Submitted:
14 June 2024
Posted:
17 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Intermolecular Binding Affinity in Drug Discovery and Design
1.2. Clinical Relevance of Semaglutide in the Management of Blood Glocuse and Weight
1.3. Ligand-Receptor Binding Affinity in Drug Design
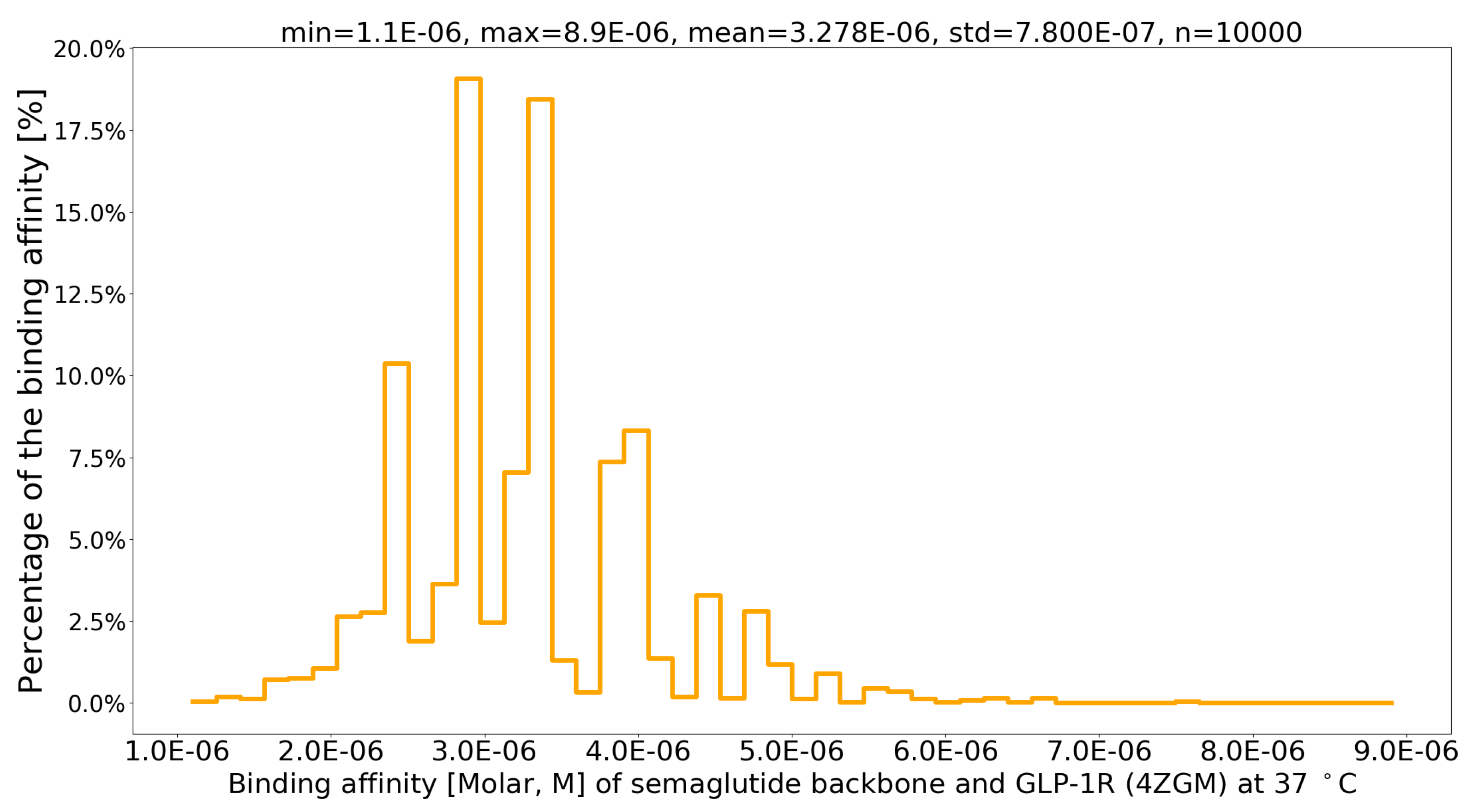
| PDB file | Protein-Protein Complex | G (kcal/mol) | Kd (M) at 37 C | Fold |
|---|---|---|---|---|
| 4ZGM [44] | semaglutide-GLP-1R [44] | -7.8 | 3.4 × 10-6 | 1 |
| sema.pdb [6] | Val27-Arg28 exchange [6] | -8.4 | 1.1 × 10-6 | 3.09 |
2. Motivation
3. Materials and Methods
| PDB ID | Structure Title (release date from newest to oldest) |
|---|---|
| 7KI0 | Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein |
| PDB ID | Structure Title (release date from newest to oldest) |
|---|---|
| 7KI0 | Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein |
| 7KI1 | Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein |
| 4ZGM | Crystal structure of Semaglutide peptide backbone in complex with the GLP-1 receptor extracellular domain |
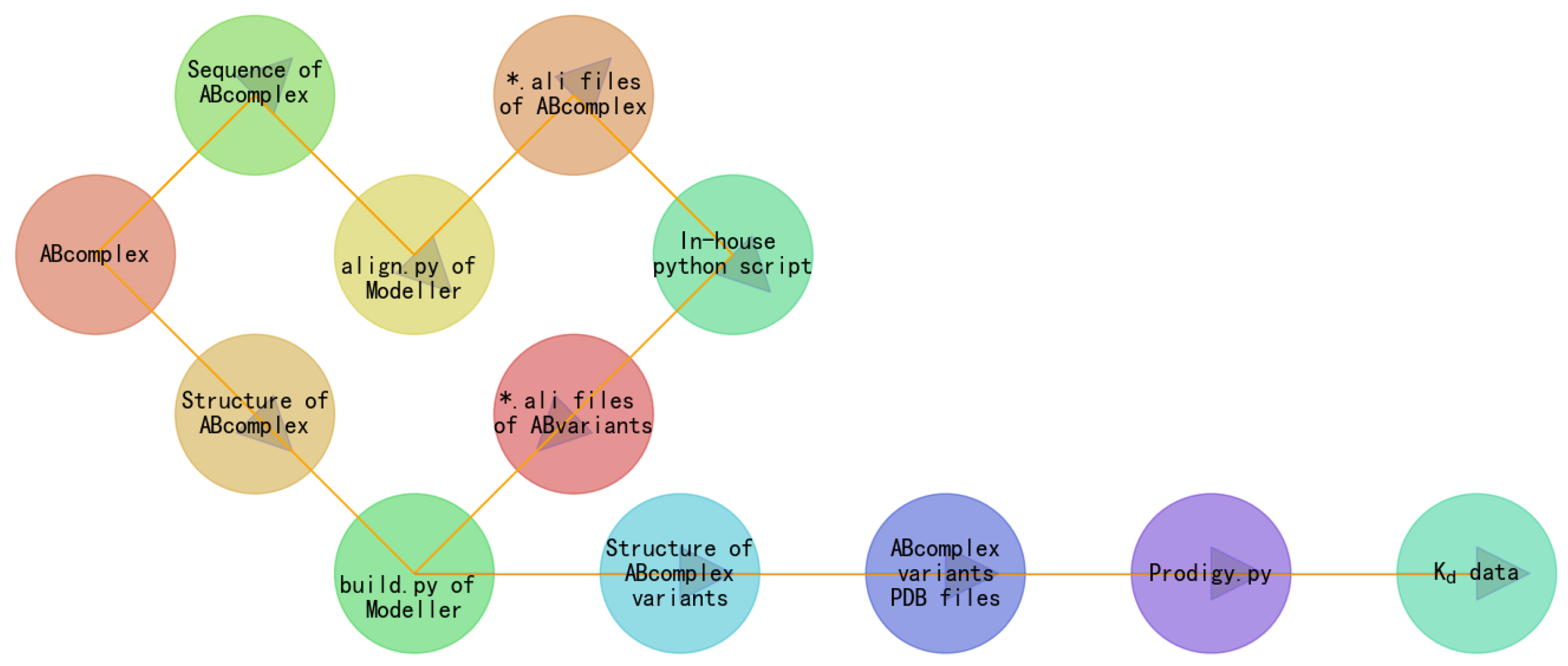
4. Results
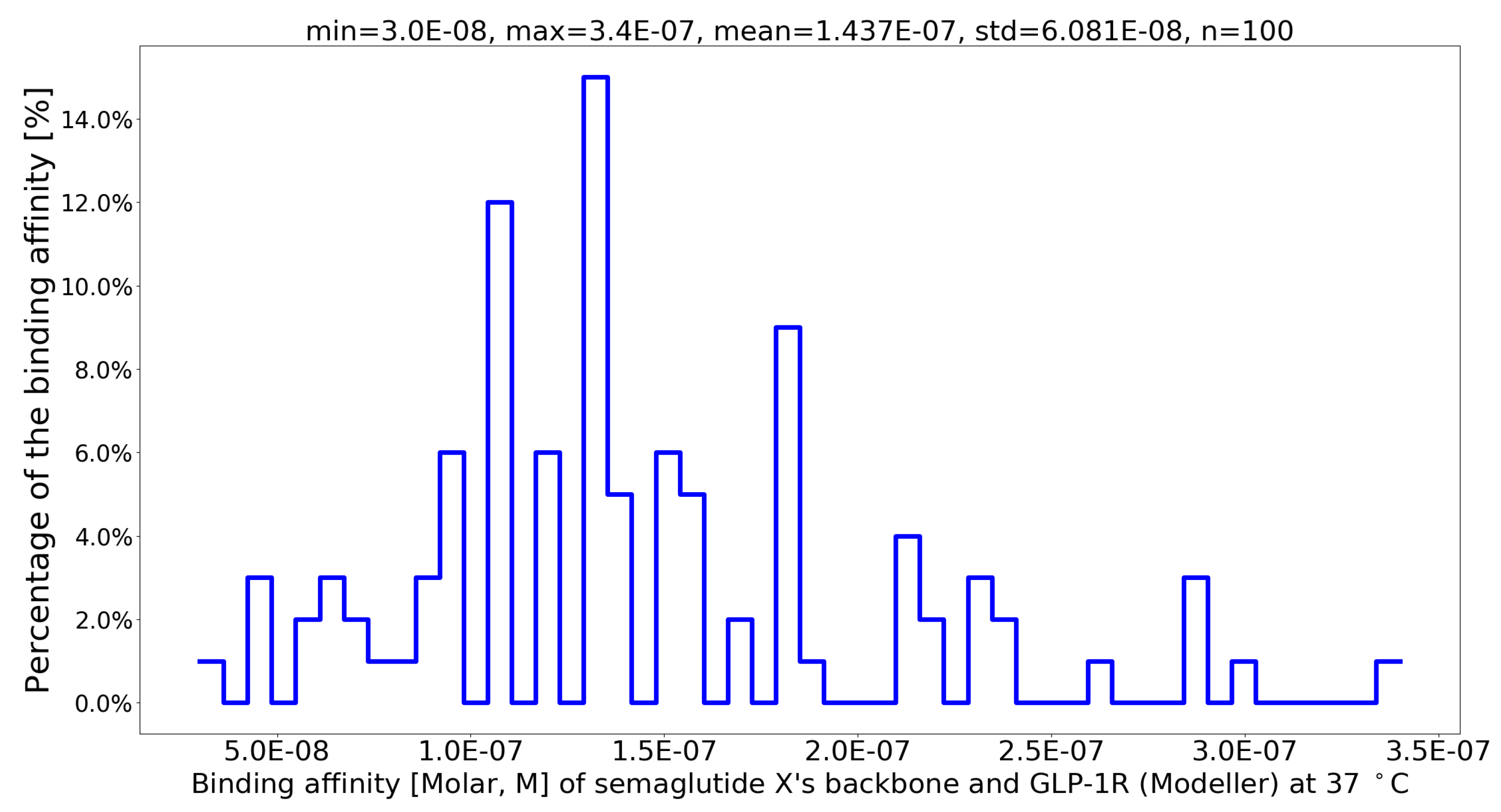
| No. | Muta1 | Muta2 | Muta3 | Muta4 | Min | Max | Mean | Std |
|---|---|---|---|---|---|---|---|---|
| 1 | G13B_A | I20B_Q | L23B_Q | V24B_N | 5.3E-08 | 2.2E-07 | 1.337E-07 | 4.778E-08 |
| 2 | G13B_A | I20B_N | L23B_R | V24B_N | 6.5E-08 | 2.4E-07 | 1.344E-07 | 4.996E-08 |
| 3 | G13B_A | I20B_N | L23B_Q | V24B_T | 6.6E-08 | 2.2E-07 | 1.376E-07 | 4.199E-08 |
| 4 | G13B_A | I20B_T | L23B_Q | V24B_N | 8.0E-08 | 3.1E-07 | 1.404E-07 | 5.478E-08 |
