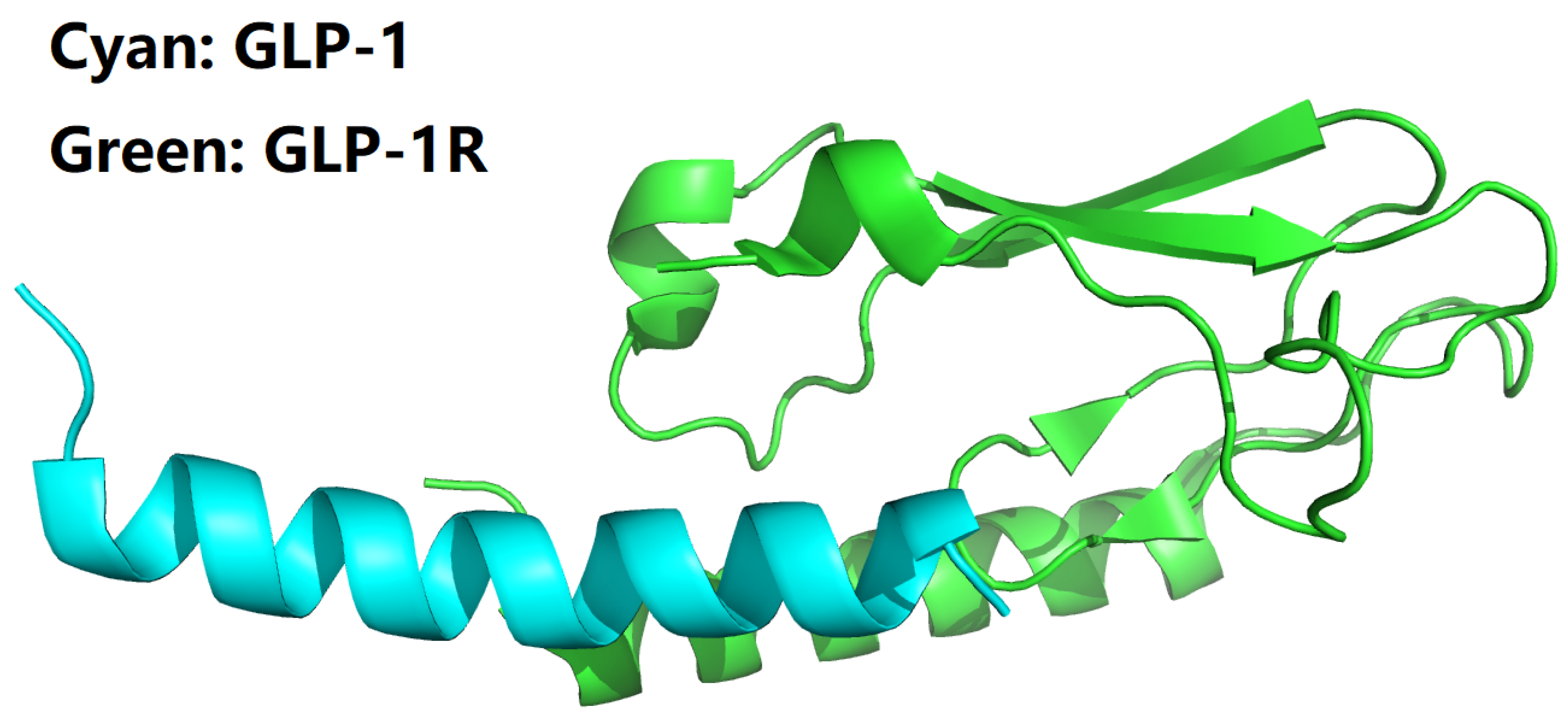
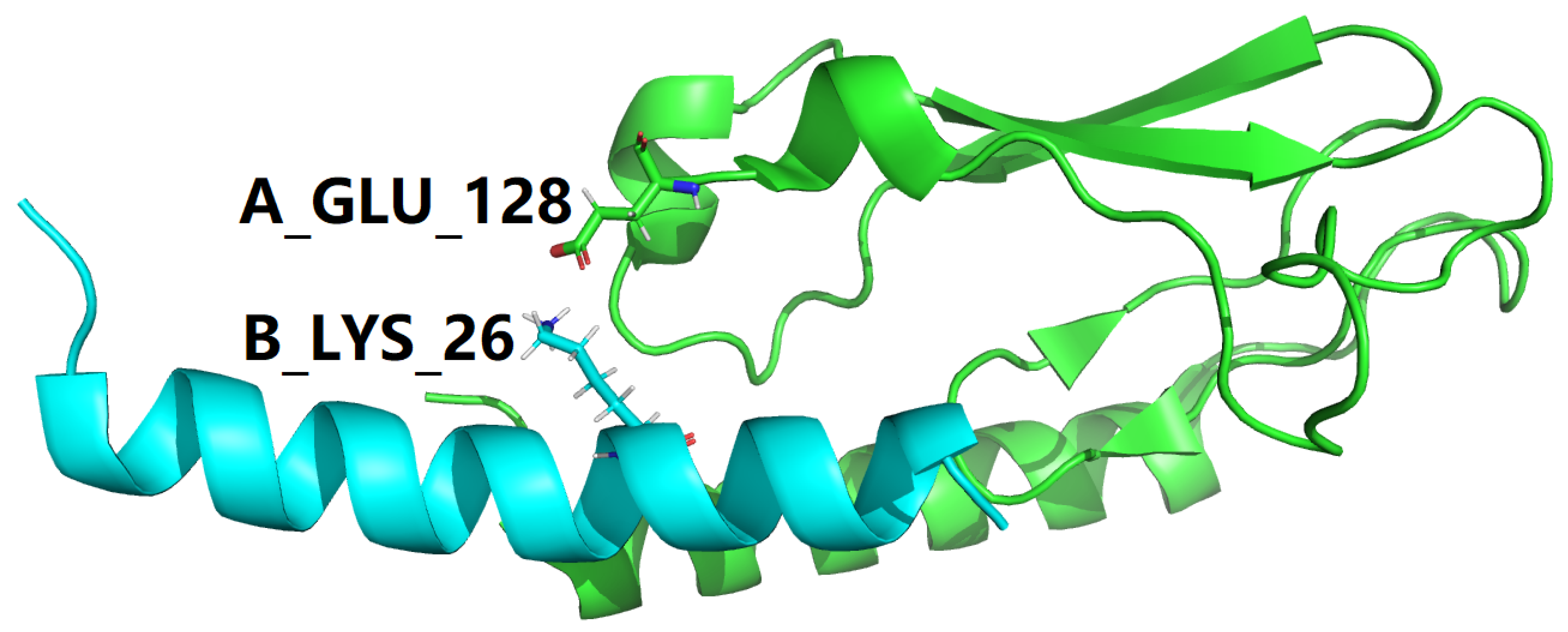
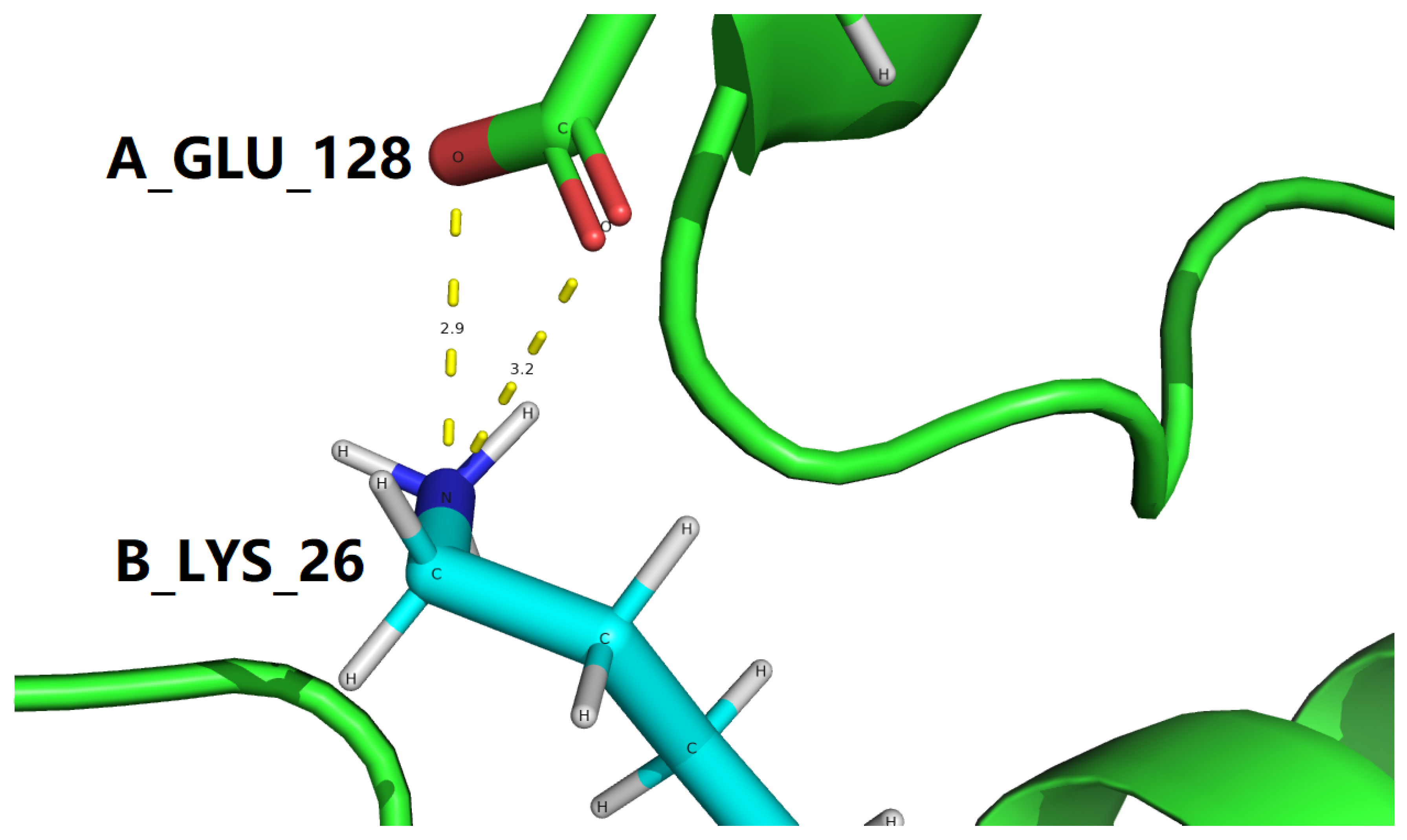
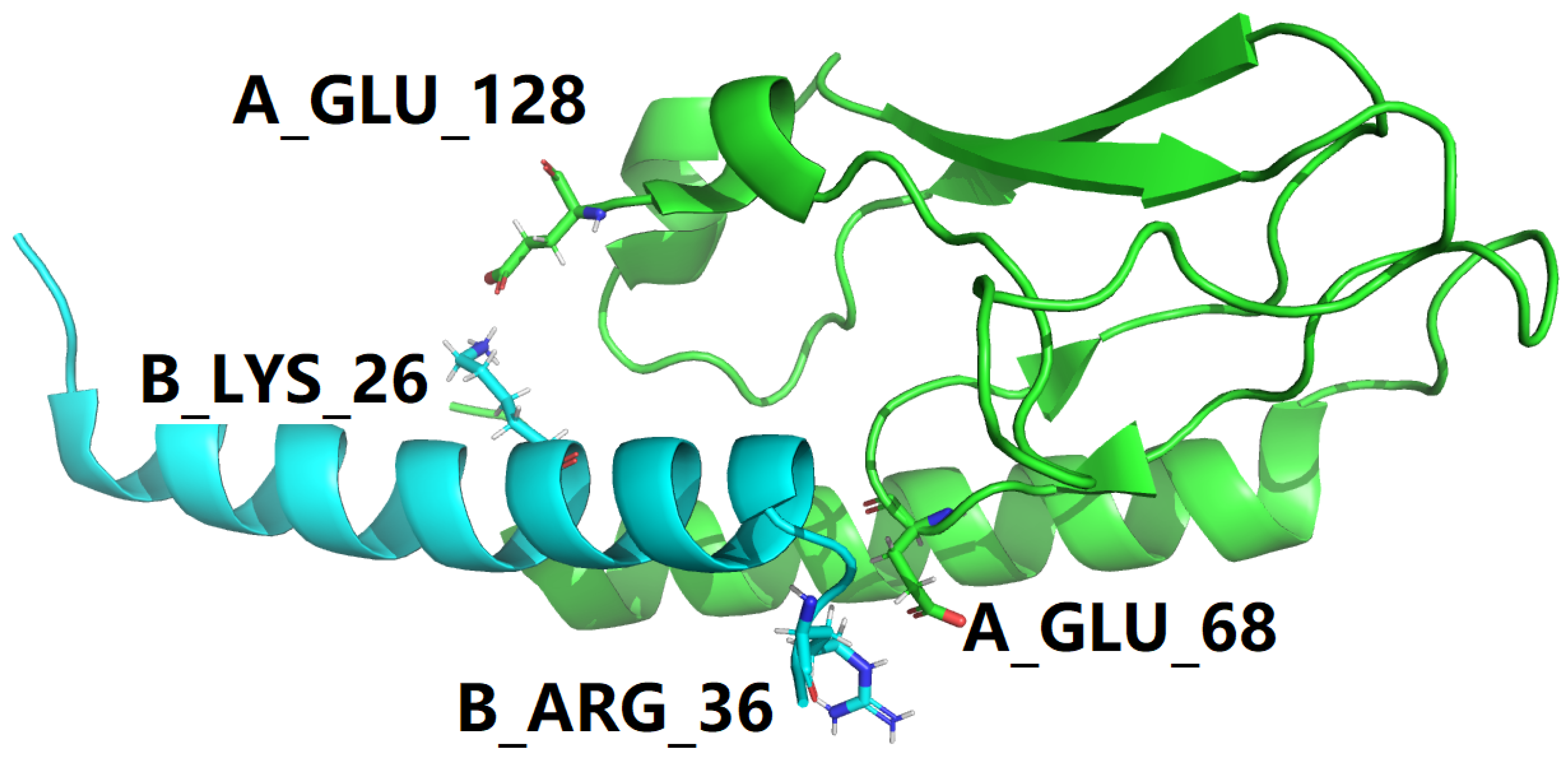
1. Introduction
Direct binding and interaction between ligands and receptors plays a fundamental role in various biological processes, including signal transduction, membrane protein trafficking, cell communication, and drug pharmacology [
1,
2,
3,
4]. Understanding the intricate details of these interactions, particularly the structural features at the binding interface of ligand and receptor, is crucial for unraveling the underlying mechanisms and designing effective therapeutics [
5,
6,
7]. Among the myriad of ligand-receptor systems, the glucagon-like peptide-1 (GLP-1) and its cognate receptor, GLP-1 receptor (GLP-1R), constitute a pivotal axis in metabolic regulation and insulin secretion [
8,
9,
10]. GLP-1, an incretin hormone secreted by enteroendocrine intestinal L-cells, exerts its physiological effects by binding to GLP-1R, a G-protein-coupled receptor predominantly expressed in pancreatic β-cells, among other tissues [
11,
12,
13]. Activation of GLP-1R initiates a signaling cascade leading to enhanced insulin release, inhibition of glucagon secretion, and regulation of satiety, making it an attractive target for the treatment of type 2 diabetes and obesity [
14,
15,
16].
