Submitted:
08 February 2024
Posted:
09 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
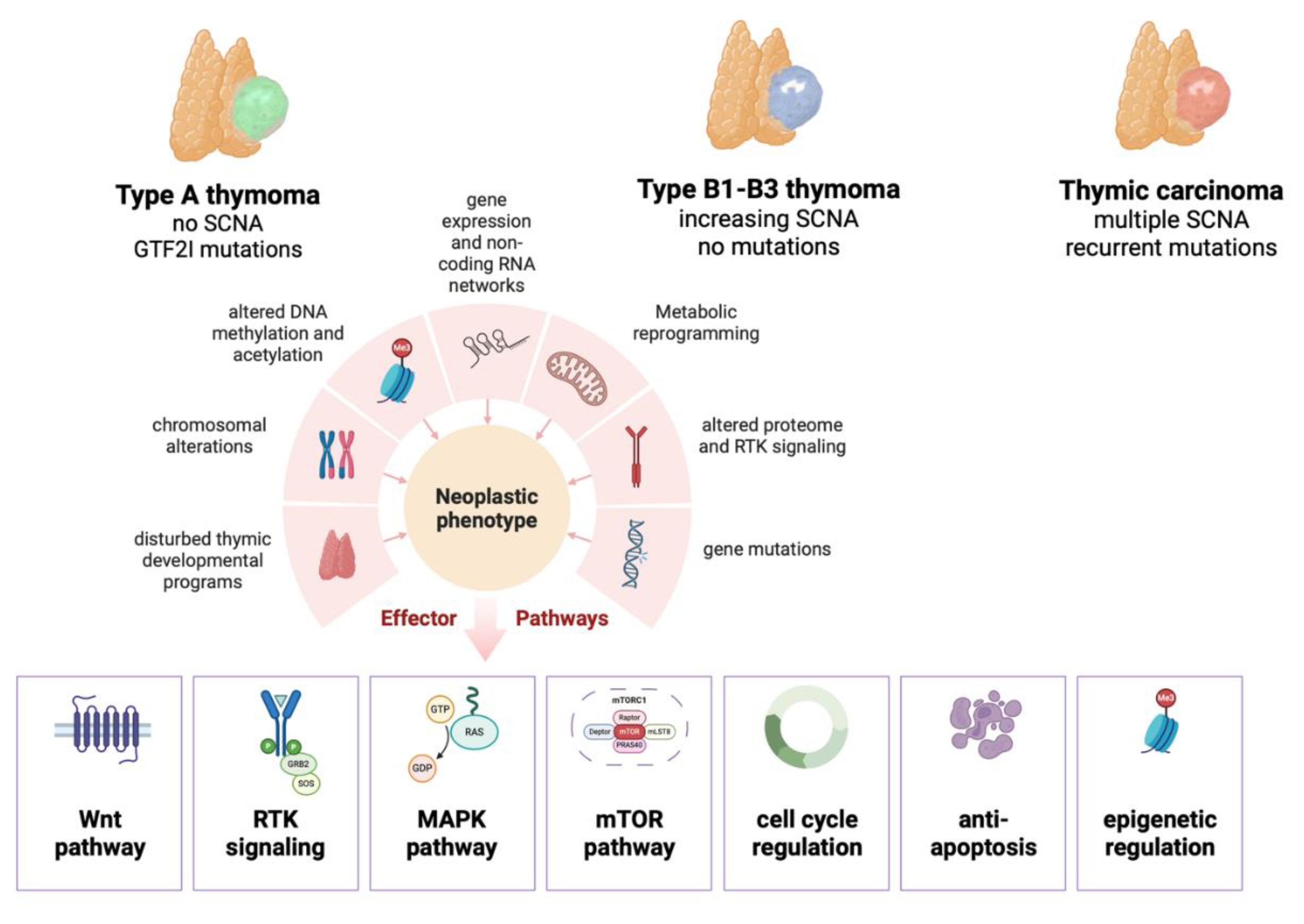
Disturbed thymic developmental programs in thymomas
Functional implications of chromosomal alterations in thymomas
Altered DNA methylation and acetylation in thymomas
Altered gene expression and non-coding RNA networks in thymomas
Metabolic reprogramming in thymomas
Proteomics and altered tyrosine kinase signaling
Conclusions and future directions
References
- Buckley, C.; Douek, D.; Newsom-Davis, J.; Vincent, A.; Willcox, N. Mature, Long-Lived Cd4+ and Cd8+ T Cells Are Generated by the Thymoma in Myasthenia Gravis. Ann. Neurol. 2001, 50, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Marx, A., F. Detterbeck, E.M. Marom, P. Ströbel, and A. Rajan. "Tumors of the Thymus: Introduction." In Who Classification of Tumours. Thoracic Tumours., edited by A. Marx and J.K.C. Chan. Lyon: International Agency for Research on Cancer, 2021.