| 5 | G13B_A | I20B_Q | L23B_Q | V24B_T | 6.8E-08 | 2.0E-07 | 1.407E-07 | 3.779E-08 |
| 6 | G13B_A | I20B_S | L23B_R | V24B_T | 6.1E-08 | 2.5E-07 | 1.408E-07 | 5.527E-08 |
| 7 | G13B_A | I20B_Q | L23B_R | V24B_N | 3.0E-08 | 3.2E-07 | 1.461E-07 | 7.095E-08 |
| 8 | G13B_A | I20B_T | L23B_R | V24B_N | 8.3E-08 | 2.1E-07 | 1.467E-07 | 3.690E-08 |
| 9 | G13B_A | I20B_N | L23B_R | V24B_Q | 6.3E-08 | 2.9E-07 | 1.487E-07 | 5.848E-08 |
| 10 | G13B_A | I20B_Q | L23B_R | V24B_Q | 8.6E-08 | 2.5E-07 | 1.489E-07 | 5.170E-08 |
| 11 | G13B_A | I20B_Q | L23B_Q | V24B_Q | 6.3E-08 | 2.4E-07 | 1.505E-07 | 5.269E-08 |
| 12 | G13B_A | I20B_S | L23B_R | V24B_N | 4.4E-08 | 3.5E-07 | 1.520E-07 | 6.568E-08 |
| 13 | G13B_A | I20B_T | L23B_R | V24B_T | 9.4E-08 | 2.2E-07 | 1.545E-07 | 4.188E-08 |
| 14 | G13B_A | I20B_N | L23B_Q | V24B_N | 7.7E-08 | 2.2E-07 | 1.559E-07 | 4.164E-08 |
| 15 | G13B_A | I20B_S | L23B_R | V24B_Q | 7.7E-08 | 3.0E-07 | 1.571E-07 | 6.401E-08 |
| 16 | G13B_A | I20B_S | F19B_Q | V24B_N | 3.5E-08 | 2.8E-07 | 1.583E-07 | 6.648E-08 |
| 17 | G13B_A | I20B_N | L23B_Q | V24B_Q | 8.2E-08 | 2.9E-07 | 1.602E-07 | 5.879E-08 |
| 18 | G13B_A | I20B_N | F19B_Q | V24B_N | 5.0E-08 | 2.9E-07 | 1.634E-07 | 7.035E-08 |
| 19 | G13B_A | I20B_T | F19B_Q | V24B_Q | 9.7E-08 | 2.9E-07 | 1.653E-07 | 4.839E-08 |
| 20 | G13B_A | I20B_N | L23B_R | V24B_T | 8.0E-08 | 3.4E-07 | 1.662E-07 | 8.233E-08 |
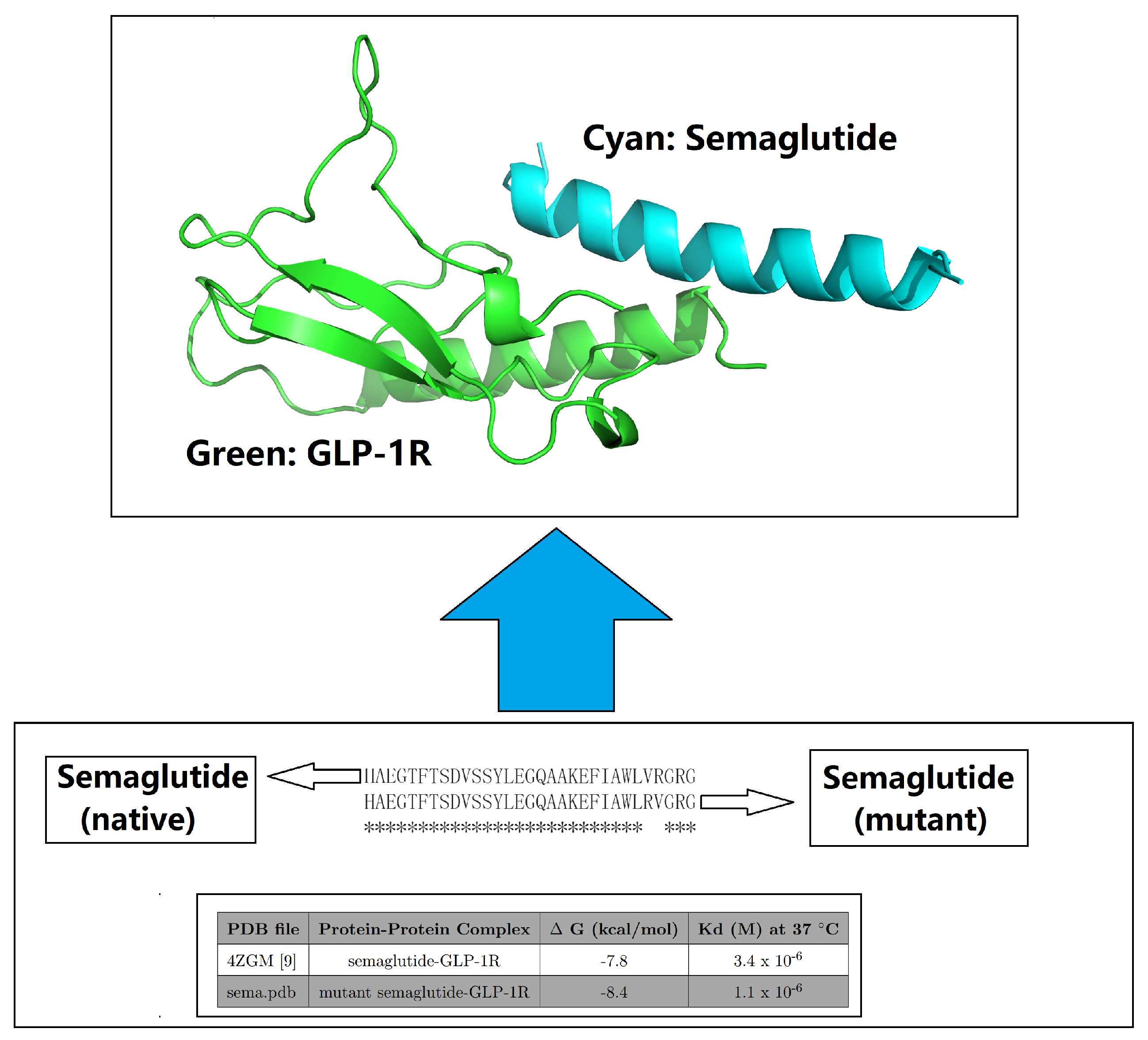
| PDB file | Protein-Protein Complex | G (kcal/mol) | Kd (M) at 37 C | Fold |
|---|---|---|---|---|
| 4ZGM [44] | semaglutide-GLP-1R [44] | -7.8 | 3.4 × 10-6 | 1 |