To date, a wide range of experimental structures have been determined for the GLP-1/GLP-1R complex, constituting the structural basis of GLP-1/GLP-1R interactions, particularly the structural electrostatic features at the binding interface [
17,
18,
19]. Here, with a set of computational structural biophysical analyses for currently available experimental GLP-1/GLP-1R complex structures, this article systematically characterized the electrostatic interactions within the GLP-1-GLP-1R complexes with two sets of criteria to identify interfacial salt bridges and hydrogen bonds. By structurally elucidating the electrostatic landscape of GLP-1/GLP-1R interactions, this article aims to provide valuable insights into the molecular basis of ligand recognition and receptor activation, with structural and biophysical implications for rational design and development of therapeutics with improved efficacy and safety targeting the GLP-1/GLP-1R axis.
2. Motivation
Thanks to the continued development of experimental structural biology and the half-a-century old Protein Data Bank (PDB) [
20,
21], a comprehensive structural biophysical (CSB) analysis becomes possible [
22,
23,
24] for specific ligand-receptor [
25,
26,
27], antigen-antibody [
28] or enzyme-substrate [
29,
30,
31] complex structures deposited in PDB, expanding our understanding of the structural and biophysical basis of their interfacial structural stability, and facilitating the design of drug analogues with improved affinity to their interacting partners [
6,
32].
Semaglutide is a human GLP-1 analogue with 94% structural homology with native human GLP-1, with 3 important modifications: an amino acid substitution at position 8 that makes it less susceptible to degradation by dipeptidyl peptidase-4; lysine acylation of the peptide backbone, with a spacer and C-18 fatty di-acid chain at position 26 that provides strong, specific binding to albumin; and another amino acid substitution at position 34, which prevents C=18 fatty di-acid binding at the wrong site [
33]. Interestingly, a semaglutide analogue was for the first time reported with a simple Val27-Arg28 exchange in its peptide backbone in 2021 [
7]. Semaglutide is a glucagon-like peptide 1 analog used for the treatment of patients with type 2 diabetes mellitus. With 94% sequence similarity to human GLP-1, semaglutide is a glucagon-like peptide-1 receptor (GLP-1R) agonist, which binds directly to GLP-1R, causing various beneficial downstream effects that reduce blood glucose. Specifically, the amino acid sequence of GLP-1 is listed in italics in fasta format as below,
>SemaglutideNative
HAEGTFTSDVSSYLEGQAAKEFIAWLVRGRG
The amino acid sequence of the semaglutide analogue with a simple Val27-Arg28 exchange in its peptide backbone is listed in italics in fasta format as below,
>SemaglutideMutant
HAEGTFTSDVSSYLEGQAAKEFIAWLRVGRG
Of further interest, a new pair of structure-stabilizing interfacial salt bridges between Glu104 of chain A (GLP-1R) and Arg27 of chain B (semaglutide) are entirely due to the Val27-Arg28 exchange in the semaglutide peptide backbone, according to the binding affinity of semaglutide and GLP-1R calculated using Prodigy [
34,
35]. Nonetheless, the design of the semaglutide analogue with a simple Val27-Arg28 exchange was on a manual basis of close naked-eye inspection of the GLP-1-GLP-1R binding interface 4ZGM [
19,
36], and hand-picked from a set of analogue candidates of semaglutide to ensure improved ligand-receptor binding affinity [
34,
35] with minimum modification (i.e., a simple Val27-Arg28 exchange) to the backbone of semaglutide. Here, this article employs a high-throughput approach for extraction of electrostatic structural features from GLP-1-GLP-1R complexes towards the construction of a GLP-1-GLP-1R-based mini general intermolecular binding affinity calculator (GIBAC) with adequate accuracy and efficiency [
2]. Overall, the motivation here stems from the pressing need within the drug discovery and design community for efficient methods to assess intermolecular binding affinities, particularly in the context of peptide-receptor interactions. By focusing on GLP-1-GLP-1R complexes, pivotal targets in diabetes and obesity [
37,
38], this article aims to showcase the utility of our miniaturized GIBAC approach in rapidly elucidating key electrostatic interactions critical for ligand-receptor binding for the purpose of discovery and design of therapeutics for diabetes and obesity with improved efficacy and safety.
3. Materials and Methods
As of March 11, 2024, a total of 44 experimental structures have been deposited in Protein Data Bank (PDB) [
20] as listed in
Table 1, according to a text query:
QUERY: Polymer Entity Description = "Glucagon-like peptide 1 receptor" of the Protein Data Bank [
20]. Among them, only two experimental structures represent the GLP-1-GLP-1R complex, with PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36], respectively, providing an accurate structural basis of the GLP-1-GLP-1R interaction specificity for subsequent comprehensive structural biophysical (CSB) analysis of the two structural models (two yellow rows in
Table 1).
Of the two experimental structures represent the GLP-1-GLP-1R complex, with PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36], two sets of amino acid sequences are listed in italics in fasta format as below,
>3IOL_1|Chain A|Glucagon-like peptide 1 receptor|Homo sapiens (9606)
GSHMRPQGATVSLWETVQKWREYRRQCQRSLTEDPPPATDLFCNRTFDEYACWPDGEPGSFVNVSCPWYLPWASSVPQGHVYRFCTAEGLWLQKDNSSLPWRDLSECEESKRGERSSPEEQLLFLY
>3IOL_2|Chain B|Glucagon|Homo sapiens (9606)
HAEGTFTSDVSSYLEGQAAKEFIAWLVKGRG
>4ZGM_1|Chain A|Glucagon-like peptide 1 receptor|Homo sapiens (9606)
RPQGATVSLWETVQKWREYRRQCQRSLTEDPPPATDLFCNRTFDEYACWPDGEPGSFVNVSCPWYLPWASSVPQGHVYRFCTAEGLWLQKDNSSLPWRDLSECEESKRGERSSPEEQLLFLY
>4ZGM_2|Chain B|Semaglutide peptide backbone; 8Aib,34R-GLP-1(7-37)-OH|Homo sapiens (9606)
HAEGTFTSDVSSYLEGQAAKEFIAWLVRGRG
With the amino acid sequence alignment as shown in Figure 1, it is quite clear that the sequences GLP-1 for 3IOL [
17,
18] and 4ZGM [
19,
36] is at position 33 or 34, i.e., an exchange of lysine and arginine, while the GLP-1R sequences for 3IOL [
17,
18] and 4ZGM [
19,
36] are in complete alignment, except for the presence of one N-terminal four residue fragment (
GSHM) in 3IOL [
17,
18] and the absence of it (
GSHM) at N-terminal in 4ZGM [
19,
36].
Figure 1.
GLP-1 amino acid sequence alignment by Clustal Omega [
39] of two experimental structures represent the GLP-1-GLP-1R complex, with PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36].
Figure 1.
