Submitted:
22 February 2024
Posted:
22 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. PCA
2.2. Data Curation
2.3. 3D Structure of Proteins
3. Results
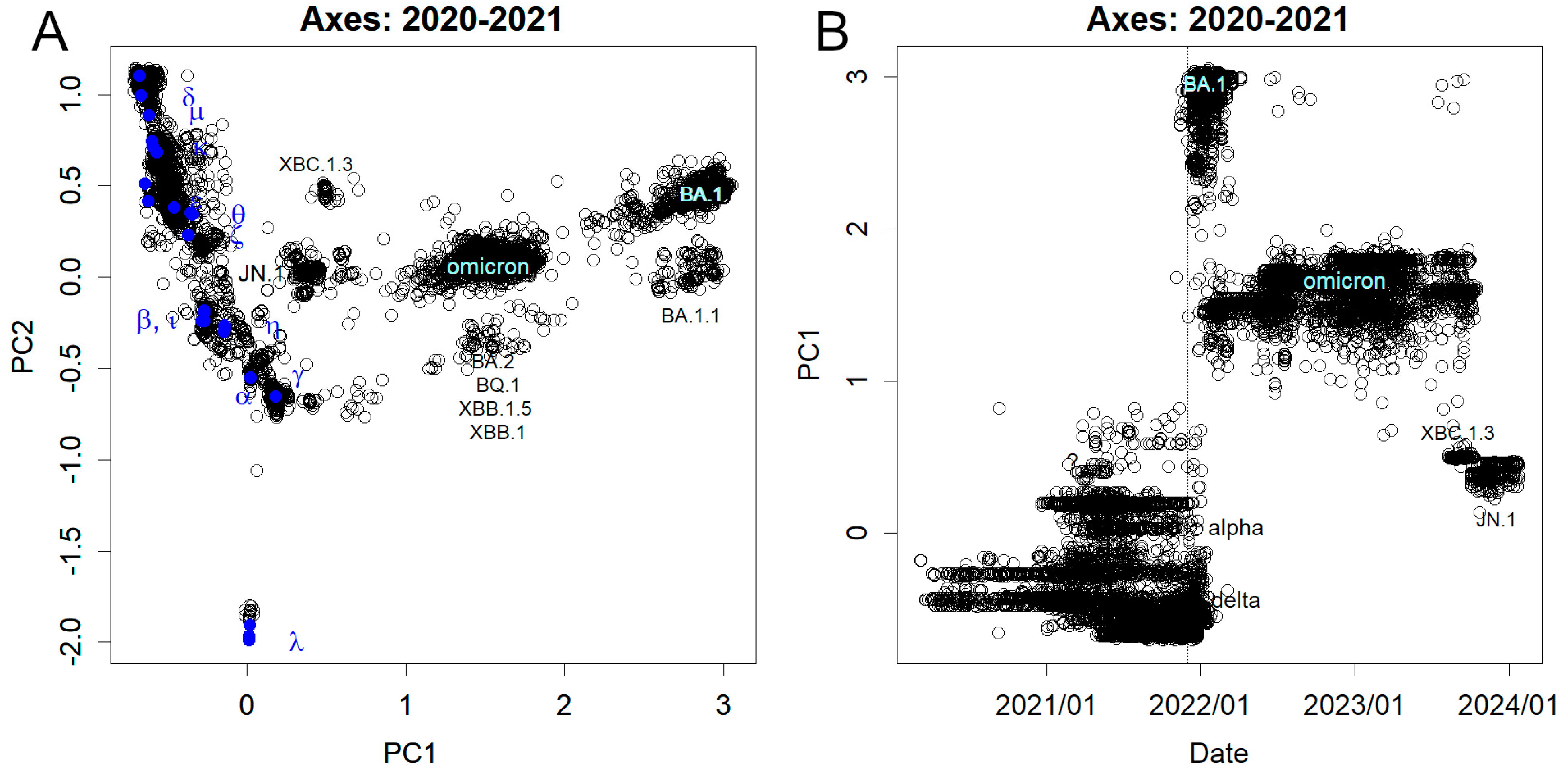
3.1. Mutations Observed from Nucleotide Sequence
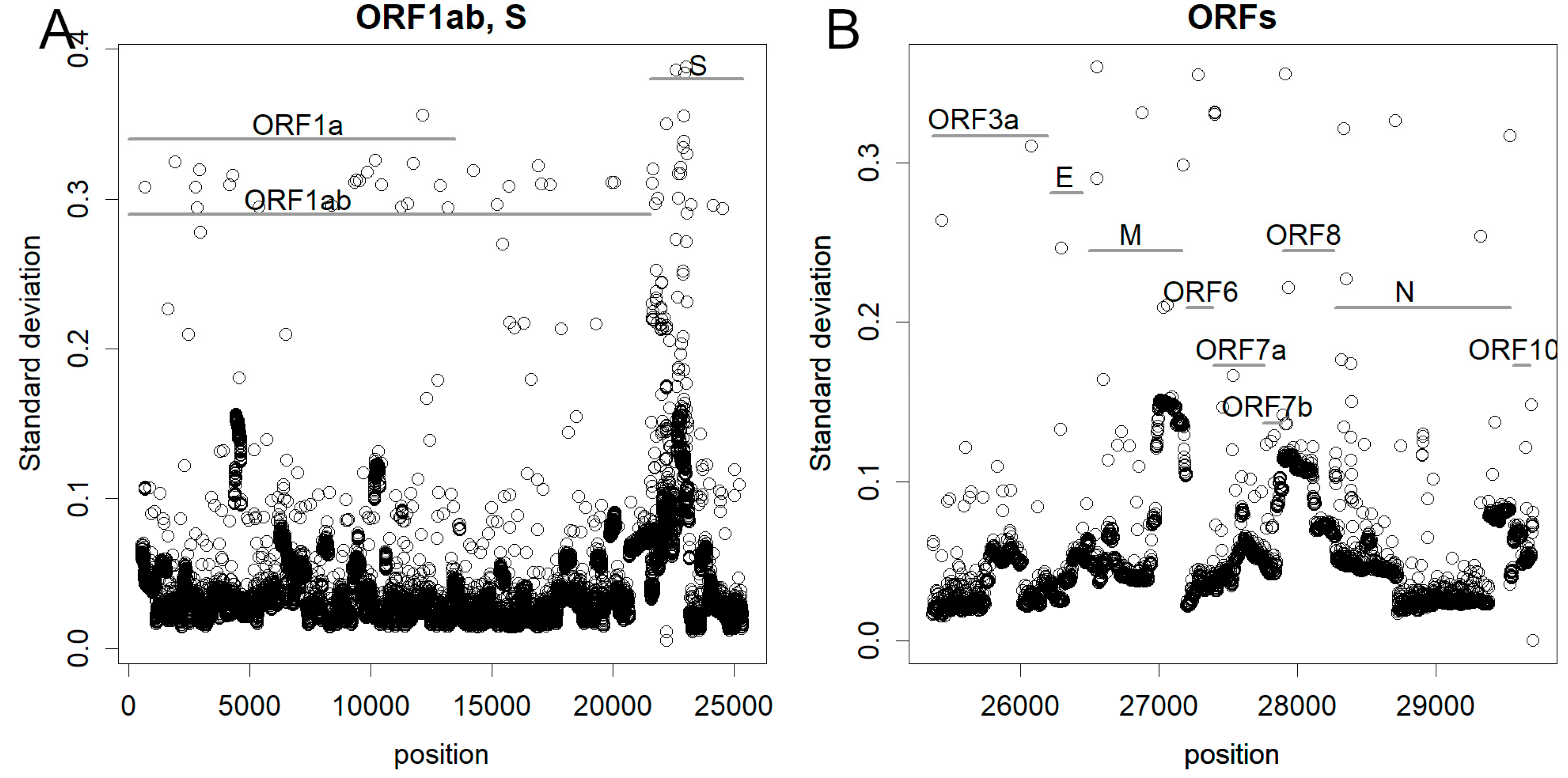
3.2. Sites of Mutation in the Genome
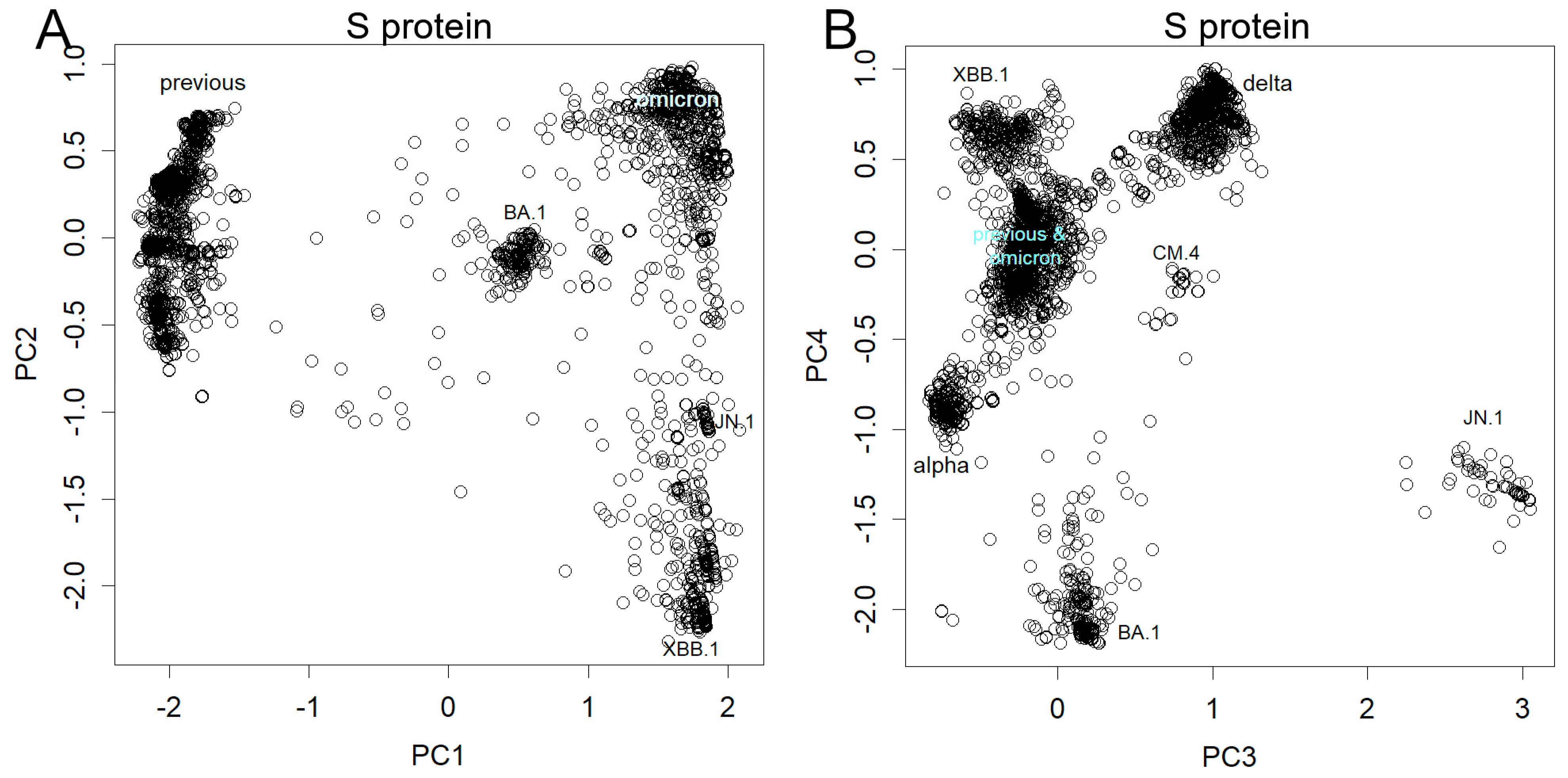
3.3. Mutations Observed in Spike Protein
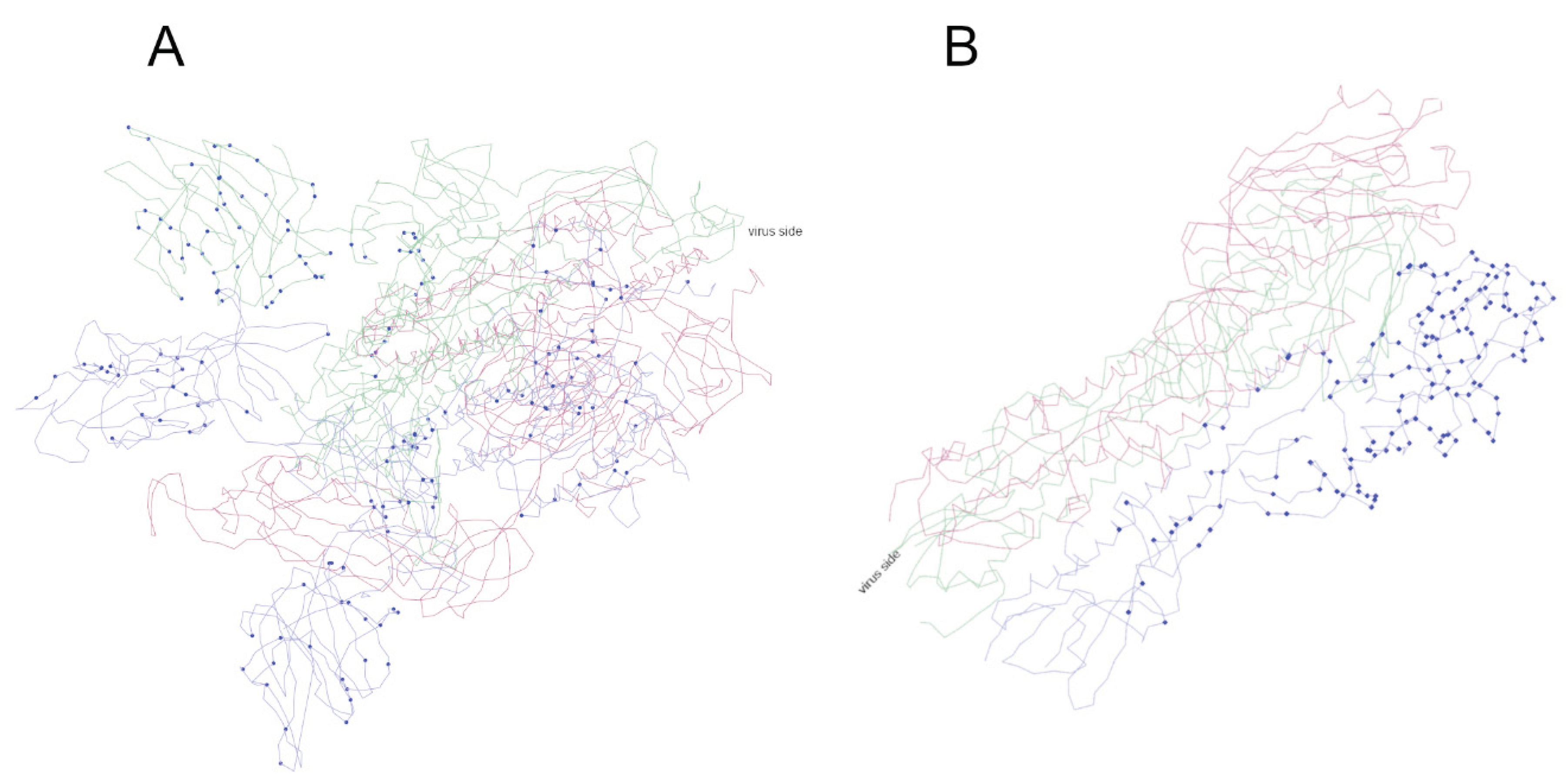
3.4. Mutated Sites in Spike Proteins in the 3D Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Coronavirus disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 23 May).
- Konishi, T. Mutations in SARS-CoV-2 are on the increase against the acquired immunity. PLoS ONE 2022, 17, e0271305. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T.; Matsukuma, S.; Fuji, H.; Nakamura, D.; Satou, N.; Okano, K. Principal Component Analysis applied directly to Sequence Matrix. Scientific Reports 2019, 9, 19297. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://cran.r-project.org/ (accessed on 19 February 2024).
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics 2015, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T. Re-evaluation of the evolution of influenza H1 viruses using direct PCA. Scientific Reports 2019, 9, 19287. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.H., Zhang, Y.Y., Li, Y.N., Ye, F.F., Guo, Y.Y., Xia, L., Zhong, X.Y., Chi, X.M., Zhou, Q. S protein of SARS-CoV-2 in the active conformation. Available online: https://doi.org/10.2210/pdb7DWZ/pdb (accessed on 19 February 2024).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Research 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Yan, R., Zhang, Y., Li, Y., Ye, F., Guo, Y., Xia, L., Zhong, X., Chi, X., Zhou, Q. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res 2021, 31, 717–719. [CrossRef] [PubMed]
- Murdoch, D.; Adler, D.; Nenadic, O.; Urbanek, S.; Chen, M.; Gebhardt, A.; Bolker, B.; Csardi, G.; Strzelecki, A.; Senger, A.; et al. rgl: 3D Visualization Using OpenGL. Available online: https://cran.r-project.org/web/packages/rgl/index.html (accessed on 19 February 2024).