| sema.pdb [6] | Val27-Arg28 exchange [6] | -8.4 | 1.1 × 10-6 | 3.09 |
| semx.pdb [54] | G13B_A I20B_Q L23B_R V24B_N [54] | -10.7 | 3.0 × 10-8 | 113.33 |
5. Conclusion
6. Discussion
| Size (s) of the synthetic structural and biophysical data set | |||||
|---|---|---|---|---|---|
| Semaglutide backbone (28 Aa) | Molecule X (100 Aa) | ||||
| g(28,1) | 560 | g(100,1) | 2000 | ||
| g(28,2) | 151200 | g(100,2) | 1980000 | ||
| g(28,3) | 26208000 | g(100,3) | 1293600000 | ||
| g(28,4) | 3276000000 | g(100,4) | 627396000000 | ||
| g(28,5) | 314496000000 | g(100,5) | 240920064000000 | ||
Ethical statement
Statement of Usage of Artificial Intelligence
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murphy, K.M.; Gould, R.J.; Largent, B.L.; Snyder, S.H. A unitary mechanism of calcium antagonist drug action. Proceedings of the National Academy of Sciences 1983, 80, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Palzkill, T. Determinants of binding affinity and specificity for the interaction of TEM-1 and SME-1 β-lactamase with β-lactamase inhibitory protein. J. Biol. Chem. 2003, 278, 45706–45712. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, H.; Wang, H.; Chen, Z.; Zhang, Z.; Chen, X.; Li, Y.; Qi, Y.; Wang, R. PLANET: A Multi-objective Graph Neural Network Model for Protein-Ligand Binding Affinity Prediction. Journal of Chemical Information and Modeling 2023. [Google Scholar] [CrossRef] [PubMed]
- Li, W. High-Throughput Extraction of Interfacial Electrostatic Features from GLP-1-GLP-1R Complex Structures: A GLP-1-GLP-1R-Based Mini GIBAC Perspective 2024. [CrossRef]
