Submitted:
30 January 2024
Posted:
31 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Indirect Light Sensing
3. Direct Light Sensing
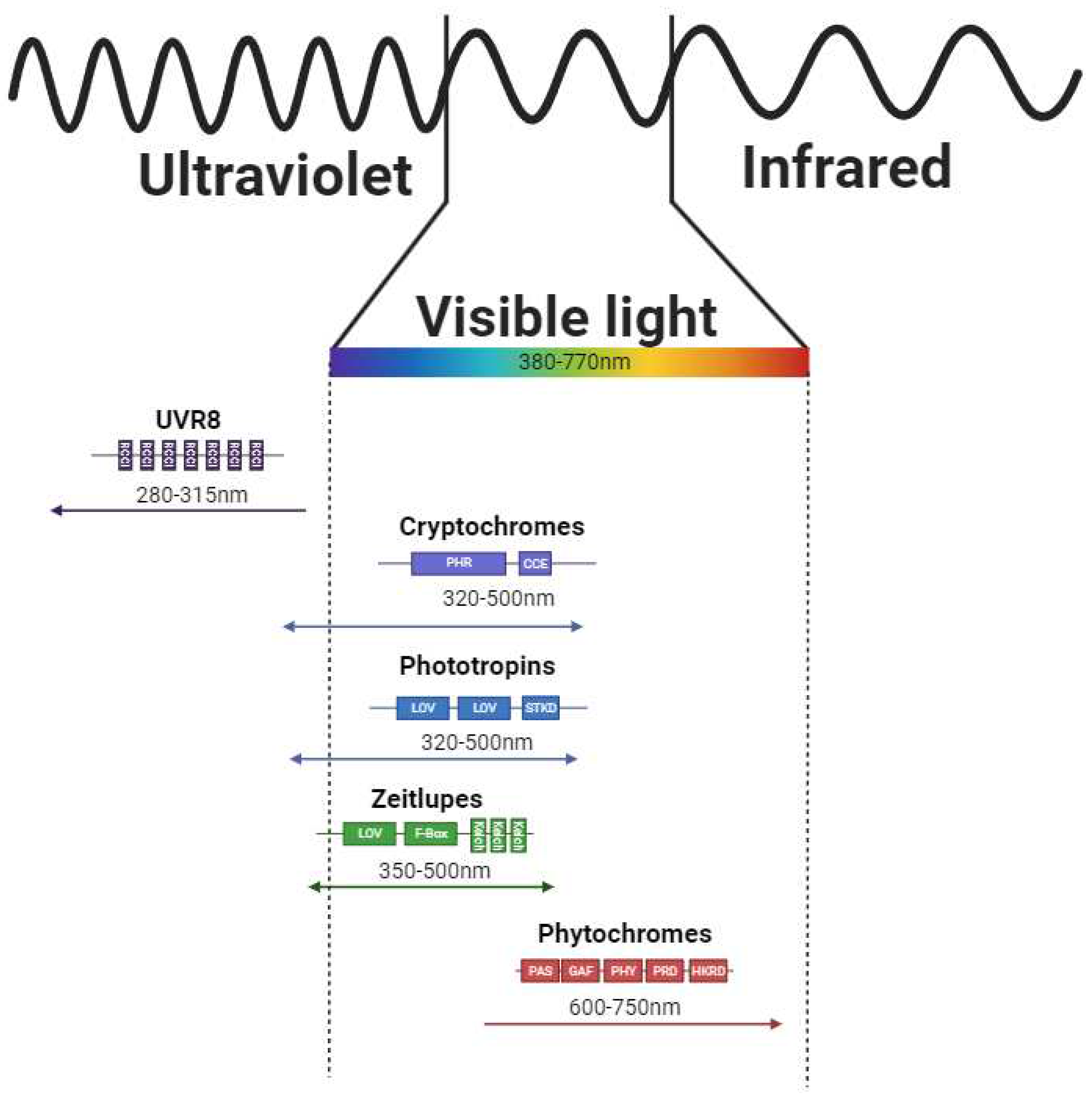
3.1. Basic Description of Photoreceptors and Mechanism of its Activation
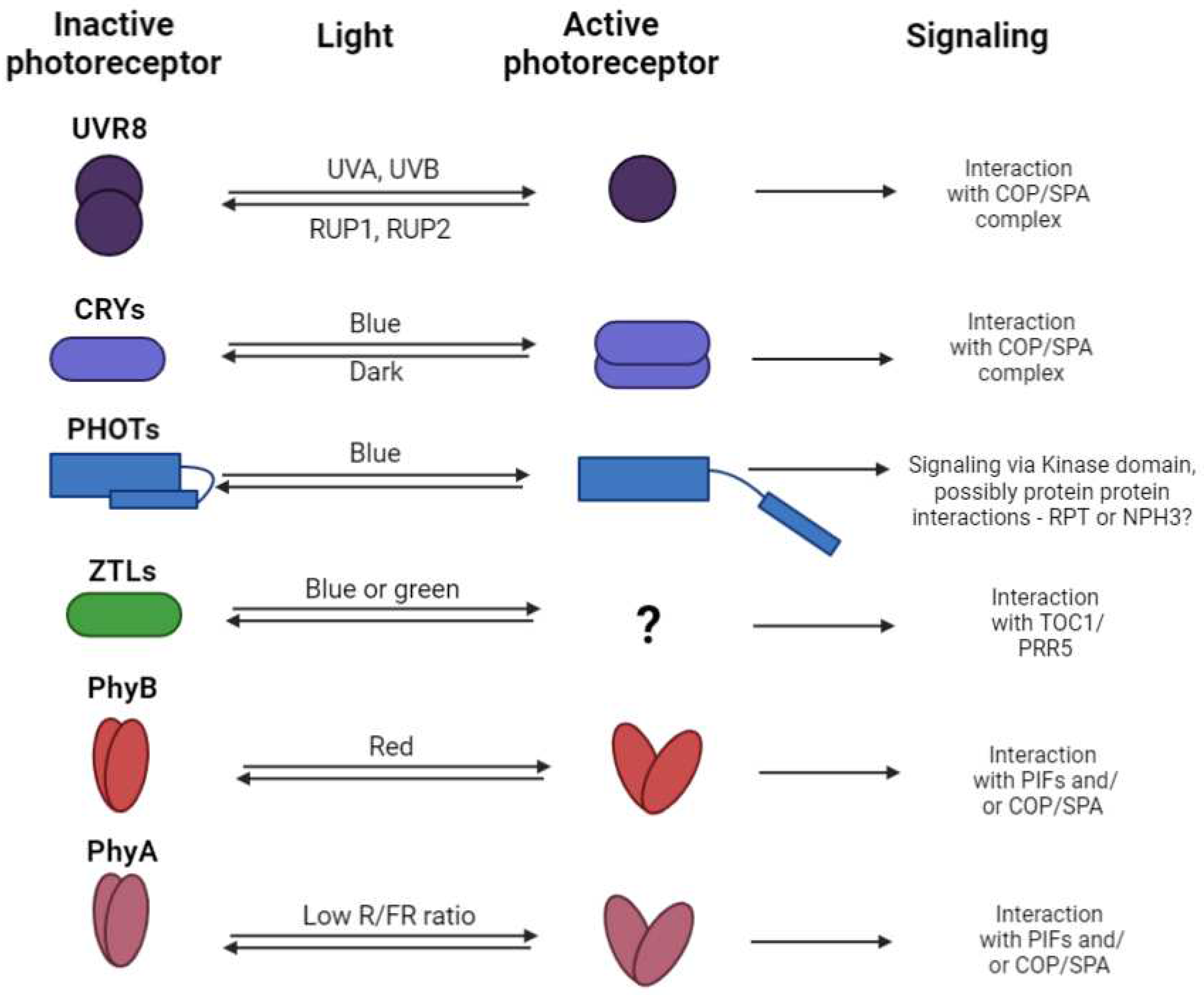
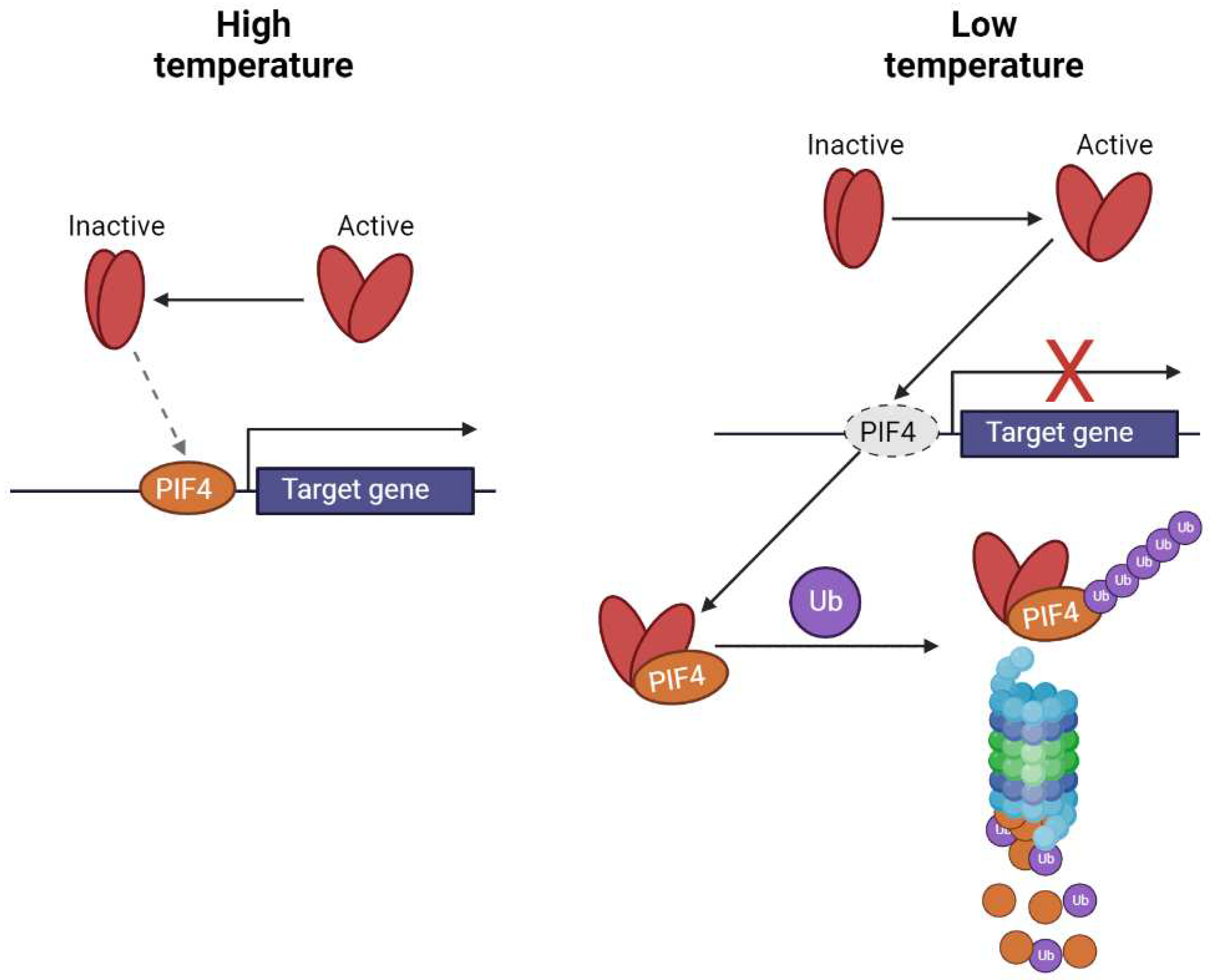
4. Photoreceptor Signaling and Thermosensing
5. COP1/SPA Complex
6. Transcription Factors Involved in PheCs Biosynthesis Control
6.1. Proteins Containing Basic Helix-Loop-Helix Motifs (So-Called bHLH)
6.2. Basic ZIPper Containing Proteins (bZIP)
6.3. Transcription Factors Containing Helix-Turn-Helix Motifs (HTH TFs)
| TF | TT8 | HY5 | HYH_1 | HYH_2 | HYH_3 | HYH_4 | TT2 | PAP1 | MYB11 | MYB12 | MYB111 | WRKY23 | WRKY36 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Motif | bHLH | bZIP | bZIP | bZIP | bZIP | bZIP | HTH | HTH | HTH | HTH | HTH | WRKY | WRKY |
| ID | AT4G09820 | AT5G11260 | AT3G17609.1 | AT3G17609.2 | AT3G17609.3 | AT3G17609.4 | AT5G35550.1 | AT1G56650.1 | AT3G62610.1 | AT5G49330.1 | AT5G49330.1 | AT2G47260.1 | AT1G69810.1 |