GLP-1 amino acid sequence alignment by Clustal Omega [
39] of two experimental structures represent the GLP-1-GLP-1R complex, with PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36].
After the atomic coordinates file for PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36] were downloaded from the PDB [
20] website, Chimera [
40] was employed to manually add hydrogen atoms to the structural model of the two structural models representing the GLP-1-GLP-1R complex structures, with file names
3IOH.pdb and
4ZGH.pdb, respectively. Afterwards, the two hydrogen-added structural models were subject to a set of comprehensive structural biophysical (CSB) analysis as described in [
22] to identify key residue-specific interactions at the GLP-1-GLP-1R binding interface and uncover the interstructural biophysics underlying the GLP-1-GLP-1R complex structure.
Specifically, the CSB analysis here [
22] consists of the structural identification of salt bridges and side chain hydrogen bonds at the binding interface of GLP-1 and GLP-1R. Given the fact that native proteins are in dynamic equilibrium with their less-structured, partially folded and/or unfolded states [
41], this article uses two sets of screening criteria for the structural identification of potential hotspots at the GLP-1-GLP-1R binding interface in the two structural models i.e., PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36].
First, the same set of criteria (referred to as the old criteria below) as in [
22] was used, i.e., the interfacial salt bridge analysis was conducted with an in-house python script only for titrateable residues (Asp, Glu, Lys, Arg and His), 4.0 Å was used as the cutoff distance for the two oppositely charged groups [
22,
42]. The hydrogen bond analysis was also conducted for only side chain nuclei with an in-house python script, and employed two geometric criteria: (a) a cutoff value of the angle formed by acceptor (A), donor (D) and hydrogen (H) (
) of 30° (b) a cutoff value of donor-acceptor distance at 3.0 Å. That is, a hydrogen bond is only considered to be formed if
is not larger than 30° and the donor-acceptor distance is not larger than 3.0 Å [
22,
42].
Afterwards, a new set of criteria (referred to as the new criteria below) was used to account for the GLP-1-GLP-1R complex structures, i.e., the interfacial salt bridge analysis was conducted with an in-house python script only for titrateable residues (Asp, Glu, Lys, Arg and His), 6.0 Å was used as the cutoff distance for the two oppositely charged groups [
22,
42]. The hydrogen bond analysis was also conducted for only side chain nuclei with an in-house python script, and employed two geometric criteria: (a) a cutoff value of the angle formed by acceptor (A), donor (D) and hydrogen (H) (
) of 50°; (b) a cutoff value of donor-acceptor distance at 5.0 Å. That is, a hydrogen bond is only considered to be formed if
is not larger than 50° and the donor-acceptor distance is not larger than 5.0 Å [
22,
42].
Here, the in-house python scripts essentially are the same as those used in [
43], except for the differences in three key parameters related to the screening criteria, i.e., the salt bridge distance cutoff in Å, cutoff angle
in ° for hydrogen bonding network screening, and the cutoff distance (in Å) of donor-acceptor for hydrogen bonding network screening.
6. Towards a GLP-1-GLP-1R-based mini GIBAC: a brief future perspective
On August 11, 2022, the concept of a general intermolecular binding affinity calculator (GIBAC) was for the first time proposed in a preprint [
48], which defined a collective set of the standards (as defined below) of a truly general intermolecular binding affinity calculator, i.e, a truly GIBAC:
a truly GIBAC needs to take genetic variations into account;
a truly GIBAC needs to work even without structural information;
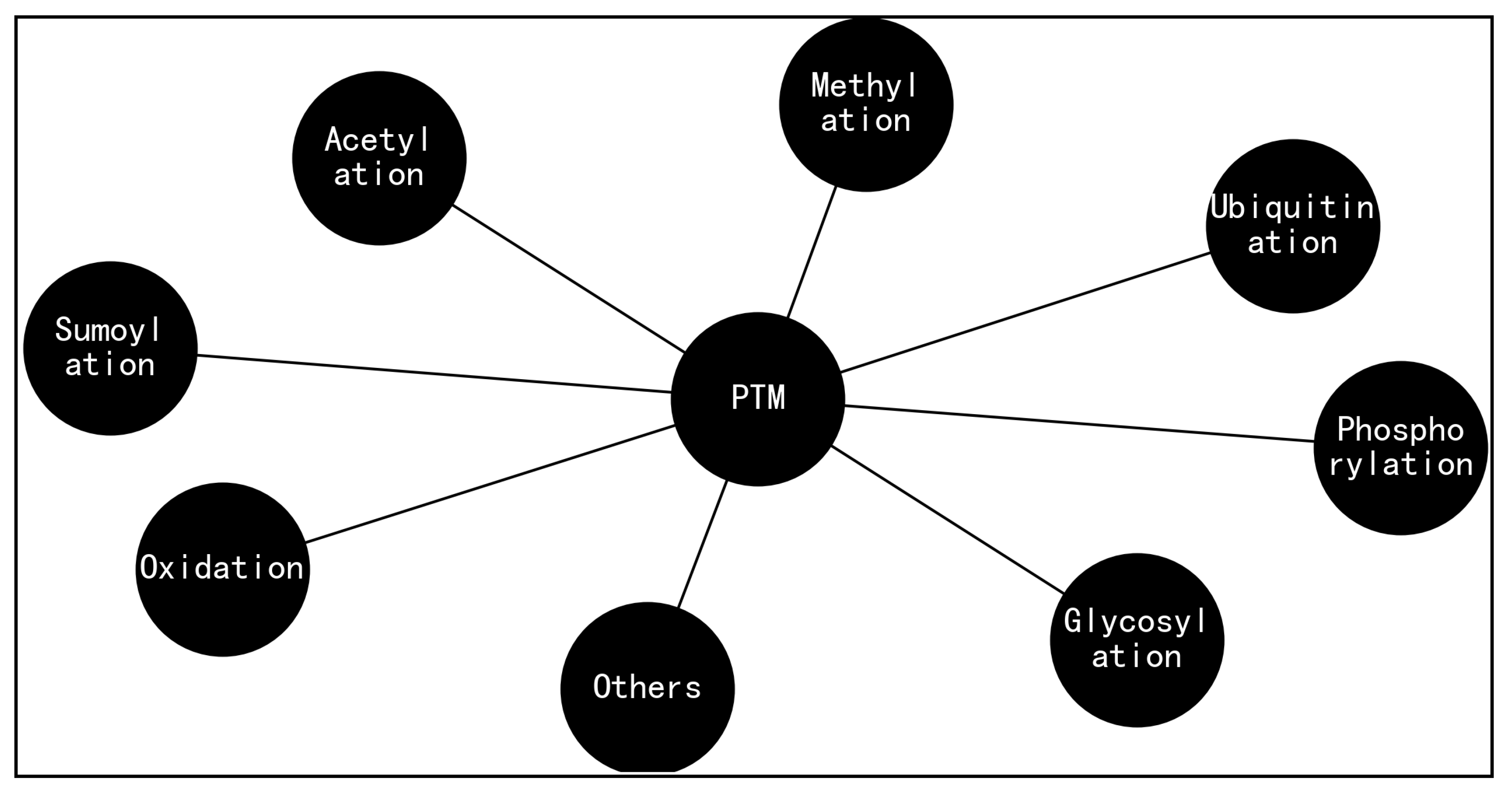
for a truly GIBAC, a variety of factors need to be taken into account, such as temperature, pH [
49,
50], site-specific protonation states (e.g., side chain pKa of protein) [
51,
52], post-translational modifications (PTMs, Figure 8) [
25,
53,
54], post-expression modifications (PEMs) [
7,
55], buffer conditions [
56], et cetera;
a truly GIBAC is able to be used the other way around, i.e., to be used as a search engine for therapeutic candidate(s). With such a GIBAC-based search engine, a list of therapeutic candidates can be retrieved and ranked according to drug-target Kd value(s), with input parameters including drug target(s) and a desired drug-target Kd value or a range of it.