- Konishi, T. Continuous mutation of SARS-CoV-2 during migration via three routes. PeerJ 2022, 10, e12681. [Google Scholar] [CrossRef]
- Konishi, T. Principal component analysis for designed experiments. BMC Bioinformatics 2015, 16 (Suppl. 18), S7. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T. A Comparative Analysis of COVID-19 Response Measures and Their Impact on Mortality Rate. COVID 2024, 4, 130–150. [Google Scholar] [CrossRef]
- Konishi, T. COVID-19 Epidemics Monitored Through the Logarithmic Growth Rate and SIR Model. J Clin Immunol Microbiol 2022, 3, 1–45. [Google Scholar] [CrossRef]
- Suryamohan, K.; Diwanji, D.; Stawiski, E.W.; Gupta, R.; Miersch, S.; Liu, J.; Chen, C.; Jiang, Y.-P.; Fellouse, F.A.; Sathirapongsasuti, J.F.; et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Communications Biology 2021, 4, 475. [Google Scholar] [CrossRef] [PubMed]
- Sano, E.; Deguchi, S.; Sakamoto, A.; Mimura, N.; Hirabayashi, A.; Muramoto, Y.; Noda, T.; Yamamoto, T.; Takayama, K. Modeling SARS-CoV-2 infection and its individual differences with ACE2-expressing human iPS cells. iScience 2021, 24, 102428. [Google Scholar] [CrossRef]
- Gao, F.X.; Wu, R.X.; Shen, M.Y.; Huang, J.J.; Li, T.T.; Hu, C.; Luo, F.Y.; Song, S.Y.; Mu, S.; Hao, Y.N.; et al. Extended SARS-CoV-2 RBD booster vaccination induces humoral and cellular immune tolerance in mice. iScience 2022, 25, 105479. [Google Scholar] [CrossRef] [PubMed]
- Gruell, H.; Vanshylla, K.; Tober-Lau, P.; Hillus, D.; Schommers, P.; Lehmann, C.; Kurth, F.; Sander, L.E.; Klein, F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nature Medicine 2022. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nature Reviews Microbiology 2021. [Google Scholar] [CrossRef] [PubMed]
- Irrgang, P.; Gerling, J.; Kocher, K.; Lapuente, D.; Steininger, P.; Habenicht, K.; Wytopil, M.; Beileke, S.; Schäfer, S.; Zhong, J.; et al. Class switch toward noninflammatory, spike-specific IgG4 antibodies after repeated SARS-CoV-2 mRNA vaccination. Science Immunology 2023, 8, eade2798. [Google Scholar] [CrossRef] [PubMed]
- Kiszel, P.; Sík, P.; Miklós, J.; Kajdácsi, E.; Sinkovits, G.; Cervenak, L.; Prohászka, Z. Class switch towards spike protein-specific IgG4 antibodies after SARS-CoV-2 mRNA vaccination depends on prior infection history. Scientific Reports 2023, 13, 13166. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Redwan, E.M.; Makis, W.; Rubio-Casillas, A. IgG4 Antibodies Induced by Repeated Vaccination May Generate Immune Tolerance to the SARS-CoV-2 Spike Protein. Vaccines (Basel) 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T. SARS-CoV-2 mutations among minks show reduced lethality and infectivity to humans. PLOS ONE 2021, 16, e0247626. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Infectious Diseases, Japan. New Coronavirus Infectious Disease Surveillance News/Weekly Report: Understand the status of outbreak trends. Available online: https://www.niid.go.jp/niid/ja/2019-ncov/2484-idsc/12015-covid19-surveillance-report.html (accessed on 19 February 2024).
- Kathy, K. 3 Things to Know About JN.1, the New Coronavirus Strain. Yale Medicine, News. Available online: https://www.yalemedicine.org/news/jn1-coronavirus-variant-covid (accessed on 22 February 2024).
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A., Jr.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. The Lancet Infectious Diseases 2024, 24, e82. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