| Target genes activated | BAN, TT8, MYBL2, DFR, GL2, MES6, MES4, TTG2 | RBCS1A, ELIP1, PHR1, LZF1, IAA7, CHS, CAB2, ABI5, IAA14, DFR, CAB1, UGT84A1, PSBD, NIA2, ELF4, LDOX, HEMA1, MYB12, HB-8 | PHR1, NIA2, ELIP1, PEX11B, CHS | PHR1, NIA2, ELIP1, PEX11B, CHS | PHR1, NIA2, ELIP1, PEX11B, CHS | PHR1, NIA2, ELIP1, PEX11B, CHS | ANS, TT8, TT2, TTG2, GL2, DFR, BAN | CHS, CHI, DFR, MYB3, TT8, UF3GT, PAP2, A5GT, UGT78D2, 5MAT, MBD2.2, GST | CHS, CHI, F3H, FLS1 | CHS, CHI, F3H, FLS1 | CHS, CHI, F3H, FLS1 | ??? | ??? |
| Target genes repressed | - | NPF6.3, FHY1, FHL | NPF6.3 | NPF6.3 | NPF6.3 | NPF6.3 | - | scpl10 | - | - | - | ??? | ??? |
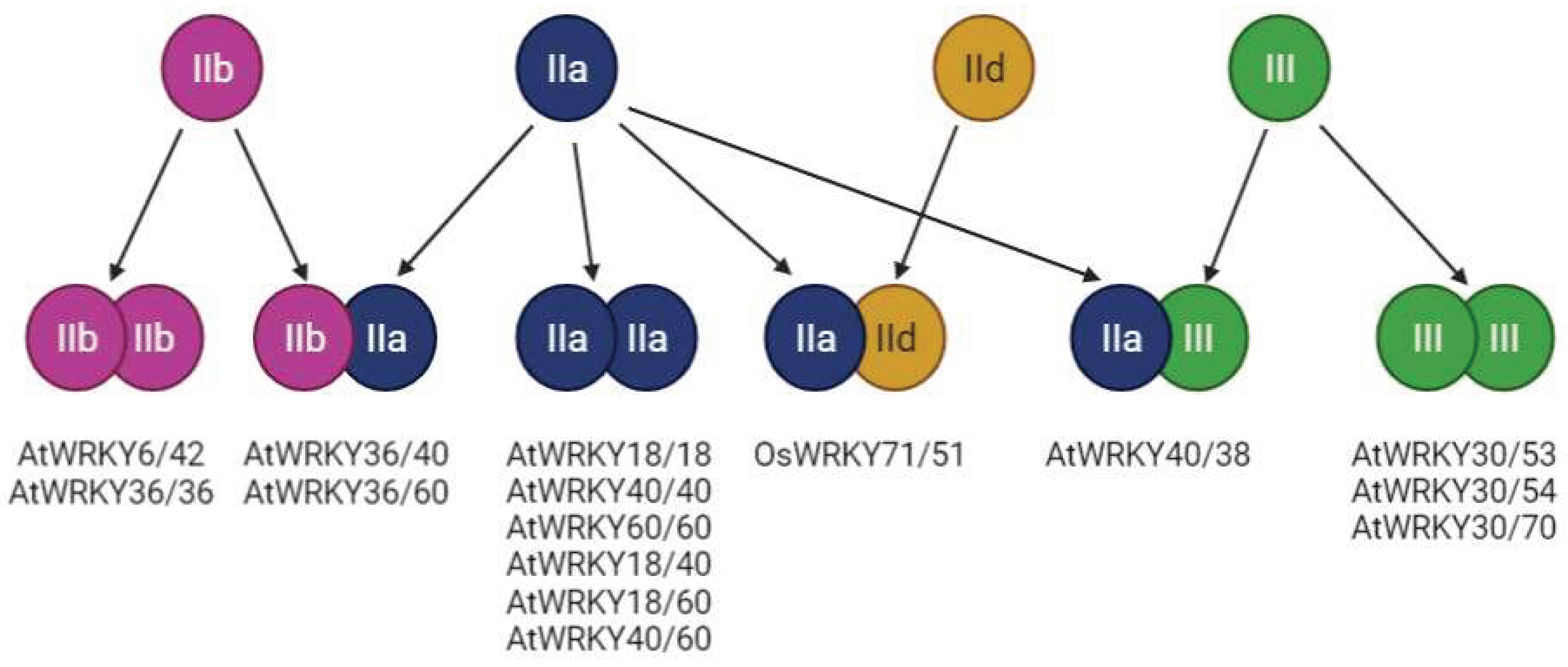
6.4. WRKY Proteins
6.5. WD-40
6.6. MYB-bHLH-WD-40 complex
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- J.J. Casal, Environmental cues affecting development, Current Opinion in Plant Biology 5 (2002) 37–42. [CrossRef]
- M. Landi, M. Zivcak, O. Sytar, M. Brestic, S.I. Allakhverdiev, Plasticity of photosynthetic processes and the accumulation of secondary metabolites in plants in response to monochromatic light environments: A review, Biochimica et Biophysica Acta (BBA) - Bioenergetics 1861 (2020) 148131. [CrossRef]