Figure 8.
A range of post-translational modifications (PTMs) for biomolecules such as protein, where side chain placement and energy minimization algorithms [
57] are useful to incorporate structural information of PTMs (Figure 8) and PEMs [
7,
55] into structural models of biomolecules such as protein, similar to the way the structure of semaglutide is modified with a C-18 fatty di-acid chain at position 26 that provides strong, specific binding to albumin [
33].
Figure 8.
A range of post-translational modifications (PTMs) for biomolecules such as protein, where side chain placement and energy minimization algorithms [
57] are useful to incorporate structural information of PTMs (Figure 8) and PEMs [
7,
55] into structural models of biomolecules such as protein, similar to the way the structure of semaglutide is modified with a C-18 fatty di-acid chain at position 26 that provides strong, specific binding to albumin [
33].
As is known, the entire space of molecular types and drug modalities is vast [
58], extending far beyond proteins and small molecules, which makes a comprehensive physics-based exploration practically impossible and unnecessary [
59]. Nonetheless, AI algorithms rely on huge amounts of data to learn and train continuously, where its quantity and quality is inextricably linked to the performance of the model [
60,
61].
As charted out previously in [
48], therefore, the construction of GIBAC requires two key ingredients, i.e.,
data and
algorithm, where
algorithm is like the engine of a car, and
data the appropriate fuel or power source of it. Here, this article argues that in addition to data and algorithm, the construction of a real GIBAC with adequate accuracy and efficiency also requires our knowledge of biophysics underlying the structure, folding, dynamics, and the direct binding and interaction between ligand and receptor, e.g., the electrostatic structural features extracted from the two experimental structures represent the GLP-1-GLP-1R complex, with PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36].
To sum up, the development of a GLP-1-GLP-1R-based mini GIBAC perspective represents a promising strategy for extracting electrostatic structural features from ligand-receptor complexes and advancing drug discovery efforts targeting the GLP-1/GLP-1R axis. By combining computational modeling with experimental validation, a GLP-1-GLP-1R-based mini GIBAC offers a powerful tool for elucidating the molecular basis of ligand recognition and receptor activation, paving the way for the development of next-generation therapeutics with improved efficacy and safety for patients with diabetes and/or obesity [
14,
15,
16].
Table 1.
Experimentally determined GLP-1R-related structures in the Protein Data Bank (PDB [
20]) as of March 11, 2024 with a
QUERY: Polymer Entity Description = "Glucagon-like peptide 1 receptor". In this table, the two structural models representing the complex structures of ligand-bound GLP-1R are highlighted in two yellow rows, i.e., PDB IDs: 3IOL [
17] and 4ZGM [
36].
Table 1.
Experimentally determined GLP-1R-related structures in the Protein Data Bank (PDB [
20]) as of March 11, 2024 with a
QUERY: Polymer Entity Description = "Glucagon-like peptide 1 receptor". In this table, the two structural models representing the complex structures of ligand-bound GLP-1R are highlighted in two yellow rows, i.e., PDB IDs: 3IOL [
17] and 4ZGM [
36].
|
PDB ID |
Structure Title |
| 8JIS |
Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex |
|
8JIP |
Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex |
| 8JIR |
Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex |
|
7X8R |
Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex |
| 7X8S |
Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex |
|
7S15 |
GLP-1 receptor bound with Pfizer small molecule agonist |
| 7RG9 |
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R in apo form |
|
7RGP |
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R bound to tirzepatide |
| 7VBH |
Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex |
|
7VBI |
Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GLP-1R-Gs complex |
| 7LLL |
Exendin-4-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein |
|
7LLY |
Oxyntomodulin-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein |
| 7S1M |
Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 1) |
|
7S3I |
Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 2) |
| 7RTB |
Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) |
|
7DUR |
Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex |
| 7EVM |
Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex |
|
7KI0 |
Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein |
| 7KI1 |
Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein |
|
7DUQ |
Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex |
| 7E14 |
Compound2_GLP-1R_OWL833_Gs complex structure |
|
7LCI |
PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex |
| 7LCJ |
PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex |
|
7LCK |
PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R) |
| 6XOX |
cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970 |
|
6X18 |
GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor |
| 6X19 |
Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor |
|
6X1A |
Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor |
| 7C2E |
GLP-1R-Gs complex structure with a small molecule full agonist |
|
6VCB |
Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator |
| 6ORV |
Non-peptide agonist (TT-OAD2) bound to the Glucagon-Like peptide-1 (GLP-1) Receptor |
|
5OTT |
Extracellular domain of GLP-1 receptor in complex with exendin-4 variant Gly2Hcs/Thr5Hcs |
| 5OTU |
Extracellular domain of GLP-1 receptor in complex with GLP-1 variant Ala8Hcs/Thr11Hcs |
|
5OTV |
Extracellular domain of GLP-1 receptor in complex with GLP-1 variant Ala8Cyc/Thr11Hcs |
| 5OTW |
Extracellular domain of GLP-1 receptor in complex with GLP-1 variant Ala8Hcs/Thr11Cys |
|
5OTX |
Extracellular domain of GLP-1 receptor in complex with GLP-1 variant Ala8Cys/Thr11Cys |
| 6GB1 |
Crystal structure of the GLP1 receptor ECD with Peptide 11 |
|
6B3J |
3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex |
| 5NX2 |
Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 Å resolution |
|
5E94 |
Antibody-bound Glucagon-like Peptide-1 receptor extracellular domain |
|
4ZGM |
Crystal structure of Semaglutide peptide backbone in complex with the GLP-1 receptor extracellular domain |
|
3IOL |
Crystal structure of Glucagon-Like Peptide-1 in complex with the extracellular domain of the Glucagon-Like Peptide-1 Receptor |
| 3C59 |
Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain |
|
3C5T |
Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain |
Table 2.
Salt bridging screening for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
Table 2.
Salt bridging screening for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
|
PDB ID |
Residue A |
Atom A |
Residue B |
Atom B |
Distance (Å) |
| 3IOH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE1 |
3.682 |
| 3IOH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE2 |
2.811 |
| 3IOH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD1 |
3.801 |
| 3IOH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD2 |
2.782 |
| 3IOH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD2 |
3.054 |
| 3IOH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD1 |
3.519 |
| 3IOH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD2 |
2.959 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.212 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.924 |
| 4ZGH |
A_LYS_38 |
NZ |
A_GLU_34 |
OE1 |
3.823 |
| 4ZGH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE1 |
3.861 |
| 4ZGH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE2 |
2.747 |
| 4ZGH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD1 |
3.779 |
| 4ZGH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD2 |
2.832 |
| 4ZGH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD2 |
3.198 |
| 4ZGH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD1 |
3.561 |
| 4ZGH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD2 |
2.953 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.409 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.771 |
Table 3.
Side chain hydrogen bonding network analysis for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number,
represents the angle formed by acceptor (A), donor (D) and hydrogen (H)
.
Table 3.
Side chain hydrogen bonding network analysis for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number,
represents the angle formed by acceptor (A), donor (D) and hydrogen (H)
.