- Marx, A.; Porubsky, S.; Belharazem, D.; Saruhan-Direskeneli, G.; Schalke, B.; Ströbel, P.; Weis, C.-A. Thymoma related myasthenia gravis in humans and potential animal models. Exp. Neurol. 2015, 270, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Radovich, M.; Pickering, C.R.; Felau, I.; Ha, G.; Zhang, H.; Jo, H.; Hoadley, K.A.; Anur, P.; Zhang, J.; McLellan, M.; et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell 2018, 33, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Venuta, F.; A Rendina, E.; O Pescarmona, E.; De Giacomo, T.; Vegna, M.L.; Fazi, P.; Flaishman, I.; Guarino, E.; Ricci, C. Multimodality Treatment of Thymoma: A Prospective Study. Ann. Thorac. Surg. 1997, 64, 1585–1592. [Google Scholar] [CrossRef]
- Petrini, I.; Meltzer, P.S.; Kim, I.-K.; Lucchi, M.; Park, K.-S.; Fontanini, G.; Gao, J.; A Zucali, P.; Calabrese, F.; Favaretto, A.; et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nat. Genet. 2014, 46, 844–849. [Google Scholar] [CrossRef]
- Ströbel, P.; Hartmann, E.; Rosenwald, A.; Kalla, J.; Ott, G.; Friedel, G.; Schalke, B.; Kasahara, M.; Tomaru, U.; Marx, A. Corticomedullary differentiation and maturational arrest in thymomas. Histopathology 2013, 64, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Giorgetti, O.B.; Nusser, A.; Boehm, T. Human thymoma-associated mutation of the GTF2I transcription factor impairs thymic epithelial progenitor differentiation in mice. Commun. Biol. 2022, 5, 1–12. [Google Scholar] [CrossRef]
- Ströbel, P.; Murumägi, A.; Klein, R.; Luster, M.; Lahti, M.; Krohn, K.; Schalke, B.; Nix, W.; Gold, R.; Rieckmann, P.; et al. Deficiency of the autoimmune regulator AIRE in thymomas is insufficient to elicit autoimmune polyendocrinopathy syndrome type 1 (APS-1). J. Pathol. 2007, 211, 563–571. [Google Scholar] [CrossRef]
- Bleul, C.C.; Corbeaux, T.; Reuter, A.; Fisch, P.; Mönting, J.S.; Boehm, T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 2006, 441, 992–996. [Google Scholar] [CrossRef]
- Rossi, S.W.; Jenkinson, W.E.; Anderson, G.; Jenkinson, E.J. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 2006, 441, 988–991. [Google Scholar] [CrossRef]
- Bautista, J.L.; Cramer, N.T.; Miller, C.N.; Chavez, J.; Berrios, D.I.; Byrnes, L.E.; Germino, J.; Ntranos, V.; Sneddon, J.B.; Burt, T.D.; et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lepletier, A.; Hun, M.L.; Hammett, M.V.; Wong, K.; Naeem, H.; Hedger, M.; Loveland, K.; Chidgey, A.P. Interplay between Follistatin, Activin A, and BMP4 Signaling Regulates Postnatal Thymic Epithelial Progenitor Cell Differentiation during Aging. Cell Rep. 2019, 27, 3887–3901. [Google Scholar] [CrossRef] [PubMed]
- Balciunaite, G.; Keller, M.P.; Balciunaite, E.; Piali, L.; Zuklys, S.; Mathieu, Y.D.; Gill, J.; Boyd, R.; Sussman, D.J.; Hollander, G.A. Wnt Glycoproteins Regulate the Expression of Foxn1, the Gene Defective in Nude Mice. Nat. Immunol. 2002, 3, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.; Patel, S.R.; Mishina, Y.; Manley, N.R. Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev. Biol. 2010, 339, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Bleul, C.C.; Boehm, T. BMP Signaling Is Required for Normal Thymus Development. J. Immunol. 2005, 175, 5213–5221. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, N.; Guo, Z.; Chi, F.; Song, Y.; Zhu, X. Wnt4 signaling is associated with the decrease of proliferation and increase of apoptosis during age-related thymic involution. Mol. Med. Rep. 2015, 12, 7568–7576. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, K.M.; Vanegas, J.R.; Brochu, S.; Shan, J.; Vainio, S.J.; Perreault, C. Wnt4 regulates thymic cellularity through the expansion of thymic epithelial cells and early thymic progenitors. Blood 2011, 118, 5163–5173. [Google Scholar] [CrossRef] [PubMed]
- Varecza, Z.; Kvell, K.; Talabér, G.; Miskei, G.; Csongei, V.; Bartis, D.; Anderson, G.; Jenkinson, E.J.; Pongracz, J.E. Multiple suppression pathways of canonical Wnt signalling control thymic epithelial senescence. Mech. Ageing Dev. 2011, 132, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Kvell, K.; Varecza, Z.; Bartis, D.; Hesse, S.; Parnell, S.; Anderson, G.; Jenkinson, E.J.; Pongracz, J.E. Wnt4 and LAP2alpha as Pacemakers of Thymic Epithelial Senescence. PLoS ONE 2010, 5, e10701. [Google Scholar] [CrossRef]