- G. Kudo, T.Y. Ida, T. Tani, Linkages Between Phenology, Pollination, Photosynthesis, and Reproduction in Deciduous Forest Understory Plants, Ecology 89 (2008) 321–331. [CrossRef]
- J. Saile, T. Wießner-Kroh, K. Erbstein, D.M. Obermüller, A. Pfeiffer, D. Janocha, J. Lohmann, A. Wachter, SNF1-RELATED KINASE 1 and TARGET OF RAPAMYCIN control light-responsive splicing events and developmental characteristics in etiolated Arabidopsis seedlings, The Plant Cell 35 (2023) 3413–3428. [CrossRef]
- K. Apel, H. Hirt, Reactive oxygen species: Metabolism, oxidative stress, and signal transduction, Annu. Rev. Plant Biol. 55 (2004) 373–399. [CrossRef]
- M. El-Esawi, L.-D. Arthaut, N. Jourdan, A. d’Harlingue, J. Link, C.F. Martino, M. Ahmad, Blue-light induced biosynthesis of ROS contributes to the signaling mechanism of Arabidopsis cryptochrome, Scientific Reports 7 (2017) 13875. [CrossRef]
- E. Horváth, T. Janda, G. Szalai, E. Páldi, In vitro salicylic acid inhibition of catalase activity in maize: differences between the isozymes and a possible role in the induction of chilling tolerance, Plant Science 163 (2002) 1129–1135. [CrossRef]