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 3IOH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.87 |
1.87 |
4.40 |
| 3IOH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.81 |
1.86 |
15.77 |
| 3IOH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.80 |
1.81 |
7.74 |
| 3IOH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH22, A_ARG_64 |
2.78 |
1.78 |
5.75 |
| 3IOH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.69 |
1.83 |
24.82 |
| 3IOH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.63 |
1.69 |
8.92 |
| 3IOH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH12, A_ARG_121 |
2.96 |
2.09 |
25.64 |
| 3IOH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.65 |
1.73 |
12.69 |
| 4ZGH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.92 |
2.08 |
10.96 |
| 4ZGH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.75 |
1.91 |
11.18 |
| 4ZGH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.99 |
2.14 |
5.73 |
| 4ZGH |
OD1, A_ASP_74 |
NE, A_ARG_64 |
HE, A_ARG_64 |
2.97 |
2.12 |
3.34 |
| 4ZGH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH21, A_ARG_64 |
2.83 |
1.98 |
7.04 |
| 4ZGH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.80 |
2.02 |
20.99 |
| 4ZGH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.82 |
1.99 |
6.37 |
| 4ZGH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH11, A_ARG_121 |
2.95 |
2.17 |
20.60 |
| 4ZGH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.67 |
1.91 |
21.21 |
| 4ZGH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
2.77 |
2.03 |
27.77 |
| 4ZGH |
OXT, B_GLY_37 |
NE1, B_TRP_31 |
HE1, B_TRP_31 |
2.92 |
2.09 |
12.63 |
Table 4.
Interfacial Salt bridging screening for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
Table 4.
Interfacial Salt bridging screening for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
|
PDB ID |
Residue A |
Atom A |
Residue B |
Atom B |
Distance (Å) |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.212 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.924 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.409 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.771 |
Table 5.
3IOH-specific side chain hydrogen bonding analysis. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 5.
3IOH-specific side chain hydrogen bonding analysis. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 3IOH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.87 |
1.87 |
4.40 |
| 3IOH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.81 |
1.86 |
15.77 |
| 3IOH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.80 |
1.81 |
7.74 |
| 3IOH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH22, A_ARG_64 |
2.78 |
1.78 |
5.75 |
| 3IOH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.69 |
1.83 |
24.82 |
| 3IOH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.63 |
1.69 |
8.92 |
| 3IOH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH12, A_ARG_121 |
2.96 |
2.09 |
25.64 |
| 3IOH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.65 |
1.73 |
12.69 |
Table 6.
4ZGH-specific side chain hydrogen bonding analysis. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 6.
4ZGH-specific side chain hydrogen bonding analysis. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 4ZGH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.92 |
2.08 |
10.96 |
| 4ZGH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.75 |
1.91 |
11.18 |
| 4ZGH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.99 |
2.14 |
5.73 |
| 4ZGH |
OD1, A_ASP_74 |
NE, A_ARG_64 |
HE, A_ARG_64 |
2.97 |
2.12 |
3.34 |
| 4ZGH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH21, A_ARG_64 |
2.83 |
1.98 |
7.04 |
| 4ZGH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.80 |
2.02 |
20.99 |
| 4ZGH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.82 |
1.99 |
6.37 |
| 4ZGH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH11, A_ARG_121 |
2.95 |
2.17 |
20.60 |
| 4ZGH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.67 |
1.91 |
21.21 |
| 4ZGH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
2.77 |
2.03 |
27.77 |
| 4ZGH |
OXT, B_GLY_37 |