- Zhang, X.; Schalke, B.; Kvell, K.; Kriegsmann, K.; Kriegsmann, M.; Graeter, T.; Preissler, G.; Ott, G.; Kurz, K.; Bulut, E.; et al. WNT4 overexpression and secretion in thymic epithelial tumors drive an autocrine loop in tumor cells in vitro. Front. Oncol. 2022, 12, 920871. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, P.; Tang, P.; Lv, P.; Li, X.; Wang, Y.; Lv, Y.; Liu, Y. Wnt4 overexpression promotes thymoma development through a JNK-mediated planar cell polarity-like pathway. Oncol. Lett. 2017, 15, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Nehls, M.; Kyewski, B.; Messerle, M.; Waldschütz, R.; Schüddekopf, K.; Smith, A.J.H.; Boehm, T. Two Genetically Separable Steps in the Differentiation of Thymic Epithelium. Science 1996, 272, 886–889. [Google Scholar] [CrossRef]
- Corbeaux, T.; Hess, I.; Swann, J.B.; Kanzler, B.; Haas-Assenbaum, A.; Boehm, T. Thymopoiesis in mice depends on a Foxn1 -positive thymic epithelial cell lineage. Proc. Natl. Acad. Sci. USA 2010, 107, 16613–16618. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wachsmuth, L.P.; Xiao, S.; Condie, B.G.; Manley, N.R. Foxn1 Overexpression Promotes Thymic Epithelial Progenitor Cell Proliferation and Mtec Maintenance, but Does Not Prevent Thymic Involution. Development 2023, 150, 8. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, P.; Liu, Y.; Lv, P.; Wang, Y.; Chen, Y. In vitro study of the effect of small interfering ribonucleic acid on the expression of FOXN1 and B cell-attracting chemokine 1 in thymoma cell lines. Thorac. Cancer 2015, 6, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, X.; Liu, Y.; Wang, Y.; Wang, H.; Lu, C.; Zhang, P. Decreased Wnt4 expression inhibits thymoma development through downregulation of FoxN1. J. Thorac. Dis. 2017, 9, 1574–1583. [Google Scholar] [CrossRef]
- Nonaka, D.; Henley, J.D.; Chiriboga, L.; Yee, H. Diagnostic Utility of Thymic Epithelial Markers CD205 (DEC205) and Foxn1 in Thymic Epithelial Neoplasms. Am. J. Surg. Pathol. 2007, 31, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Goto, T.; Hirotsu, Y.; Yokoyama, Y.; Nakagomi, T.; Otake, S.; Amemiya, K.; Oyama, T.; Mochizuki, H.; Omata, M. Primary Driver Mutations in GTF2I Specific to the Development of Thymomas. Cancers 2020, 12, 2032. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Kim, I.-K.; Bian, J.; Polyzos, A.; Di Giammartino, D.C.; Zhang, Y.-W.; Luo, J.; Hernandez, M.O.; Kedei, N.; Cam, M.; et al. A Knock-In Mouse Model of Thymoma With the GTF2I L424H Mutation. J. Thorac. Oncol. 2022, 17, 1375–1386. [Google Scholar] [CrossRef]
- Kim, I.-K.; Rao, G.; Zhao, X.; Fan, R.; Avantaggiati, M.L.; Wang, Y.; Zhang, Y.-W.; Giaccone, G. Mutant GTF2I induces cell transformation and metabolic alterations in thymic epithelial cells. Cell Death Differ. 2020, 27, 2263–2279. [Google Scholar] [CrossRef]
- Cavallo, F.; Troglio, F.; Faga, G.; Fancelli, D.; Shyti, R.; Trattaro, S.; Zanella, M.; G. D'Agostino; Hughes, J.M.; Cera, M.R.; Pasi, M.; Gabriele, M.; Lazzarin, M.; Mihailovich, M.; Kooy, F.; Rosa, A.; Mercurio, C.; Varasi, M.; Testa, G. "High-Throughput Screening Identifies Histone Deacetylase Inhibitors That Modulate Gtf2i Expression in 7q11.23 Microduplication Autism Spectrum Disorder Patient-Derived Cortical Neurons. Mol. Autism. 2020, 11, 88. [Google Scholar] [PubMed]
- Adamo, A.; Atashpaz, S.; Germain, P.-L.; Zanella, M.; D'Agostino, G.; Albertin, V.; Chenoweth, J.; Micale, L.; Fusco, C.; Unger, C.; et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 2014, 47, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Zettl, A.; Ströbel, P.; Wagner, K.; Katzenberger, T.; Ott, G.; Rosenwald, A.; Peters, K.; Krein, A.; Semik, M.; Müller-Hermelink, H.-K.; et al. Recurrent Genetic Aberrations in Thymoma and Thymic Carcinoma. Am. J. Pathol. 2000, 157, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I.; Meltzer, P.S.; A Zucali, P.; Luo, J.; Lee, C.; Santoro, A.; Lee, H.S.; Killian, K.J.; Wang, Y.; Tsokos, M. Copy number aberrations of BCL2 and CDKN2A/B identified by array-CGH in thymic epithelial tumors. Cell Death Dis. 2012, 3, e351. [Google Scholar] [CrossRef]
- Copy Number Aberrations of Bcl2 and Cdkn2a/B Identified by Array-Cgh in Thymic Epithelial Tumors. Cell Death Dis. 2012, 3, e351. [CrossRef] [PubMed]
- Sherr, C.J. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001, 2, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Girard, N.; Basse, C.; Schrock, A.; Ramkissoon, S.; Killian, K.; Ross, J.S. Comprehensive Genomic Profiling of 274 Thymic Epithelial Tumors Unveils Oncogenic Pathways and Predictive Biomarkers. Oncol. 2022, 27, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, K.; Shukuya, T.; Greenstein, R.; Kaplan, B.; Wakelee, H.; Ross, J.; Miura, K.; Furuta, K.; Kato, S.; Suh, J.; et al. Genomic characterization of thymic epithelial tumors in a real-world dataset. ESMO Open 2023, 8, 101627. [Google Scholar] [CrossRef] [PubMed]