- S. Riegler, L. Servi, M.R. Scarpin, M.A. Godoy Herz, M.G. Kubaczka, P. Venhuizen, C. Meyer, J.O. Brunkard, M. Kalyna, A. Barta, E. Petrillo, Light regulates alternative splicing outcomes via the TOR kinase pathway, Cell Reports 36 (2021) 109676. [CrossRef]
- K. McCready, V. Spencer, M. Kim, The Importance of TOR Kinase in Plant Development, Frontiers in Plant Science 11 (2020). https://www.frontiersin.org/articles/10.3389/fpls.2020.00016 (accessed January 12, 2024).
- M. Jamsheer K, P. Awasthi, A. Laxmi, The social network of target of rapamycin complex 1 in plants, Journal of Experimental Botany 73 (2022) 7026–7040. [CrossRef]
- The UniProt Consortium, UniProt: a worldwide hub of protein knowledge, Nucleic Acids Research 47 (2019) D506–D515. [CrossRef]
- AT4G29130(HXK1), (n.d.). https://www.arabidopsis.org/servlets/TairObject?name=AT4G29130&type=locus (accessed January 12, 2024).
- R. Wu, X. Lin, J. He, A. Min, L. Pang, Y. Wang, Y. Lin, Y. Zhang, W. He, M. Li, Y. Zhang, Y. Luo, X. Wang, H. Tang, Q. Chen, Hexokinase1: A glucose sensor involved in drought stress response and sugar metabolism depending on its kinase activity in strawberry, Frontiers in Plant Science 14 (2023). https://www.frontiersin.org/articles/10.3389/fpls.2023.1069830 (accessed January 12, 2024).