- Trosset, J.Y.; Cavé, C. In Silico Drug-Target Profiling. In Target Identification and Validation in Drug Discovery; Springer New York, 2019; pp. 89–103.
- Li, W. Strengthening Semaglutide-GLP-1R Binding Affinity via a Val27-Arg28 Exchange in the Peptide Backbone of Semaglutide: A Computational Structural Approach. Journal of Computational Biophysics and Chemistry 2021, 20, 495–499. [Google Scholar] [CrossRef]
- Noble, D.; Blundell, T.L.; Kohl, P. Progress in biophysics and molecular biology: A brief history of the journal. Progress in Biophysics and Molecular Biology 2018, 140, 1–4. [Google Scholar] [CrossRef]
- Tsien, R.W.; Hess, P.; McCleskey, E.W.; Rosenberg, R.L. Calcium channels: Mechanisms of Selectivity, Permeation, and Block. Annual Review of Biophysics and Biophysical Chemistry 1987, 16, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Greenidge, P.A.; Kramer, C.; Mozziconacci, J.C.; Wolf, R.M. MM/GBSA Binding Energy Prediction on the PDBbind Data Set: Successes, Failures, and Directions for Further Improvement. Journal of Chemical Information and Modeling 2012, 53, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Yang, Q.; Du, Y.; Feng, G.; Liu, Z.; Li, Y.; Wang, R. Comparative Assessment of Scoring Functions: The CASF-2016 Update. Journal of Chemical Information and Modeling 2018, 59, 895–913. [Google Scholar] [CrossRef]
- Freire, E. Biophysical methods for the determination of protein-ligand binding constants. Annual review of biophysics 2009, 38, 123–142. [Google Scholar]
- Johnson, C.M. Isothermal Calorimetry. In Protein-Ligand Interactions; Springer US, 2021; pp. 135–159.