NE1, B_TRP_31 |
HE1, B_TRP_31 |
2.92 |
2.09 |
12.63 |
Table 7.
4ZGH-specific interfacial side chain hydrogen bonding analysis, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 7.
4ZGH-specific interfacial side chain hydrogen bonding analysis, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 4ZGH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
2.77 |
2.03 |
27.77 |
Table 8.
Salt bridging screening with the
new criteria as defined in the section of Materials and Methods for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
Table 8.
Salt bridging screening with the
new criteria as defined in the section of Materials and Methods for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
|
PDB ID |
Residue A |
Atom A |
Residue B |
Atom B |
Distance (Å) |
| 3IOH |
A_LYS_38 |
NZ |
A_GLU_34 |
OE1 |
5.019 |
| 3IOH |
A_LYS_38 |
NZ |
A_GLU_34 |
OE2 |
5.563 |
| 3IOH |
A_ARG_43 |
NH1 |
A_GLU_68 |
OE1 |
5.108 |
| 3IOH |
A_ARG_44 |
NH1 |
A_GLU_41 |
OE1 |
5.031 |
| 3IOH |
A_ARG_44 |
NH1 |
A_GLU_41 |
OE2 |
4.860 |
| 3IOH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE1 |
3.682 |
| 3IOH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE2 |
2.811 |
| 3IOH |
A_ARG_48 |
NH1 |
A_GLU_41 |
OE2 |
5.664 |
| 3IOH |
A_ARG_64 |
NH1 |
A_ASP_53 |
OD1 |
5.728 |
| 3IOH |
A_ARG_64 |
NH1 |
A_ASP_53 |
OD2 |
4.864 |
| 3IOH |
A_ARG_64 |
NH1 |
A_ASP_74 |
OD1 |
5.246 |
| 3IOH |
A_ARG_64 |
NH1 |
A_ASP_74 |
OD2 |
4.902 |
| 3IOH |
A_ARG_64 |
NH2 |
A_ASP_53 |
OD2 |
5.347 |
| 3IOH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD1 |
3.801 |
| 3IOH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD2 |
2.782 |
| 3IOH |
A_HIS_99 |
ND1 |
A_GLU_125 |
OE1 |
4.292 |
| 3IOH |
A_HIS_99 |
ND1 |
A_GLU_125 |
OE2 |
5.603 |
| 3IOH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD1 |
4.865 |
| 3IOH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD2 |
3.054 |
| 3IOH |
A_ARG_102 |
NH2 |
A_ASP_67 |
OD2 |
5.183 |
| 3IOH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD1 |
3.519 |
| 3IOH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD2 |
2.959 |
| 3IOH |
A_ARG_121 |
NH2 |
A_ASP_67 |
OD1 |
5.721 |
| 3IOH |
A_ARG_121 |
NH2 |
A_ASP_67 |
OD2 |
5.061 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.212 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.924 |
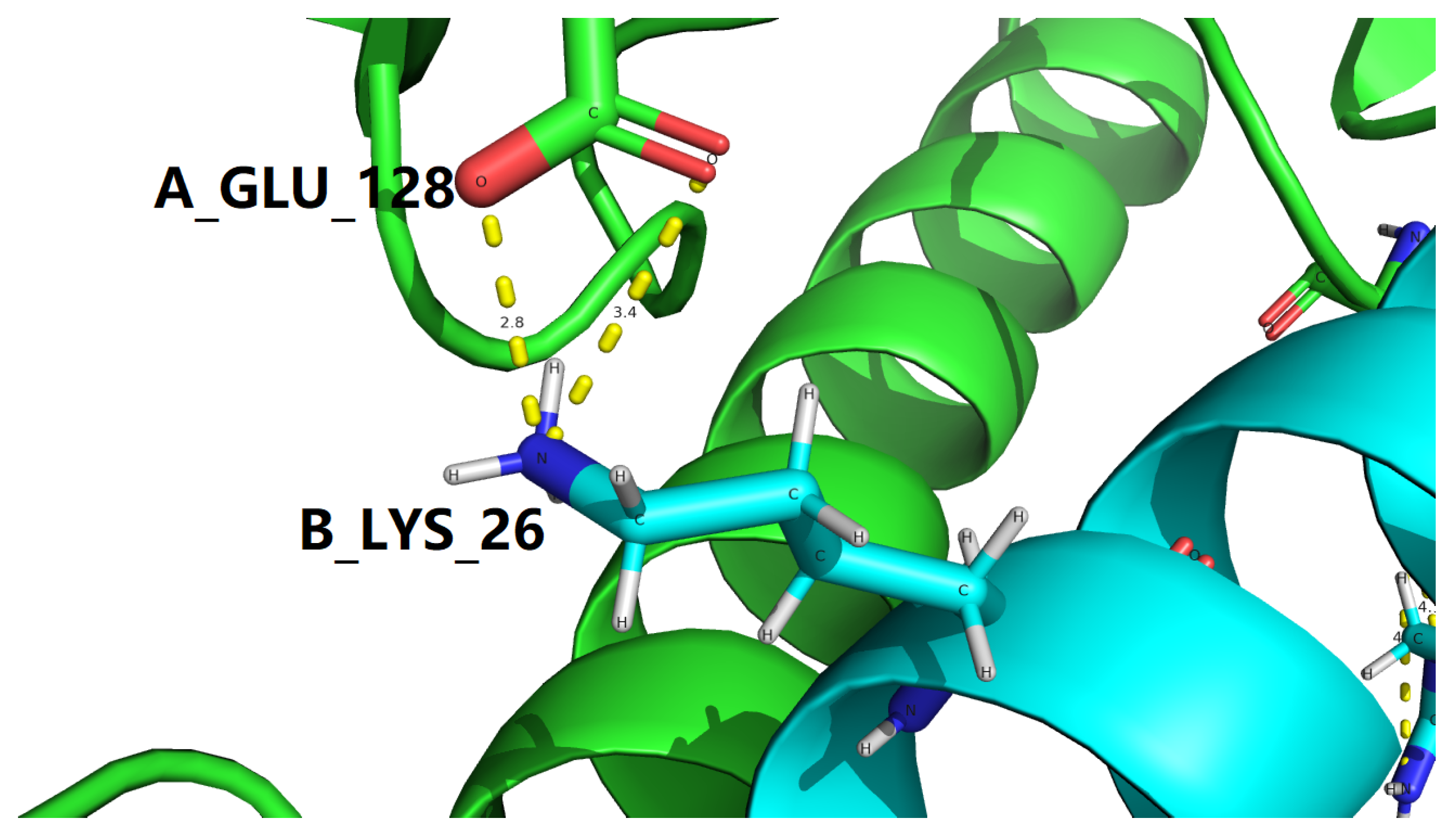
| 4ZGH |
A_LYS_38 |
NZ |
A_GLU_34 |
OE1 |
3.823 |
| 4ZGH |
A_LYS_38 |
NZ |
A_GLU_34 |
OE2 |
5.820 |
| 4ZGH |
A_ARG_43 |
NH1 |
A_GLU_68 |
OE1 |
5.144 |
| 4ZGH |
A_ARG_44 |
NH1 |
A_GLU_41 |
OE1 |
5.098 |
| 4ZGH |
A_ARG_44 |
NH1 |
A_GLU_41 |
OE2 |
4.740 |
| 4ZGH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE1 |
3.861 |
| 4ZGH |
A_ARG_44 |
NH2 |
A_GLU_41 |
OE2 |
2.747 |
| 4ZGH |
A_ARG_48 |
NH1 |
A_GLU_41 |
OE2 |
5.698 |
| 4ZGH |
A_ARG_64 |
NH1 |
A_ASP_53 |
OD1 |
5.693 |
| 4ZGH |
A_ARG_64 |
NH1 |
A_ASP_53 |
OD2 |
4.947 |
| 4ZGH |
A_ARG_64 |
NH1 |
A_ASP_74 |
OD1 |
5.111 |
| 4ZGH |
A_ARG_64 |
NH1 |
A_ASP_74 |
OD2 |
4.906 |
| 4ZGH |
A_ARG_64 |
NH2 |
A_ASP_53 |
OD2 |
5.424 |
| 4ZGH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD1 |
3.779 |
| 4ZGH |
A_ARG_64 |
NH2 |
A_ASP_74 |
OD2 |
2.832 |
| 4ZGH |
A_HIS_99 |
ND1 |
A_GLU_125 |
OE1 |
4.316 |
| 4ZGH |
A_HIS_99 |
ND1 |
A_GLU_125 |
OE2 |
5.690 |
| 4ZGH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD1 |
4.996 |
| 4ZGH |
A_ARG_102 |
NH1 |
A_ASP_67 |
OD2 |
3.198 |
| 4ZGH |
A_ARG_102 |
NH2 |
A_ASP_67 |
OD2 |
5.343 |
| 4ZGH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD1 |
3.561 |
| 4ZGH |
A_ARG_121 |
NH1 |
A_ASP_67 |
OD2 |
2.953 |
| 4ZGH |
A_ARG_121 |
NH2 |
A_ASP_67 |
OD1 |
5.760 |
| 4ZGH |
A_ARG_121 |
NH2 |
A_ASP_67 |
OD2 |
5.107 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.409 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.771 |
| 4ZGH |
B_ARG_34 |
NH1 |
B_GLU_27 |
OE1 |
5.552 |
| 4ZGH |
B_ARG_34 |
NH2 |
B_GLU_27 |
OE1 |
5.072 |
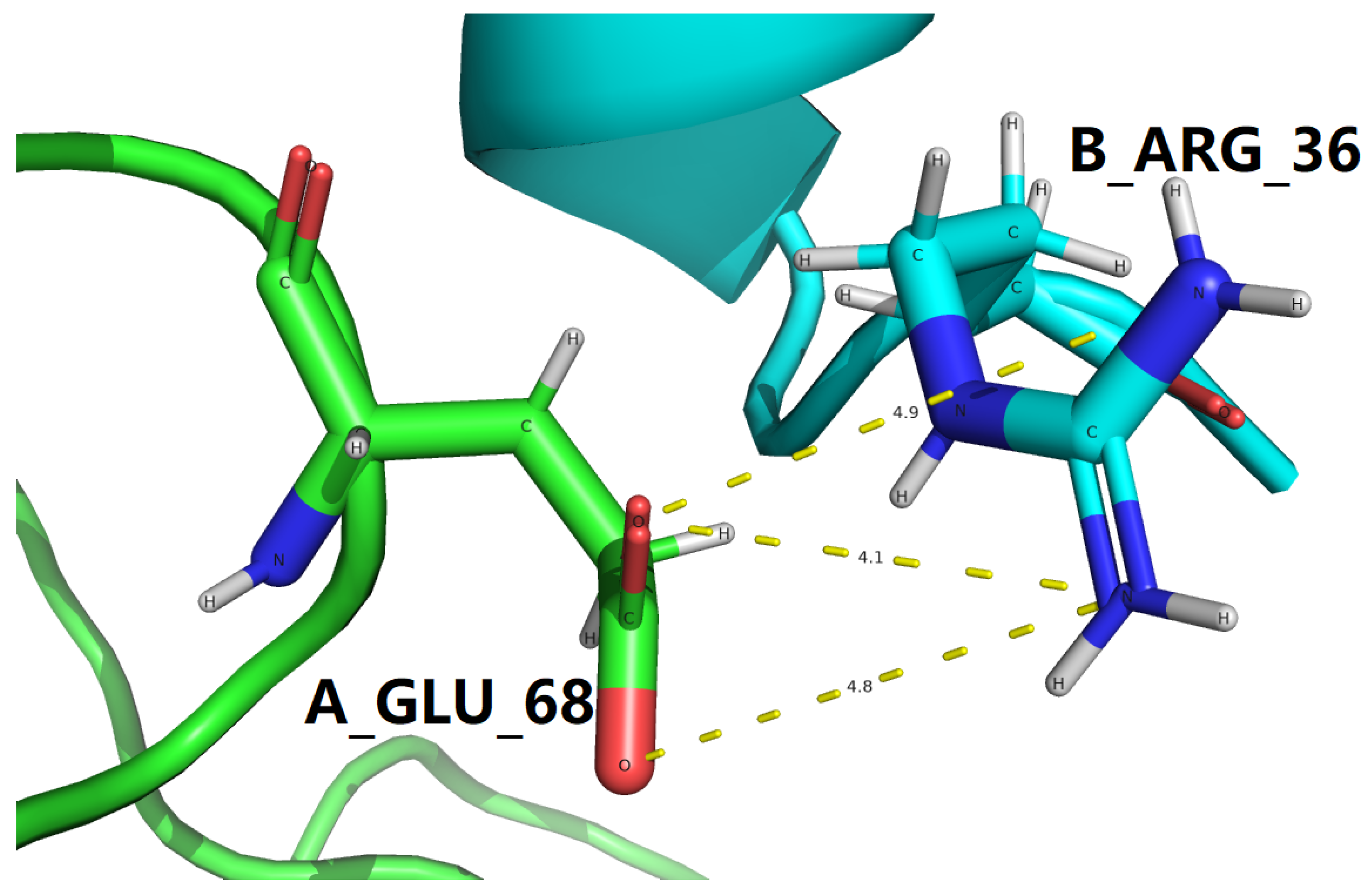
| 4ZGH |
B_ARG_36 |
NH1 |
A_GLU_68 |
OE1 |
4.147 |
| 4ZGH |
B_ARG_36 |
NH1 |
A_GLU_68 |
OE2 |
4.835 |
| 4ZGH |
B_ARG_36 |
NH2 |
A_GLU_68 |
OE1 |
4.893 |
Table 9.
Interfacial salt bridging screening with the