- B. Peixoto, E. Baena-González, Management of plant central metabolism by SnRK1 protein kinases, Journal of Experimental Botany 73 (2022) 7068–7082. [CrossRef]
- B. Wang, X. Zhao, Y. Zhao, J. Shanklin, Q. Zhao, C.-J. Liu, Arabidopsis SnRK1 negatively regulates phenylpropanoid metabolism via Kelch domain-containing F-box proteins, New Phytologist 229 (2021) 3345–3359. [CrossRef]
- S. Hulsmans, M. Rodriguez, B.D. Coninck, F. Rolland, The SnRK1 Energy Sensor in Plant Biotic Interactions, Trends in Plant Science 21 (2016) 648–661. [CrossRef]
- J.B. Rossel, I.W. Wilson, B.J. Pogson, Global Changes in Gene Expression in Response to High Light in Arabidopsis, Plant Physiology 130 (2002) 1109–1120. [CrossRef]
- S.I. Zandalinas, S. Sengupta, D. Burks, R.K. Azad, R. Mittler, Identification and characterization of a core set of ROS wave-associated transcripts involved in the systemic acquired acclimation response of Arabidopsis to excess light, The Plant Journal 98 (2019) 126–141. [CrossRef]
- B. Moore, L. Zhou, F. Rolland, Q. Hall, W.-H. Cheng, Y.-X. Liu, I. Hwang, T. Jones, J. Sheen, Role of the Arabidopsis Glucose Sensor HXK1 in Nutrient, Light, and Hormonal Signaling, Science 300 (2003) 332–336. [CrossRef]
- O. Avidan, T.A. Moraes, V. Mengin, R. Feil, F. Rolland, M. Stitt, J.E. Lunn, In vivo protein kinase activity of SnRK1 fluctuates in Arabidopsis rosettes during light-dark cycles, Plant Physiology 192 (2023) 387–408. [CrossRef]
- S. Shokrian Hajibehzad, S.S. Silva, N. Peeters, E. Stouten, G. Buijs, S. Smeekens, M. Proveniers, Arabidopsis thaliana rosette habit is controlled by combined light and energy signaling converging on transcriptional control of the TALE homeobox gene ATH1, New Phytologist 239 (2023) 1051–1067. [CrossRef]
- C. Orth, N. Niemann, L. Hennig, L.-O. Essen, A. Batschauer, Hyperactivity of the Arabidopsis cryptochrome (cry1) L407F mutant is caused by a structural alteration close to the cry1 ATP-binding site, Journal of Biological Chemistry 292 (2017) 12906–12920. [CrossRef]
- L. Lopez, C. Fasano, G. Perrella, P. Facella, Cryptochromes and the Circadian Clock: The Story of a Very Complex Relationship in a Spinning World, Genes 12 (2021) 672. [CrossRef]
- C. Lin, T. Todo, The cryptochromes, Genome Biology 6 (2005) 220. [CrossRef]
- J.M. Barrero, A.B. Downie, Q. Xu, F. Gubler, A Role for Barley CRYPTOCHROME1 in Light Regulation of Grain Dormancy and Germination, The Plant Cell 26 (2014) 1094–1104. [CrossRef]
- N. Tissot, R. Ulm, Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis, Nature Communications 11 (2020) 1323. [CrossRef]
- L. Ma, Z. Guan, Q. Wang, X. Yan, J. Wang, Z. Wang, J. Cao, D. Zhang, X. Gong, P. Yin, Structural insights into the photoactivation of Arabidopsis CRY2, Nature Plants 6 (2020) 1432–1438. [CrossRef]
- M. Palayam, J. Ganapathy, A.M. Guercio, L. Tal, S.L. Deck, N. Shabek, Structural insights into photoactivation of plant Cryptochrome-2, Communications Biology 4 (2021) 1–11. [CrossRef]
- K. Shao, X. Zhang, X. Li, Y. Hao, X. Huang, M. Ma, M. Zhang, F. Yu, H. Liu, P. Zhang, The oligomeric structures of plant cryptochromes, Nature Structural & Molecular Biology 27 (2020) 480–488. [CrossRef]
- J.M. Christie, Phototropin Blue-Light Receptors, Annual Review of Plant Biology 58 (2007) 21–45. [CrossRef]
- J.M. Christie, T.E. Swartz, R.A. Bogomolni, W.R. Briggs, Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function, The Plant Journal 32 (2002) 205–219. [CrossRef]
- S. Inoue, A. Takemiya, K. Shimazaki, Phototropin signaling and stomatal opening as a model case, Current Opinion in Plant Biology 13 (2010) 587–593. [CrossRef]
- Y. Nakasone, M. Ohshima, K. Okajima, S. Tokutomi, M. Terazima, Photoreaction Dynamics of Full-Length Phototropin from Chlamydomonas reinhardtii, The Journal of Physical Chemistry B 123 (2019) 10939–10950. [CrossRef]
- A. Motchoulski, E. Liscum, Arabidopsis NPH3: A NPH1 Photoreceptor-Interacting Protein Essential for Phototropism, Science 286 (1999) 961–964. [CrossRef]
- S. Inada, M. Ohgishi, T. Mayama, K. Okada, T. Sakai, RPT2 Is a Signal Transducer Involved in Phototropic Response and Stomatal Opening by Association with Phototropin 1 in Arabidopsis thaliana, Plant Cell 16 (2004) 887–896. [CrossRef]