- Rubin, S.M. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 2013, 38, 12–19. [Google Scholar] [CrossRef]
- Jiang, Y.-Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.-D.; Liu, Y.-R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440. [Google Scholar] [CrossRef]
- Hirose, Y.; Kondo, K.; Takizawa, H.; Nagao, T.; Nakagawa, Y.; Fujino, H.; Toba, H.; Kenzaki, K.; Sakiyama, S.; Tangoku, A. Aberrant methylation of tumour-related genes in thymic epithelial tumours. Lung Cancer 2009, 64, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, H.; Fujii, Y.; Sakaguchi, M.; Tanaka, H.; Yoon, H.E.; Komoto, Y.; Inoue, M.; Miyoshi, S.; Matsuda, H. P16ink4, Prb, P53 and Cyclin D1 Expression and Hypermethylation of Cdkn2 Gene in Thymoma and Thymic Carcinoma. Int. J. Cancer 1997, 73, 639–644. [Google Scholar] [CrossRef]
- Lal, A.; Kim, H.H.; Abdelmohsen, K.; Kuwano, Y.; Pullmann, R.; Srikantan, S.; Subrahmanyam, R.; Martindale, J.L.; Yang, X.; Ahmed, F.; et al. p16INK4a Translation Suppressed by miR-24. PLoS ONE 2008, 3, e1864. [Google Scholar] [CrossRef] [PubMed]
- Aesif, S.W.; Aubry, M.C.; Yi, E.S.; Kloft-Nelson, S.M.; Jenkins, S.M.; Spears, G.M.; Greipp, P.T.; Sukov, W.R.; Roden, A.C. Loss of p16 INK4A Expression and Homozygous CDKN2A Deletion Are Associated with Worse Outcome and Younger Age in Thymic Carcinomas. J. Thorac. Oncol. 2017, 12, 860–871. [Google Scholar] [CrossRef]
- Stefanaki, K.; Rontogianni, C. H. Kouvidou, S. Bolioti. Expression of P53, Mdm2, P21/Waf1 and Bcl-2 Proteins in Thymomas. Histopathology 1997, 30, 549–55. [Google Scholar] [CrossRef] [PubMed]
- Pan, C. C.; Chen, L. S. Wang. Expression of Apoptosis-Related Markers and Her-2/Neu in Thymic Epithelial Tumours. Histopathology 2003, 43, 165–72. [Google Scholar] [CrossRef]
- Khoury, T.; Arshad, A.; Bogner, P.; Ramnath, N.; Zhang, S.; Chandrasekhar, R.; Wilding, G.; Alrawi, S.; Tan, D. Apoptosis-Related (Survivin, Bcl-2), Tumor Suppressor Gene (p53), Proliferation (Ki-67), and Non-Receptor Tyrosine Kinase (Src) Markers Expression and Correlation With Clinicopathologic Variables in 60 Thymic Neoplasms. Chest 2009, 136, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Lozada-Nur, F. HIV: common early oral markers. J. Calif. Dent. Assoc. 1989, 17, 36–41. [Google Scholar]
- Chen, F.; Yan, J.; Chang, K.; Lai, W.; Chen, R.; Jin, Y. Immunohistochemical localization of Mcl-1 and bcl-2 proteins in thymic epithelial tumours. Histopathology 1996, 29, 541–547. [Google Scholar] [CrossRef]
- Huang, B.; Belharazem, D.; Li, L.; Kneitz, S.; Schnabel, P.A.; Rieker, R.J.; Körner, D.; Nix, W.; Schalke, B.; Müller-Hermelink, H.K.; et al. Anti-Apoptotic Signature in Thymic Squamous Cell Carcinomas – Functional Relevance of Anti-Apoptotic BIRC3 Expression in the Thymic Carcinoma Cell Line 1889c. Front. Oncol. 2013, 3. [Google Scholar] [CrossRef]
- Belharazem, D., A. Grass, C. Paul, M. Vitacolonna, B. Schalke, R. J. Rieker, D. Korner, P. Jungebluth, K. Simon-Keller, P. Hohenberger, E. M. Roessner, K. Wiebe, T. Grater, T. Kyriss, G. Ott, P. Geserick, M. Leverkus, P. Strobel, and A. Marx. Increased Cflip Expression in Thymic Epithelial Tumors Blocks Autophagy Via Nf-Kappab Signalling. Oncotarget 2017, 8, 89580–94. [Google Scholar] [PubMed]
- Muller, D.; Mazzeo, R.; Koch. Functional Apoptosis Profiling Identifies Mcl-1 and Bcl-Xl as Prognostic Markers and Therapeutic Targets in Advanced Thymomas and Thymic Carcinomas. BMC Med. 2021, 19, 300. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Ren, R.; Fang, X. Identification and Characterization of Non-Coding RNAs in Thymoma. Med. Sci. Monit. 2021, 27, e929727–1. [Google Scholar] [CrossRef] [PubMed]
- Massoth, L. R., Y. P. Hung, D. Dias-Santagata, M. Onozato, N. Shah, E. Severson, D. Duncan, B. J. Gillespie, N. F. Williams, J. S. Ross, J. A. Vergilio, S. K. Harkins, K. Glomski, V. Nardi, L. R. Zukerberg, R. P. Hasserjian, A. Louissaint, Jr., and E. A. Williams. Pan-Cancer Landscape Analysis Reveals Recurrent Kmt2a-Maml2 Gene Fusion in Aggressive Histologic Subtypes of Thymoma. JCO Precis. Oncol. 2020, 4. [Google Scholar]