- Velázquez-Coy, A.; Ohtaka, H.; Nezami, A.; Muzammil, S.; Freire, E. Isothermal Titration Calorimetry. Current Protocols in Cell Biology 2004, 23. [Google Scholar]
- Sauer, U.G. Surface plasmon resonance-a label-free tool for cellular analysis. Journal of biotechnology 2008, 133, 101–108. [Google Scholar]
- Ernst, R.R.; Bodenhausen, G.; Wokaun, A. Principles of nuclear magnetic resonance in one and two dimensions. Principles of nuclear magnetic resonance in one and two dimensions 1990, 14. [Google Scholar]
- Fuji, H.; Qi, F.; Qu, L.; Takaesu, Y.; Hoshino, T. Prediction of Ligand Binding Affinity to Target Proteins by Molecular Mechanics Theoretical Calculation. Chemical and Pharmaceutical Bulletin 2017, 65, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Zhou, H.X. Calculation of Protein-Ligand Binding Affinities. Annual Review of Biophysics and Biomolecular Structure 2007, 36, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Li, W. Designing rt-PA Analogs to Release its Trapped Thrombolytic Activity. Journal of Computational Biophysics and Chemistry 2021, 20, 719–727. [Google Scholar] [CrossRef]
- Jubb, H.C.; Pandurangan, A.P.; Turner, M.A.; Ochoa-Montaño, B.; Blundell, T.L.; Ascher, D.B. Mutations at protein-protein interfaces: Small changes over big surfaces have large impacts on human health. Progress in Biophysics and Molecular Biology 2017, 128, 3–13. [Google Scholar] [CrossRef]
- Kulkarni-Kale, U.; Raskar-Renuse, S.; Natekar-Kalantre, G.; Saxena, S.A. Antigen-Antibody Interaction Database AgAbDb: A Compendium of Antigen-Antibody Interactions. In Methods in Molecular Biology; Springer New York, 2014; pp. 149–164.