- T. Sakai, T. Wada, S. Ishiguro, K. Okada, RPT2: A Signal Transducer of the Phototropic Response in Arabidopsis, The Plant Cell 12 (2000) 225–236. [CrossRef]
- S.K. Gupta, M. Sharma, F. Deeba, V. Pandey, Plant Response, in: UV-B Radiation, John Wiley & Sons, Ltd, 2017: pp. 217–258. [CrossRef]
- S.-G. Kong, K. Okajima, Diverse photoreceptors and light responses in plants, Journal of Plant Research 129 (2016) 111–114. [CrossRef]
- M. Legris, Y.Ç. Ince, C. Fankhauser, Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants, Nature Communications 10 (2019) 5219. [CrossRef]
- Paik, E. Huq, Plant photoreceptors: Multi-functional sensory proteins and their signaling networks, Seminars in Cell & Developmental Biology 92 (2019) 114–121. [CrossRef]
- A. Pudasaini, B.D. Zoltowski, Zeitlupe Senses Blue-Light Fluence To Mediate Circadian Timing in Arabidopsis thaliana, Biochemistry 52 (2013) 7150–7158. [CrossRef]
- A. Pudasaini, J.S. Shim, Y.H. Song, H. Shi, T. Kiba, D.E. Somers, T. Imaizumi, B.D. Zoltowski, Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis, ELife 6 (2017) e21646. [CrossRef]
- S. Ito, Y.H. Song, T. Imaizumi, LOV Domain-Containing F-Box Proteins: Light-Dependent Protein Degradation Modules in Arabidopsis, Molecular Plant 5 (2012) 573–582. [CrossRef]
- J.M. Christie, A.S. Arvai, K.J. Baxter, M. Heilmann, A.J. Pratt, A. O’Hara, S.M. Kelly, M. Hothorn, B.O. Smith, K. Hitomi, G.I. Jenkins, E.D. Getzoff, Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges, Science 335 (2012) 1492–1496. [CrossRef]
- T. Mathes, M. Heilmann, A. Pandit, J. Zhu, J. Ravensbergen, M. Kloz, Y. Fu, B.O. Smith, J.M. Christie, G.I. Jenkins, J.T.M. Kennis, Proton-Coupled Electron Transfer Constitutes the Photoactivation Mechanism of the Plant Photoreceptor UVR8, Journal of the American Chemical Society 137 (2015) 8113–8120. [CrossRef]
- X. Li, H. Ren, M. Kundu, Z. Liu, F.W. Zhong, L. Wang, J. Gao, D. Zhong, A leap in quantum efficiency through light harvesting in photoreceptor UVR8, Nature Communications 11 (2020) 4316. [CrossRef]
- V.E. Tossi, J.J. Regalado, J. Iannicelli, L.E. Laino, H.P. Burrieza, A.S. Escandón, S.I. Pitta-Álvarez, Beyond Arabidopsis: Differential UV-B Response Mediated by UVR8 in Diverse Species, Frontiers in Plant Science 10 (2019). [CrossRef]
- N.C. Rockwell, Y.-S. Su, J.C. Lagarias, PHYTOCHOME STRUCTURE AND SIGNALING MECHANISMS, Annual Review of Plant Biology 57 (2006) 837–858. [CrossRef]
- J. Li, G. Li, H. Wang, X.W. Deng, Phytochrome Signaling Mechanisms, The Arabidopsis Book 2011 (2011). [CrossRef]
- K.A. Franklin, T. Allen, G.C. Whitelam, Phytochrome A is an irradiance-dependent red light sensor, The Plant Journal 50 (2007) 108–117. [CrossRef]
- M. Legris, C. Klose, E.S. Burgie, C.C.R. Rojas, M. Neme, A. Hiltbrunner, P.A. Wigge, E. Schäfer, R.D. Vierstra, J.J. Casal, Phytochrome B integrates light and temperature signals in Arabidopsis, Science 354 (2016) 897–900. [CrossRef]
- Y. Qiu, M. Li, R.J.-A. Kim, C.M. Moore, M. Chen, Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA, Nature Communications 10 (2019) 140. [CrossRef]
- R. Bianchetti, B. De Luca, L.A. de Haro, D. Rosado, D. Demarco, M. Conte, L. Bermudez, L. Freschi, A.R. Fernie, L.V. Michaelson, R.P. Haslam, M. Rossi, F. Carrari, Phytochrome-Dependent Temperature Perception Modulates Isoprenoid Metabolism, Plant Physiology 183 (2020) 869–882. [CrossRef]
- V.N. Pham, P.K. Kathare, E. Huq, Phytochromes and Phytochrome Interacting Factors, Plant Physiology 176 (2018) 1025–1038. [CrossRef]
- T. Sakamoto, S. Kimura, Plant Temperature Sensors, Sensors 18 (2018) 4365. [CrossRef]
- B.Y.W. Chung, M. Balcerowicz, M. Di Antonio, K.E. Jaeger, F. Geng, K. Franaszek, P. Marriott, I. Brierley, A.E. Firth, P.A. Wigge, An RNA thermoswitch regulates daytime growth in Arabidopsis, Nature Plants 6 (2020) 522–532. [CrossRef]
- S. Hayes, J. Schachtschabel, M. Mishkind, T. Munnik, S.A. Arisz, Hot topic: Thermosensing in plants, Plant, Cell & Environment 44 (2021) 2018–2033. [CrossRef]
- Y. Fujii, H. Tanaka, N. Konno, Y. Ogasawara, N. Hamashima, S. Tamura, S. Hasegawa, Y. Hayasaki, K. Okajima, Y. Kodama, Phototropin perceives temperature based on the lifetime of its photoactivated state, Proceedings of the National Academy of Sciences 114 (2017) 9206–9211. [CrossRef]