new criteria as defined in the section of Materials and Methods for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
Table 9.
Interfacial salt bridging screening with the
new criteria as defined in the section of Materials and Methods for two GLP-1-GLP-1R complex structures (PDB IDs: 3IOL [
17,
18] and 4ZGM [
19,
36]). In this table, the residue naming scheme is
Chain ID_residue name_residue number.
|
PDB ID |
Residue A |
Atom A |
Residue B |
Atom B |
Distance (Å) |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.212 |
| 3IOH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.924 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE1 |
3.409 |
| 4ZGH |
B_LYS_26 |
NZ |
A_GLU_128 |
OE2 |
2.771 |
| 4ZGH |
B_ARG_36 |
NH1 |
A_GLU_68 |
OE1 |
4.147 |
| 4ZGH |
B_ARG_36 |
NH1 |
A_GLU_68 |
OE2 |
4.835 |
| 4ZGH |
B_ARG_36 |
NH2 |
A_GLU_68 |
OE1 |
4.893 |
Table 10.
3IOH-specific side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 10.
3IOH-specific side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 3IOH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.87 |
1.87 |
4.40 |
| 3IOH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.81 |
1.86 |
15.77 |
| 3IOH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.80 |
1.81 |
7.74 |
| 3IOH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH22, A_ARG_64 |
2.78 |
1.78 |
5.75 |
| 3IOH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.69 |
1.83 |
24.82 |
| 3IOH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.63 |
1.69 |
8.92 |
| 3IOH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH12, A_ARG_121 |
2.96 |
2.09 |
25.64 |
| 3IOH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.65 |
1.73 |
12.69 |
Table 11.
4ZGH-specific side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 11.
4ZGH-specific side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 4ZGH |
OE1, A_GLU_41 |
NE, A_ARG_44 |
HE, A_ARG_44 |
2.92 |
2.08 |
10.96 |
| 4ZGH |
OE2, A_GLU_41 |
NH2, A_ARG_44 |
HH21, A_ARG_44 |
2.75 |
1.91 |
11.18 |
| 4ZGH |
OE2, A_GLU_41 |
NE2, A_GLN_45 |
HE21, A_GLN_45 |
2.99 |
2.14 |
5.73 |
| 4ZGH |
OD1, A_ASP_74 |
NE, A_ARG_64 |
HE, A_ARG_64 |
2.97 |
2.12 |
3.34 |
| 4ZGH |
OD2, A_ASP_74 |
NH2, A_ARG_64 |
HH21, A_ARG_64 |
2.83 |
1.98 |
7.04 |
| 4ZGH |
OD2, A_ASP_67 |
NE1, A_TRP_72 |
HE1, A_TRP_72 |
2.80 |
2.02 |
20.99 |
| 4ZGH |
OE2, A_GLU_125 |
OH, A_TYR_101 |
HH, A_TYR_101 |
2.82 |
1.99 |
6.37 |
| 4ZGH |
OD2, A_ASP_67 |
NH1, A_ARG_121 |
HH11, A_ARG_121 |
2.95 |
2.17 |
20.60 |
| 4ZGH |
OD1, A_ASP_122 |
OG, A_SER_124 |
HG, A_SER_124 |
2.67 |
1.91 |
21.21 |
| 4ZGH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
2.77 |
2.03 |
27.77 |
| 4ZGH |
OXT, B_GLY_37 |
NE1, B_TRP_31 |
HE1, B_TRP_31 |
2.92 |
2.09 |
12.63 |
Table 12.
3IOH-specific interfacial side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 12.
3IOH-specific interfacial side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
|
PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 3IOH |
OG1, A_THR_29 |
OG, B_SER_18 |
HG, B_SER_18 |
3.26 |
2.42 |
24.61 |
| 3IOH |
OE1, A_GLU_128 |
NZ, B_LYS_26 |
HZ1, B_LYS_26 |
3.21 |
2.54 |
41.12 |
| 3IOH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ1, B_LYS_26 |
2.92 |
2.28 |
42.31 |
Table 13.
4ZGH-specific interfacial side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
Table 13.
4ZGH-specific interfacial side chain hydrogen bonding analysis with the new criteria as defined in the section of Materials and Methods. In this table, the residue naming scheme is Chain ID_residue name_residue number, represents the angle formed by acceptor (A), donor (D) and hydrogen (H) .
| PDB |
Acceptor (A) |
Donor (D) |
Hydrogen (H) |
D-A (Å) |
H-A (Å) |
|
| 4ZGH |
OE1, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
3.41 |
2.69 |
31.93 |
| 4ZGH |
OE2, A_GLU_128 |
NZ, B_LYS_26 |
HZ2, B_LYS_26 |
2.77 |
2.03 |
27.77 |
| 4ZGH |
OE1, A_GLU_68 |
NE, B_ARG_36 |
HE, B_ARG_36 |
2.75 |
2.13 |
37.73 |
| 4ZGH |
OE2, A_GLU_68 |
NE, B_ARG_36 |
HE, B_ARG_36 |
4.14 |
3.30 |
9.50 |
| 4ZGH |
OE2, A_GLU_68 |
NH1, B_ARG_36 |
HH11, B_ARG_36 |
4.83 |
4.13 |
31.95 |