- Manso, T.; Folch, G.; Giudicelli, V.; Jabado-Michaloud, J.; Kushwaha, A.; Ngoune, V.N.; Georga, M.; Papadaki, A.; Debbagh, C.; Pégorier, P.; Bertignac, M.; Hadi-Saljoqi, S.; Chentli, I.; Cherouali, K.; Aouinti, S.; Hamwi, A.E.; Albani, A.; Elhassani, M.E.; Viart, B.; Goret, A.; Tran, A.; Sanou, G.; Rollin, M.; Duroux, P.; Kossida, S. IMGT® databases, related tools and web resources through three main axes of research and development. Nucleic Acids Research 2021, 50, D1262–D1272. [Google Scholar] [CrossRef]
- Li, W. Towards a General Intermolecular Binding Affinity Calculator 2022.
- Li, W.; Vottevor, G. Towards a Truly General Intermolecular Binding Affinity Calculator for Drug Discovery & Design 2023.
- Li, W. A one-dimensional semaglutide-GLP-1R-based mini static GIBAC 2024. [CrossRef]
- Marijic, J.; Neelankavil, J.P. Semaglutide: A New Medical Swiss Army Knife? Journal of Cardiothoracic and Vascular Anesthesia 2024, 38, 871–873. [Google Scholar] [CrossRef]
- Cowart, K. Oral Semaglutide: First-in-Class Oral GLP-1 Receptor Agonist for the Treatment of Type 2 Diabetes Mellitus. Annals of Pharmacotherapy 2019, 54, 478–485. [Google Scholar] [CrossRef]
- Knudsen, L.B.; Lau, J. The Discovery and Development of Liraglutide and Semaglutide. Frontiers in Endocrinology 2019, 10, 1–32. [Google Scholar]
- Wang, W.; Volkow, N.D.; Berger, N.A.; Davis, P.B.; Kaelber, D.C.; Xu, R. Association of semaglutide with risk of suicidal ideation in a real-world cohort. Nature Medicine 2024, 30, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Li, W. Designing Insulin Analogues with Lower Binding Affinity to Insulin Receptor than That of Insulin Icodec 2024. [CrossRef]
- Li, W. Delving Deep into the Structural Aspects of the BPro28-BLys29 Exchange in Insulin Lispro: A Structural Biophysical Lesson 2020.
- Kosiborod, M.N.; Petrie, M.C.; Borlaug, B.A.; Butler, J.; Davies, M.J.; Hovingh, G.K.; Kitzman, D.W.; Møller, D.V.; Treppendahl, M.B.; Verma, S.; Jensen, T.J.; Liisberg, K.; Lindegaard, M.L.; Abhayaratna, W.; Ahmed, F.Z.; Ben-Gal, T.; Chopra, V.; Ezekowitz, J.A.; Fu, M.; Ito, H.; Lelonek, M.; Melenovský, V.; Merkely, B.; Núñez, J.; Perna, E.; Schou, M.; Senni, M.; Sharma, K.; van der Meer, P.; Von Lewinski, D.; Wolf, D.; Shah, S.J. Semaglutide in Patients with Obesity-Related Heart Failure and Type 2 Diabetes. New England Journal of Medicine 2024, 390, 1394–1407. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.P.H. Semaglutide in weight management: Author’s reply. The Lancet 2019, 394, 1226–1227. [Google Scholar] [CrossRef] [PubMed]
- Bucheit, J.D.; Pamulapati, L.G.; Carter, N.; Malloy, K.; Dixon, D.L.; Sisson, E.M. Oral Semaglutide: A Review of the First Oral Glucagon-Like Peptide 1 Receptor Agonist. Diabetes Technology & Therapeutics 2020, 22, 10–18. [Google Scholar]
- Kanters, S.; Wilkinson, L.; Vrazic, H.; Sharma, R.; Lopes, S.; Popoff, E.; Druyts, E. Comparative efficacy of once-weekly semaglutide versus SGLT-2 inhibitors in patients inadequately controlled with one to two oral antidiabetic drugs: a systematic literature review and network meta-analysis. BMJ Open 2019, 9, e023458. [Google Scholar] [CrossRef]
- Granhall, C.; Donsmark, M.; Blicher, T.M.; Golor, G.; Sondergaard, F.L.; Thomsen, M.; Bakdal, T.A. Safety and Pharmacokinetics of Single and Multiple Ascending Doses of the Novel Oral Human GLP-1 Analogue, Oral Semaglutide, in Healthy Subjects and Subjects with Type 2 Diabetes. Clinical Pharmacokinetics 2018, 58, 781–791. [Google Scholar] [CrossRef]
- Garg, S.K.; Kaur, G.; Haider, Z.; Rodriquez, E.; Beatson, C.; Snell-Bergeon, J. Efficacy of Semaglutide in Overweight and Obese Patients with Type 1 Diabetes. Diabetes Technology & Therapeutics 2024, 26, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Europe, T.L.R.H. Semaglutide and beyond a turning point in obesity pharmacotherapy. The Lancet Regional Health Europe 2024, 37, 100860. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nature Structural & Molecular Biology 2003, 10, 980–980. [Google Scholar]
- Li, W. Half-a-century Burial of ρ, θ and φ in PDB 2021. [CrossRef]
- Li, W. Visualising the Experimentally Uncharted Territories of Membrane Protein Structures inside Protein Data Bank 2020.