- M. Pooam, N. Dixon, M. Hilvert, P. Misko, K. Waters, N. Jourdan, S. Drahy, S. Mills, D. Engle, J. Link, M. Ahmad, Effect of temperature on the Arabidopsis cryptochrome photocycle, Physiologia Plantarum 172 (2021) 1653–1661. [CrossRef]
- K.M.W. Findlay, G.I. Jenkins, Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions, Plant, Cell & Environment 39 (2016) 1706–1714. [CrossRef]
- P.A. Salomé, In the Heat of the Moment: ZTL-Mediated Protein Quality Control at High Temperatures, The Plant Cell 29 (2017) 2685–2686. [CrossRef]
- M. Noguchi, Y. Kodama, Temperature Sensing in Plants: On the Dawn of Molecular Thermosensor Research, Plant and Cell Physiology 63 (2022) 737–743. [CrossRef]
- Ponnu, U. Hoecker, Illuminating the COP1/SPA Ubiquitin Ligase: Fresh Insights Into Its Structure and Functions During Plant Photomorphogenesis, Frontiers in Plant Science 12 (2021). [CrossRef]
- N. Pires, L. Dolan, Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants, Molecular Biology and Evolution 27 (2010) 862–874. [CrossRef]
- Y. Hao, X. Zong, P. Ren, Y. Qian, A. Fu, Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis, International Journal of Molecular Sciences 22 (2021) 7152. [CrossRef]
- Y. Qian, T. Zhang, Y. Yu, L. Gou, J. Yang, J. Xu, E. Pi, Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses, Frontiers in Plant Science 12 (2021). https://www.frontiersin.org/articles/10.3389/fpls.2021.677611 (accessed October 11, 2022).
- N. Nesi, I. Debeaujon, C. Jond, G. Pelletier, M. Caboche, L. Lepiniec, The TT8 Gene Encodes a Basic Helix-Loop-Helix Domain Protein Required for Expression of DFR and BAN Genes in Arabidopsis Siliques, Plant Cell 12 (2000) 1863–1878.
- M. Devic, J. Guilleminot, I. Debeaujon, N. Bechtold, E. Bensaude, M. Koornneef, G. Pelletier, M. Delseny, The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development, The Plant Journal 19 (1999) 387–398. [CrossRef]
- D.-Y. Xie, S.B. Sharma, N.L. Paiva, D. Ferreira, R.A. Dixon, Role of Anthocyanidin Reductase, Encoded by BANYULS in Plant Flavonoid Biosynthesis, Science 299 (2003) 396–399. [CrossRef]
- L.G.G. Corrêa, D.M. Riaño-Pachón, C.G. Schrago, R.V. dos Santos, B. Mueller-Roeber, M. Vincentz, The Role of bZIP Transcription Factors in Green Plant Evolution: Adaptive Features Emerging from Four Founder Genes, PLOS One 3 (2008) e2944. [CrossRef]
- Y. Yu, Y. Qian, M. Jiang, J. Xu, J. Yang, T. Zhang, L. Gou, E. Pi, Regulation Mechanisms of Plant Basic Leucine Zippers to Various Abiotic Stresses, Frontiers in Plant Science 11 (2020). https://www.frontiersin.org/articles/10.3389/fpls.2020.01258 (accessed October 11, 2022).
- M.S. Alves, S.P. Dadalto, A.B. Gonçalves, G.B. De Souza, V.A. Barros, L.G. Fietto, Plant bZIP Transcription Factors Responsive to Pathogens: A Review, International Journal of Molecular Sciences 14 (2013) 7815–7828. [CrossRef]
- S.N. Gangappa, J.F. Botto, The Multifaceted Roles of HY5 in Plant Growth and Development, Molecular Plant 9 (2016) 1353–1365. [CrossRef]
- N.H. Nguyen, HY5, an integrator of light and temperature signals in the regulation of anthocyanins biosynthesis in Arabidopsis, AIMS Molecular Science 7 (2020) 70–81. [CrossRef]
- D. Xu, Y. Jiang, J. Li, F. Lin, M. Holm, X.W. Deng, BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation, Proceedings of the National Academy of Sciences 113 (2016) 7655–7660. [CrossRef]
- M. Holm, L.-G. Ma, L.-J. Qu, X.-W. Deng, Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis, Genes & Development 16 (2002) 1247–1259. [CrossRef]
- Y. Zhang, S. Zheng, Z. Liu, L. Wang, Y. Bi, Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings, Journal of Plant Physiology 168 (2011) 367–374. [CrossRef]
- C. Dubos, R. Stracke, E. Grotewold, B. Weisshaar, C. Martin, L. Lepiniec, MYB transcription factors in Arabidopsis, Trends in Plant Science 15 (2010) 573–581. [CrossRef]
- A. Katiyar, S. Smita, S.K. Lenka, R. Rajwanshi, V. Chinnusamy, K.C. Bansal, Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis, BMC Genomics 13 (2012) 544. [CrossRef]
- N. Nesi, C. Jond, I. Debeaujon, M. Caboche, L. Lepiniec, The Arabidopsis TT2 Gene Encodes an R2R3 MYB Domain Protein That Acts as a Key Determinant for Proanthocyanidin Accumulation in Developing Seed, Plant Cell 13 (2001) 2099–2114. [CrossRef]