- Milne, T.A.; Briggs, S.D.; Brock, H.W.; Martin, M.E.; Gibbs, D.; Allis, C.D.; Hess, J.L. MLL Targets SET Domain Methyltransferase Activity to Hox Gene Promoters. Mol. Cell 2002, 10, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Wächter, K.; Kowarz, E.; Marschalek, R. Functional characterisation of different MLL fusion proteins by using inducible Sleeping Beauty vectors. Cancer Lett. 2014, 352, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, N.; Suzukawa, K.; Shimizu, S.; Shinagawa, A.; Takei, N.; Taki, T.; Hayashi, Y.; Kojima, H.; Kawakami, Y.; Nagasawa, T. Identification of a novel fusion gene MLL-MAML2 in secondary acute myelogenous leukemia and myelodysplastic syndrome with inv(11)(q21q23). Genes Chromosom. Cancer 2007, 46, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Salame, H.; Mckey, R.; Ballout, M.; Saad, W. The First Reported Case of Neurotrophic Tyrosine Receptor Kinase Fusion-Positive Thymoma Treated Successfully With Entrectinib. Cureus 2021, 13, e20588. [Google Scholar] [CrossRef]
- Chen, C.; Yin, N.; Yin, B.; Lu, Q. DNA methylation in thoracic neoplasms. Cancer Lett. 2011, 301, 7–16. [Google Scholar] [CrossRef]
- Nicolì, V.; Coppedè, F. Epigenetics of Thymic Epithelial Tumors. Cancers 2023, 15, 360. [Google Scholar] [CrossRef]
- Psilopatis, I.; Pergaris, A.; Vrettou, K.; Theocharis, S.; Troungos, C. Thymic Epithelial Neoplasms: Focusing on the Epigenetic Alterations. Int. J. Mol. Sci. 2022, 23, 4045. [Google Scholar] [CrossRef]
- Gaiser, T.; Hirsch, D.; Porth, I.; Sahm, F.; Ströbel, P.; von Deimling, A.; Marx, A. DNA-Methylation Analysis as a Tool for Thymoma Classification. Cancers 2022, 14, 5876. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Meng, Y.; Niu, Y.; Li, S.; Liu, H.; He, J.; Zhang, Y.; Liang, N.; Liu, L.; Mao, X.; et al. Genome-wide DNA methylation profile of thymomas and potential epigenetic regulation of thymoma subtypes. Oncol. Rep. 2019, 41, 2762–2774. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yuan, Y.; Xiao, H.; Dai, J.; Ye, Y.; Zhang, Q.; Zhang, Z.; Jiang, Y.; Luo, J.; Hu, J.; et al. Discovery and validation of DNA methylation markers for overall survival prognosis in patients with thymic epithelial tumors. Clin. Epigenetics 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yin, B.; Wei, Q.; Li, D.; Hu, J.; Yu, F.; Lu, Q. Aberrant DNA Methylation in Thymic Epithelial Tumors. Cancer Investig. 2009, 27, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349. [Google Scholar] [CrossRef] [PubMed]
- Palamaris, K.; Tzimou, L.-M.; Levidou, G.; Masaoutis, C.; Theochari, I.; Rontogianni, D.; Theocharis, S. Histone Deacetylases (HDACs): Promising Biomarkers and Potential Therapeutic Targets in Thymic Epithelial Tumors. Int. J. Mol. Sci. 2023, 24, 4263. [Google Scholar] [CrossRef] [PubMed]
- Arjonen, A.; Mäkelä, R.; Härmä, V.; Rintanen, N.; Kuopio, T.; Kononen, J.; Rantala, J.K. Image-based ex vivo drug screen to assess targeted therapies in recurrent thymoma. Lung Cancer 2020, 145, 27–32. [Google Scholar] [CrossRef]
- Giaccone, G.; Rajan, A.; Berman, A.; Kelly, R.J.; Szabo, E.; Lopez-Chavez, A.; Trepel, J.; Lee, M.-J.; Cao, L.; Espinoza-Delgado, I.; et al. Phase II Study of Belinostat in Patients With Recurrent or Refractory Advanced Thymic Epithelial Tumors. J. Clin. Oncol. 2011, 29, 2052–2059. [Google Scholar] [CrossRef]
- Thomas, A.; Rajan, E.; Szabo, Y. Tomita, C. A. Carter. A Phase I/Ii Trial of Belinostat in Combination with Cisplatin, Doxorubicin, and Cyclophosphamide in Thymic Epithelial Tumors: A Clinical and Translational Study. Clin. Cancer Res. 2014, 20, 5392–402. [Google Scholar] [CrossRef]
- Badve, S.; Goswami, C.; Gökmen–Polar, Y.; Nelson, R.P.; Henley, J.; Miller, N.; Zaheer, N.A.; Sledge, G.W.; Li, L.; Kesler, K.A.; et al. Molecular Analysis of Thymoma. PLoS ONE 2012, 7, e42669. [Google Scholar] [CrossRef] [PubMed]
- Gokmen-Polar, Y., R. W. Cook, C. P. Goswami, J. Wilkinson, D. Maetzold, J. F. Stone, K. M. Oelschlager, I. T. Vladislav, K. L. Shirar, K. A. Kesler, P. J. Loehrer, Sr., and S. Badve. A Gene Signature to Determine Metastatic Behavior in Thymomas. PLoS ONE 2013, 8, e66047. [Google Scholar]
- Girard, N.; Shen, R.; Guo, T.; Zakowski, M.F.; Heguy, A.; Riely, G.J.; Huang, J.; Lau, C.; Lash, A.E.; Ladanyi, M.; Viale, A.; Antonescu, C.R.; Travis, W.D.; Rusch, V.W.; Kris, M.G.; Pao, W. Comprehensive Genomic Analysis Reveals Clinically Relevant Molecular Distinctions between Thymic Carcinomas and Thymomas." Clin. Cancer Res. 2009, 15, 6790–9.