- Li, W. A Local Spherical Coordinate System Approach to Protein 3D Structure Description 2020.
- Li, W. A Reversible Spherical Geometric Conversion of Protein Backbone Structure Coordinate Matrix to Three Independent Vectors of ρ, θ and φ 2024. [CrossRef]
- Li, W. Structurally Observed Electrostatic Features of the COVID-19 Coronavirus-Related Experimental Structures inside Protein Data Bank: A Brief Update 2020.
- Lau, J.; Bloch, P.; Schäffer, L.; Pettersson, I.; Spetzler, J.; Kofoed, J.; Madsen, K.; Knudsen, L.B.; McGuire, J.; Steensgaard, D.B.; Strauss, H.M.; Gram, D.X.; Knudsen, S.M.; Nielsen, F.S.; Thygesen, P.; Reedtz-Runge, S.; Kruse, T. Discovery of the Once-Weekly Glucagon-Like Peptide-1 (GLP-1) Analogue Semaglutide. Journal of Medicinal Chemistry 2015, 58, 7370–7380. [Google Scholar] [CrossRef]
- Ahrén, B.; Atkin, S.L.; Charpentier, G.; Warren, M.L.; Wilding, J.P.H.; Birch, S.; Holst, A.G.; Leiter, L.A. Semaglutide induces weight loss in subjects with type 2 diabetes regardless of baseline BMI or gastrointestinal adverse events in the SUSTAIN 1 to 5 trials. Diabetes, Obesity and Metabolism 2018, 20, 2210–2219. [Google Scholar] [CrossRef] [PubMed]
- Aroda, V.R.; Rosenstock, J.; Terauchi, Y.; Altuntas, Y.; Lalic, N.M.; Villegas, E.C.M.; Jeppesen, O.K.; Christiansen, E.; Hertz, C.L.; Haluzík, M. PIONEER 1: Randomized Clinical Trial of the Efficacy and Safety of Oral Semaglutide Monotherapy in Comparison With Placebo in Patients With Type 2 Diabetes. Diabetes Care 2019, 42, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Blüher, M.; Rosenstock, J.; Hoefler, J.; Manuel, R.; Hennige, A.M. Dose–response effects on HbA1c and bodyweight reduction of survodutide, a dual glucagon/GLP-1 receptor agonist, compared with placebo and open-label semaglutide in people with type 2 diabetes: a randomised clinical trial. Diabetologia 2023, 67, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.L.; Beutel, T.R.; Trujillo, J.M. Oral semaglutide in type 2 diabetes. Journal of Diabetes and its Complications 2020, 34, 107520. [Google Scholar] [CrossRef] [PubMed]
- Pieber, T.R.; Bode, B.; Mertens, A.; Cho, Y.M.; Christiansen, E.; Hertz, C.L.; Wallenstein, S.O.R.; Buse, J.B.; Akın, S.; Aladağ, N.; Arif, A.A.; Aronne, L.J.; Aronoff, S.; Ataoglu, E.; Baik, S.H.; Bays, H.; Beckett, P.L.; Berker, D.; Bilz, S.; Bode, B.; Braun, E.W.; Buse, J.B.; Canani, L.H.S.; Cho, Y.M.; Chung, C.H.; Colin, I.; Condit, J.; Cooper, J.; Delgado, B.; Eagerton, D.C.; Ebrashy, I.N.E.; Hefnawy, M.H.M.F.E.; Eliaschewitz, F.G.; Finneran, M.P.; Fischli, S.; Fließer-Görzer, E.; Geohas, J.; Godbole, N.A.; Golay, A.; de Lapertosa, S.G.; Gross, J.L.; Gulseth, H.L.; Helland, F.; Høivik, H.O.; Issa, C.; Kang, E.S.; Keller, C.; Khalil, S.H.A.; Kim, N.H.; Kim, I.J.; Klaff, L.J.; Laimer, M.; LaRocque, J.C.; Lederman, S.N.; Lee, K.W.; Litchfield, W.R.; Manning, M.B.; Mertens, A.; Morawski, E.J.; Murray, A.V.; Nicol, P.R.; O’Connor, T.M.; Oğuz, A.; Ong, S.; özdemir, A.; Palace, E.M.; Palchick, B.A.; Pereles-Ortiz, J.; Pieber, T.; Prager, R.; Preumont, V.; Riffer, E.; Rista, L.; Rudofsky, G.; Sarı, R.; Scheen, A.; Schultes, B.; Seo, J.A.; Shelbaya, S.A.; Sivalingam, K.; Sorli, C.H.; Stäuble, S.; Streja, D.A.; T’Sjoen, G.; Tetiker, T.; Gaal, L.V.; Vercammen, C.; Warren, M.L.; Weinstein, D.L.; Weiss, D.; White, A.; Winnie, M.; Wium, C.; Yavuz, D. Efficacy and safety of oral semaglutide with flexible dose adjustment versus sitagliptin in type 2 diabetes (PIONEER 7): a multicentre, open-label, randomised, phase 3a trial. The Lancet Diabetes & Endocrinology 2019, 7, 528–539. [Google Scholar]
- Zhang, X.; Belousoff, M.; Danev, R.; Sexton, P.; Wootten, D. Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein, 2021. [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Methods in Molecular Biology; Springer US, 2020; pp. 239–255.
