology 26, no. 2 [25 January 2016]: R54–57. [CrossRef]
- Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms JB, Wieland FT. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J Cell Biol. 1996 Dec;135[5]:1239-48. [CrossRef] [PubMed]
- Pérez-Pulido AJ, Asencio-Cortés G, Brokate-Llanos AM, Brea-Calvo G, Rodríguez-Griñolo R, Garzón A, Muñoz MJ. Serial co-expression analysis of host factors from SARS-CoV viruses highly converges with former high-throughput screenings and proposes key regulators. Brief Bioinform. 2021 Mar 22;22(2):1038-1052. [CrossRef] [PubMed]
- Tiwari R, Mishra AR, Gupta A, Nayak D. Structural similarity-based prediction of host factors associated with SARS-CoV-2 infection and pathogenesis. J Biomol Struct Dyn. 2022 Aug;40(13):5868-5879. Epub 2021 Jan 28. [CrossRef] [PubMed]
- Fuller J, Álvarez-Rodríguez B, Todd EJAA, Mankouri J, Hewson R, Barr JN. Hazara Nairovirus Requires COPI Components in both Arf1-Dependent and Arf1-Independent Stages of Its Replication Cycle. J Virol. 2020 Aug 17;94(17):e00766-20. [CrossRef] [PubMed]
- Drake MT, Zhu Y, Kornfeld S. The assembly of AP-3 adaptor complex-containing clathrin-coated vesicles on synthetic liposomes. Mol Biol Cell. 2000 Nov;11[11]:3723-36. [CrossRef] [PubMed]
- Duden R. ER-to-Golgi transport: COP I and COP II function [Review]. Mol Membr Biol. 2003 Jul-Sep;20[3]:197-207. [CrossRef] [PubMed]
- Bonifacino, J., Lippincott-Schwartz, J. Coat proteins: shaping membrane transport. Nat Rev Mol Cell Biol 4, 409–414 [2003]. [CrossRef]
- van Vliet, C., Thomas, E. C., Merino-Trigo, A., Teasdale, R. D. & Gleeson, P. A. Intracellular sorting and transport of proteins. Progress in Biophysics and Molecular Biology 83, 1–45 (2003).
- Cui, L., Li, H., Xi, Y. et al. Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy. Mol Biomed 3, 29 (2022). [CrossRef]
- Darsow T, Burd CG, Emr SD. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J Cell Biol. 1998 Aug 24;142[4]:913-22. [CrossRef] [PubMed]
- Broniarczyk J, Massimi P, Bergant M, Banks L. Human Papillomavirus Infectious Entry and Trafficking Is a Rapid Process. J Virol. 2015 Sep;89[17]:8727-32. Epub 2015 Jun 10. [CrossRef] [PubMed]
- Bergant Marušič M, Ozbun MA, Campos SK, Myers MP, Banks L. 2012. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic 13:455–467.
- Zhang W, Kazakov T, Popa A, DiMaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires γ-secretase activity. mBio 5:e01777-14.
- Siddiqa A, Broniarczyk J, Banks L. ‘Papillomaviruses and Endocytic Trafficking’. International Journal of Molecular Sciences 19, no. 9 [September 2018]: 2619. [CrossRef]
- Fuller J, Álvarez-Rodríguez B, Todd EJAA, Mankouri J, Hewson R, Barr JN. Hazara Nairovirus Requires COPI Components in both Arf1-Dependent and Arf1-Independent Stages of Its Replication Cycle. J Virol. 2020 Aug 17;94(17):e00766-20. [CrossRef] [PubMed]
- Harwood MC, Woo TT, Takeo Y, DiMaio D, Tsai B..HPV is a cargo for the COPI sorting complex during virus entry.Sci. Adv.9,eadc9830(2023). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