- Sasaki, H.; Ide, N.; Fukai, I.; Kiriyama, M.; Yamakawa, Y.; Fujii, Y. Gene expression analysis of human thymoma correlates with tumor stage. Int. J. Cancer 2002, 101, 342–347. [Google Scholar] [CrossRef]
- Liang, N.; Liu, L.; Huang, C.; Liu, H.; Guo, C.; Li, J.; Wang, W.; Li, N.; Lin, R.; Wang, T.; et al. Transcriptomic and Mutational Analysis Discovering Distinct Molecular Characteristics Among Chinese Thymic Epithelial Tumor Patients. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Radovich, M.; Solzak, J.P.; A Hancock, B.; Conces, M.L.; Atale, R.; Porter, R.F.; Zhu, J.; Glasscock, J.; A Kesler, K.; Badve, S.S.; et al. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type A and AB thymomas. Br. J. Cancer 2016, 114, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, P.; Zhao, J.; Yang, L.; Wang, W. Identification of Molecular Characteristics and New Prognostic Targets for Thymoma by Multiomics Analysis. BioMed Res. Int. 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Wang, X.; Lin, P.; Li, Y.; Chen, G.; Yang, H.; He, Y.; Li, Q.; Liu, R. Identification of potential agents for thymoma by integrated analyses of differentially expressed tumour-associated genes and molecular docking experiments. Exp. Ther. Med. 2019, 18, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Wang, S.; Zhang, J.; Yan, Y.; Wang, C.; Yang, C.; Guan, Z.; Wang, C. Alteration in gene expression profiles of thymoma: Genetic differences and potential novel targets. Thorac. Cancer 2019, 10, 1129–1135. [Google Scholar] [CrossRef]
- Yu, L.; Ke, J.; Du, X.; Yu, Z.; Gao, D. Genetic characterization of thymoma. Sci. Rep. 2019, 9, 2369. [Google Scholar] [CrossRef]
- Lee, H. S. , Jang, H.J.; Shah, R.; Yoon, D.; Hamaji, M.; Wald, O.; Lee, J.S.; Sugarbaker, D.J.; Burt, B.M. Genomic Analysis of Thymic Epithelial Tumors Identifies Novel Subtypes Associated with Distinct Clinical Features. Clin Cancer Res 2017, 23, 4855–64. [Google Scholar] [CrossRef] [PubMed]
- Iaiza, A.; Tito, C.; Ganci, F.; Sacconi, A.; Gallo, E.; Masciarelli, S.; Fontemaggi, G.; Fatica, A.; Melis, E.; Petrozza, V.; et al. Long Non-Coding RNAs in the Cell Fate Determination of Neoplastic Thymic Epithelial Cells. Front. Immunol. 2022, 13, 867181. [Google Scholar] [CrossRef] [PubMed]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Luo, X.; Li, H.; Zhang, L.; Su, F.; Hou, S.; Yin, J.; Zhang, W.; Zou, L. Identification of differentially expressed circular RNAs associated with thymoma. Thorac. Cancer 2021, 12, 1312–1319. [Google Scholar] [CrossRef]
- Su, Y.; Chen, Y.; Tian, Z.; Lu, C.; Chen, L.; Ma, X. lncRNAs classifier to accurately predict the recurrence of thymic epithelial tumors. Thorac. Cancer 2020, 11, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Jin, S.; Pan, X.; Wang, G.; Ye, L.; Tao, H.; Wen, H.; Liu, Y.; Xie, Q. Identification of Long Non-Coding RNAs for Predicting Prognosis Among Patients with Thymoma. Clin. Lab. 2018, 64, 1193–1198. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Song, H. A Disparate Role of Rp11-424c20.2/Uhrf1 Axis through Control of Tumor Immune Escape in Liver Hepatocellular Carcinoma and Thymoma. Aging 2019, 11, 6422–39. [Google Scholar] [CrossRef]
- Obata, Y.; Furusawa, Y.; A Endo, T.; Sharif, J.; Takahashi, D.; Atarashi, K.; Nakayama, M.; Onawa, S.; Fujimura, Y.; Takahashi, M.; et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat. Immunol. 2014, 15, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Lu, T. X.; Rothenberg, M.E. Microrna. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. miRNAs and cancer: An epigenetics view. Mol. Asp. Med. 2012, 34, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Enkner, F.; Pichlhöfer, B.; Zaharie, A.T.; Krunic, M.; Holper, T.M.; Janik, S.; Moser, B.; Schlangen, K.; Neudert, B.; Walter, K.; et al. Molecular Profiling of Thymoma and Thymic Carcinoma: Genetic Differences and Potential Novel Therapeutic Targets. Pathol. Oncol. Res. 2016, 23, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, H.; Zhang, X.; Ma, H. Loxl1-As1 Promotes Thymoma and Thymic Carcinoma Progression by Regulating Mir-525-5p-Hspa9. Oncol. Rep. 2021, 45. [Google Scholar] [CrossRef]
- Tito, C.; Ganci, F.; Sacconi, A.; Masciarelli, S.; Fontemaggi, G.; Pulito, C.; Gallo, E.; Laquintana, V.; Iaiza, A.; De Angelis, L.; et al. LINC00174 is a novel prognostic factor in thymic epithelial tumors involved in cell migration and lipid metabolism. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ganci, F.; Vico, C.; Korita, E.; Sacconi, A.; Gallo, E.; Mori, F.; Cambria, A.; Russo, E.; Anile, M.; Vitolo, D.; et al. MicroRNA expression profiling of thymic epithelial tumors. Lung Cancer 2014, 85, 197–204. [Google Scholar] [CrossRef]
- Tan, S., and J. Chen. Si-Malat1 Attenuates Thymic Cancer Cell Proliferation and Promotes Apoptosis Via the Mir-145-5p/Hmga2 Pathway. Oncol. Lett. 2021, 22, 585. [Google Scholar] [CrossRef] [PubMed]