- Vangone, A.; Bonvin, A.M. Contacts-based prediction of binding affinity in protein-protein complexes. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, p. btw514.
- Li, W. An Exhaustive Exploration of the Semaglutide-GLP-1R Sequence Space towards the Design of Semaglutide Analogues with Elevated Binding Affinity to GLP-1R 2024. [CrossRef]
- Li, W. In Silico Generation of Structural and Intermolecular Binding Affinity Data with Reasonable Accuracy: Expanding Horizons in Drug Discovery and Design 2024. [CrossRef]
- Li, W.; Vottevor, G. Towards a Truly General Intermolecular Binding Affinity Calculator for Drug Discovery & Design 2023. [CrossRef]
- Wang, Y.; Wang, L.; Shen, Y.; Wang, Y.; Yuan, H.; Wu, Y.; Gu, Q. Protein Conformation Generation via Force-Guided SE(3) Diffusion Models, 2024. [CrossRef]
- Callaway, E. The entire protein universe: AI predicts shape of nearly every known protein. Nature 2022. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; Lepore, R.; Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Hasani, H.J.; Barakat, K. Homology Modeling: an Overview of Fundamentals and Tools. International Review on Modelling and Simulations (IREMOS) 2017, 10, 129. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. Journal of Computational Chemistry 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.B.; Burch, J.D.; McKay, D.; Bustamante, C.; Crackower, M.A.; Wu, H. Could AlphaFold revolutionize chemical therapeutics? Nature Structural & Molecular Biology 2021, 28, 771–772. [Google Scholar]
- Gircha, A.I.; Boev, A.S.; Avchaciov, K.; Fedichev, P.O.; Fedorov, A.K. Hybrid quantum-classical machine learning for generative chemistry and drug design. Scientific Reports 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Roggia, M.; Natale, B.; Amendola, G.; Maro, S.D.; Cosconati, S. Streamlining Large Chemical Library Docking with Artificial Intelligence: the PyRMD2Dock Approach. Journal of Chemical Information and Modeling 2023. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Scientific Reports 2023, 13. [Google Scholar] [CrossRef]
- Zheng, Z.; Merz, K.M. Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. Journal of chemical theory and computation 2017, 13, 4751–4767. [Google Scholar]
- Deng, N.J.; Zheng, Q.; Liu, J.; Hao, G.F. Predicting protein–ligand binding affinity with a random matrix framework. PLoS computational biology 2012, 8, e1002775. [Google Scholar]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nature structural biology 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Li, W. Characterising the interaction between caenopore-5 and model membranes by NMR spectroscopy and molecular dynamics simulations. PhD thesis, University of Auckland, 2016.
- Canzar, S.; Toussaint, N.C.; Klau, G.W. An exact algorithm for side-chain placement in protein design. Optimization Letters 2011, 5, 393–406. [Google Scholar] [CrossRef]
- Herget, S.; Ranzinger, R.; Maass, K.; Lieth, C.W. GlycoCT—a unifying sequence format for carbohydrates. Carbohydrate Research 2008, 343, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.M.; Moreno, P.; Fabregat, A.; Hermjakob, H.; Steinbeck, C.; Apweiler, R.; Wakelam, M.J.O.; Vizcaíno, J.A. LipidHome: A Database of Theoretical Lipids Optimized for High Throughput Mass Spectrometry Lipidomics. PLoS ONE 2013, 8, e61951. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Subramaniam, S. Template-based combinatorial enumeration of virtual compound libraries for lipids. Journal of Cheminformatics 2012, 4. [Google Scholar] [CrossRef]
- Weiss, M. Design of ultra-stable insulin analogues for the developing world. Journal of Health Specialties 2013, 1, 59. [Google Scholar] [CrossRef]
- Subramaniam, S.; Kleywegt, G.J. A paradigm shift in structural biology. Nature Methods 2022, 19, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Bonvin, A.M.J.J. 50 years of PDB: a catalyst in structural biology. Nature Methods 2021, 18, 448–449. [Google Scholar] [CrossRef] [PubMed]
- Vakili, M.G.; Gorgulla, C.; Nigam, A.; Bezrukov, D.; Varoli, D.; Aliper, A.; Polykovsky, D.; Das, K.M.P.; Snider, J.; Lyakisheva, A.; Mansob, A.H.; Yao, Z.; Bitar, L.; Radchenko, E.; Ding, X.; Liu, J.; Meng, F.; Ren, F.; Cao, Y.; Stagljar, I.; Aspuru-Guzik, A.; Zhavoronkov, A. Quantum Computing-Enhanced Algorithm Unveils Novel Inhibitors for KRAS, 2024. [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Molecular Diversity 2021, 25, 1315–1360. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