- Y. Yang, R. Sulpice, A. Himmelbach, M. Meinhard, A. Christmann, E. Grill, Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection, Proceedings of the National Academy of Sciences of the United States of America 103 (2006) 6061–6066. [CrossRef]
- M.-Z. Shi, D.-Y. Xie, Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions, Planta 231 (2010) 1385–1400. [CrossRef]
- Y. Zhang, Y.-P. Yan, Z.-Z. Wang, The Arabidopsis PAP1 Transcription Factor Plays an Important Role in the Enrichment of Phenolic Acids in Salvia miltiorrhiza, Journal of Agricultural and Food Chemistry 58 (2010) 12168–12175. [CrossRef]
- X. Li, M.-J. Gao, H.-Y. Pan, D.-J. Cui, M.Y. Gruber, Purple Canola: Arabidopsis PAP1 Increases Antioxidants and Phenolics in Brassica napus Leaves, Journal of Agricultural and Food Chemistry 58 (2010) 1639–1645. [CrossRef]
- T. Mitsunami, M. Nishihara, I. Galis, K.M. Alamgir, Y. Hojo, K. Fujita, N. Sasaki, K. Nemoto, T. Sawasaki, G. Arimura, Overexpression of the PAP1 Transcription Factor Reveals a Complex Regulation of Flavonoid and Phenylpropanoid Metabolism in Nicotiana tabacum Plants Attacked by Spodoptera litura, PLOS ONE 9 (2014) e108849. [CrossRef]
- A. Youssef, Y. Laizet, M.A. Block, E. Maréchal, J.-P. Alcaraz, T.R. Larson, D. Pontier, J. Gaffé, M. Kuntz, Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress, The Plant Journal 61 (2010) 436–445. [CrossRef]
- J. Jin, F. Tian, D.-C. Yang, Y.-Q. Meng, L. Kong, J. Luo, G. Gao, PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants, Nucleic Acids Research 45 (2017) D1040–D1045. [CrossRef]
- J. Jiang, S. Ma, N. Ye, M. Jiang, J. Cao, J. Zhang, WRKY transcription factors in plant responses to stresses, Journal of Integrative Plant Biology 59 (2017) 86–101. [CrossRef]
- S. Guillaumie, R. Mzid, V. Méchin, C. Léon, I. Hichri, A. Destrac-Irvine, C. Trossat-Magnin, S. Delrot, V. Lauvergeat, The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco, Plant Molecular Biology 72 (2010) 215–234. [CrossRef]
- U.J. Phukan, G.S. Jeena, R.K. Shukla, WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants, Frontiers in Plant Science 7 (2016) 760. [CrossRef]
- W. Grunewald, I. De Smet, D.R. Lewis, C. Löfke, L. Jansen, G. Goeminne, R. Vanden Bossche, M. Karimi, B. De Rybel, B. Vanholme, T. Teichmann, W. Boerjan, M.C.E. Van Montagu, G. Gheysen, G.K. Muday, J. Friml, T. Beeckman, Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis, Proceedings of the National Academy of Sciences of the United States of America 109 (2012) 1554–1559. [CrossRef]
- M. Aamir, V.K. Singh, M. Meena, R.S. Upadhyay, V.K. Gupta, S. Singh, Structural and Functional Insights into WRKY3 and WRKY4 Transcription Factors to Unravel the WRKY–DNA (W-Box) Complex Interaction in Tomato (Solanum lycopersicum L.). A Computational Approach, Frontiers in Plant Science 8 (2017) 819. [CrossRef]
- L. Chen, Y. Song, S. Li, L. Zhang, C. Zou, D. Yu, The role of WRKY transcription factors in plant abiotic stresses, Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819 (2012) 120–128. [CrossRef]
- Y. Chi, Y. Yang, Y. Zhou, J. Zhou, B. Fan, J.-Q. Yu, Z. Chen, Protein–Protein Interactions in the Regulation of WRKY Transcription Factors, Molecular Plant 6 (2013) 287–300. [CrossRef]
- Y. Yang, T. Liang, L. Zhang, K. Shao, X. Gu, R. Shang, N. Shi, X. Li, P. Zhang, H. Liu, UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis, Nature Plants 4 (2018) 98–107. [CrossRef]
- S. van Nocker, P. Ludwig, The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function, BMC Genomics 4 (2003) 50. [CrossRef]
- A.K. Mishra, S. Puranik, M. Prasad, Structure and regulatory networks of WD40 protein in plants, Journal of Plant Biochemistry and Biotechnology 21 (2012) 32–39. [CrossRef]
- Y. Pan, H. Shi, Stabilizing the Transcription Factors by E3 Ligase COP1, Trends in Plant Science 22 (2017) 999–1001. [CrossRef]
- S. Laubinger, K. Fittinghoff, U. Hoecker, The SPA Quartet: A Family of WD-Repeat Proteins with a Central Role in Suppression of Photomorphogenesis in Arabidopsis, Plant Cell 16 (2004) 2293–2306. [CrossRef]
- A. Lloyd, A. Brockman, L. Aguirre, A. Campbell, A. Bean, A. Cantero, A. Gonzalez, Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation, Plant and Cell Physiology 58 (2017) 1431–1441. [CrossRef]
- S. Li, Transcriptional control of flavonoid biosynthesis, Plant Signaling & Behavior 9 (2014) e27522. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).