- Iaiza, A.; Tito, C.; Ianniello, Z.; Ganci, F.; Laquintana, V.; Gallo, E.; Sacconi, A.; Masciarelli, S.; De Angelis, L.; Aversa, S.; et al. METTL3-dependent MALAT1 delocalization drives c-Myc induction in thymic epithelial tumors. Clin. Epigenetics 2021, 13, 1–15. [Google Scholar] [CrossRef]
- Damron, B. The Relationship of Maximum or Intermediate Coccidiostat Levels to Broiler Chick Water Intake. Poult. Sci. 1994, 73, 33–36. [Google Scholar] [CrossRef]
- Miller, J.W.; Faubert, B.M.; Mathews, T.P.; Waters, J.K.; DeBerardinis, R.J.; Kernstine, K.H. Metabolic signatures of thymomas: potential biomarkers and treatment targets. Eur. J. Cardio-Thoracic Surg. 2023, 65. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Huang, Y.; Ye, X.; Zhang, Y.; Yao, Q.; Wang, J.; Yang, X.; Yu, C.; Guo, Y.; Zhang, X.; et al. Comprehensive study of clinicopathological and immune cell infiltration and lactate dehydrogenase expression in patients with thymic epithelial tumours. Int. Immunopharmacol. 2024, 126, 111205. [Google Scholar] [CrossRef] [PubMed]
- Alwahsh, M.; Knitsch, R. Marchan, J. Lambert. Metabolic Profiling of Thymic Epithelial Tumors Hints to a Strong Warburg Effect, Glutaminolysis and Precarious Redox Homeostasis as Potential Therapeutic Targets. Cancers 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zeng, B.; Zhu, H.; Ma, R.; Yuan, P.; Chen, Z.; Su, C.; Liu, Z.; Yao, X.; Lawrence, A.; et al. Role of glycosphingolipid biosynthesis coregulators in malignant progression of thymoma. Int. J. Biol. Sci. 2023, 19, 4442–4456. [Google Scholar] [CrossRef]
- Tang, E.; Zhou, Y.; Liu, S.; Zhang, Z.; Zhang, R.; Huang, D.; Gao, T.; Zhang, T.; Xu, G. Metabolomic and Transcriptomic Profiling Identified Significant Genes in Thymic Epithelial Tumor. Metabolites 2022, 12, 567. [Google Scholar] [CrossRef]
- Regina Todeschini, A., and S.I. Hakomori. Functional Role of Glycosphingolipids and Gangliosides in Control of Cell Adhesion, Motility, and Growth, through Glycosynaptic Microdomains. Biochim. Biophys. Acta 2008, 1780, 421–33. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. I. "Glycosynaptic Microdomains Controlling Tumor Cell Phenotype through Alteration of Cell Growth, Adhesion, and Motility. " FEBS Lett. 2010, 584, 1901–6. [Google Scholar] [CrossRef]
- Li, J., Y. Long. Comprehensive Landscape of the St3gal Family Reveals the Significance of St3gal6-As1/St3gal6 Axis on Egfr Signaling in Lung Adenocarcinoma Cell Invasion. Front. Cell Dev. Biol. 2022, 10, 931132. [Google Scholar] [CrossRef] [PubMed]
- Bremmer, F.; Bohnenberger, H.; Findeisen, P.; Welter, S.; von Hammerstein-Equord, A.; Hinterthaner, M.; Müller, D.; Küffer, S.; Okada, S.; Marx, A.; et al. Proteomic analysis identifies argininosuccinate synthetase 1 and special AT-rich sequence binding protein 1 as reliable markers for the immunohistochemical distinction between WHO types A and B3 thymomas. Histopathology 2023, 83, 607–616. [Google Scholar] [CrossRef]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef]
- Zhao, T.; Wu, J.; Liu, X.; Zhang, L.; Chen, G.; Lu, H. Diagnosis of thymic epithelial tumor subtypes by a quantitative proteomic approach. Analyst 2018, 143, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Branson, O.E.; Shilo, K.; Hitchcock, C.L.; Freitas, M.A. Proteomic Signatures of Thymomas. PLoS ONE 2016, 11, e0166494. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.-L.; Fang, W.-T.; Feng, J.; Zhang, J.; Yang, X.-H.; Gu, Z.-T.; Zhu, L.; Sha, H.-F. Proteome analysis and tissue array for profiling protein markers associated with type B thymoma subclassification. . 2012, 125, 2811–2818. [Google Scholar] [PubMed]
- Lai, L.-C.; Sun, Q.-L.; Chen, Y.-A.; Hsiao, Y.-W.; Lu, T.-P.; Tsai, M.-H.; Zhu, L.; Chuang, E.Y.; Fang, W. Using proteomic profiling to characterize protein signatures of different thymoma subtypes. BMC Cancer 2019, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Liu, P.; Qi, G. Exploring Potential Biomarkers of Early Thymoma based on Serum Proteomics. Protein Pept. Lett. 2024, 31, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, G.; Liu, Y. Proteomics analysis of serum from thymoma patients. Sci. Rep. 2023, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Qi, G.; Dong, H.; Liu, Z.; Ma, M.; Liu, P. Identification of Potential Serum Protein Biomarkers in Thymoma with Myasthenia Gravis After Docetaxel Treatment. Neurol. Ther. 2023, 12, 559–570. [Google Scholar] [CrossRef]
- Tong, T.; Zhang, J.; Jia, L.; Liang, P.; Wang, N. Integrated proteomics and metabolomics analysis reveals hubs protein and network alterations in myasthenia gravis. Aging 2022, 14, 5417–5426. [Google Scholar] [CrossRef]
- Hayashi, Y., N. Ishii. Thymoma: Tumour Type Related to Expression of Epidermal Growth Factor (Egf), Egf-Receptor, P53, V-Erb B and Ras P21. Virchows Arch. 1995, 426, 43–50. [Google Scholar] [CrossRef]
- Ionescu, D.N.; Sasatomi, E.; Cieply, K.; Nola, M.; Dacic, S. Protein expression and gene amplification of epidermal growth factor receptor in thymomas. Cancer 2005, 103, 630–636. [Google Scholar] [CrossRef]
- E Gilhus, N.; Jones, M.; Turley, H.; Gatter, K.C.; Nagvekar, N.; Newsom-Davis, J.; Willcox, N. Oncogene proteins and proliferation antigens in thymomas: increased expression of epidermal growth factor receptor and Ki67 antigen. J. Clin. Pathol. 1995, 48, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Pescarmona, E.; Pisacane, A.; Pignatelli, E.; Baroni, C. Expression of epidermal and nerve growth factor receptors in human thymus and thymomas. Histopathology 1993, 23, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Henley, J.; Koukoulis, G.; Loehrer, P. Epidermal growth factor receptor expression in invasive thymoma. J. Cancer Res. Clin. Oncol. 2002, 128, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Abedallaa, N.; Taranchon-Clermont, E.; Bluthgen, V.; Lindsay, C.; Besse, B.; de Montpréville, V.T. CD52, CD22, CD26, EG5 and IGF-1R expression in thymic malignancies. Lung Cancer 2017, 108, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Girard, N.; Teruya-Feldstein, J.; Payabyab, E.C.; Riely, G.J.; Rusch, V.W.; Kris, M.G.; Zakowski, M.F. Insulin-Like Growth Factor-1 Receptor Expression in Thymic Malignancies. J. Thorac. Oncol. 2010, 5, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Zucali, P.A.; Petrini, I.; Lorenzi, E.; Merino, M.; Cao, L.; Di Tommaso, L.; Lee, H.S.; Incarbone, M.; Walter, B.A.; Simonelli, M.; et al. Insulin-like growth factor-1 receptor and phosphorylated AKT-serine 473 expression in 132 resected thymomas and thymic carcinomas. Cancer 2010, 116, 4686–4695. [Google Scholar] [CrossRef] [PubMed]
- Festenstein, H. MOLECULAR FEATURES OF THE H-2 CLASS I AND Qa ANTIGENS EXPRESSED ON GROSS VIRUS INDUCED AKR LEUKAEMIAS. Int. J. Immunogenetics 1989, 16, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Avci, N., G. Cecener. Molecular Markers for Patients with Thymic Malignancies: Not Feasible at Present? Asian Pac. J. Cancer Prev. 2014, 15, 3457–60. [Google Scholar] [CrossRef] [PubMed]
- Pfister, F.; Hussain, H.; Belharazem, D.; Busch, S.; Simon-Keller, K.; Becker, D.; Pfister, E.; Rieker, R.; Ströbel, P.; Marx, A. Vascular architecture as a diagnostic marker for differentiation of World Health Organization thymoma subtypes and thymic carcinoma. Histopathology 2017, 70, 693–703. [Google Scholar] [CrossRef]
- Lattanzio, R.; La Sorda, R.; Facciolo, F.; Sioletic, S.; Lauriola, L.; Martucci, R.; Gallo, E.; Palmieri, G.; Evoli, A.; Alessandrini, G.; et al. Thymic epithelial tumors express vascular endothelial growth factors and their receptors as potential targets of antiangiogenic therapy: A tissue micro array-based multicenter study. Lung Cancer 2014, 85, 191–196. [Google Scholar] [CrossRef]
- Angirekula, M.; Chang, S.Y.; Jenkins, S.M.; Greipp, P.T.; Sukov, W.R.; Marks, R.S.; Olivier, K.R.; Cassivi, S.D.; Roden, A.C. CD117, BAP1, MTAP, and TdT Is a Useful Immunohistochemical Panel to Distinguish Thymoma from Thymic Carcinoma. Cancers 2022, 14, 2299. [Google Scholar] [CrossRef] [PubMed]
- Kornstein, M.J.; Rosai, J. CD5 Labeling of Thymic Carcinomas and Other Nonlymphoid Neoplasms. Am. J. Clin. Pathol. 1998, 109, 722–726. [Google Scholar] [CrossRef]
- Jeong, J.-H.; Pyo, J.-S.; Kim, N.-Y.; Kang, D.-W. Diagnostic Roles of Immunohistochemistry in Thymic Tumors: Differentiation between Thymic Carcinoma and Thymoma. Diagnostics 2020, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Tiseo, M.; Damato, A.; Longo, L.; Barbieri, F.; Bertolini, F.; Stefani, A.; Migaldi, M.; Gnetti, L.; Camisa, R.; Bordi, P.; et al. Analysis of a panel of druggable gene mutations and of ALK and PD-L1 expression in a series of thymic epithelial tumors (TETs). Lung Cancer 2017, 104, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Aisner, S. C., S. Dahlberg. Epidermal Growth Factor Receptor, C-Kit, and Her2/Neu Immunostaining in Advanced or Recurrent Thymic Epithelial Neoplasms Staged According to the 2004 World Health Organization in Patients Treated with Octreotide and Prednisone: An Eastern Cooperative Oncology Group Study. J. Thorac. Oncol. 2010, 5, 885–92. [Google Scholar] [PubMed]
- Mimae, T.; Tsuta, K.; Kondo, T.; Nitta, H.; Grogan, T.M.; Okada, M.; Asamura, H.; Tsuda, H. Protein expression and gene copy number changes of receptor tyrosine kinase in thymomas and thymic carcinomas. Ann. Oncol. 2012, 23, 3129–3137. [Google Scholar] [CrossRef] [PubMed]
- Küffer, S.; Grabowski, J.; Okada, S.; Sojka, N.; Welter, S.; von Hammerstein-Equord, A.; Hinterthaner, M.; Cordes, L.; von Hahn, X.; Müller, D.; et al. Phosphoproteomic Analysis Identifies TYRO3 as a Mediator of Sunitinib Resistance in Metastatic Thymomas. Cancers 2022, 14, 4762. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, S.; Wang, Y.; Chen, Y.; Zhang, P.; Liu, Y.; Zhang, H.; Tao, Z.; Xiong, K. High expression of KITLG is a new hallmark activating the MAPK pathway in type A and AB thymoma. Thorac. Cancer 2020, 11, 1944–1954. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




