Submitted:
03 January 2024
Posted:
17 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Culture Conditions of N. oceanica
2.2. Total RNA Extraction
2.3. Illumina Novaseq 6000 Sequencing and mRNA Sequencing Library Preparation
2.4. Transcriptome Assembly and Functional Annotation
2.5. Differential Expression Analysis and Functional Enrichment
3. Results and Discussion
3.1. Physiological Changes after Urea Addition
3.2. Molecular-Level Response of N. oceanica to Urea
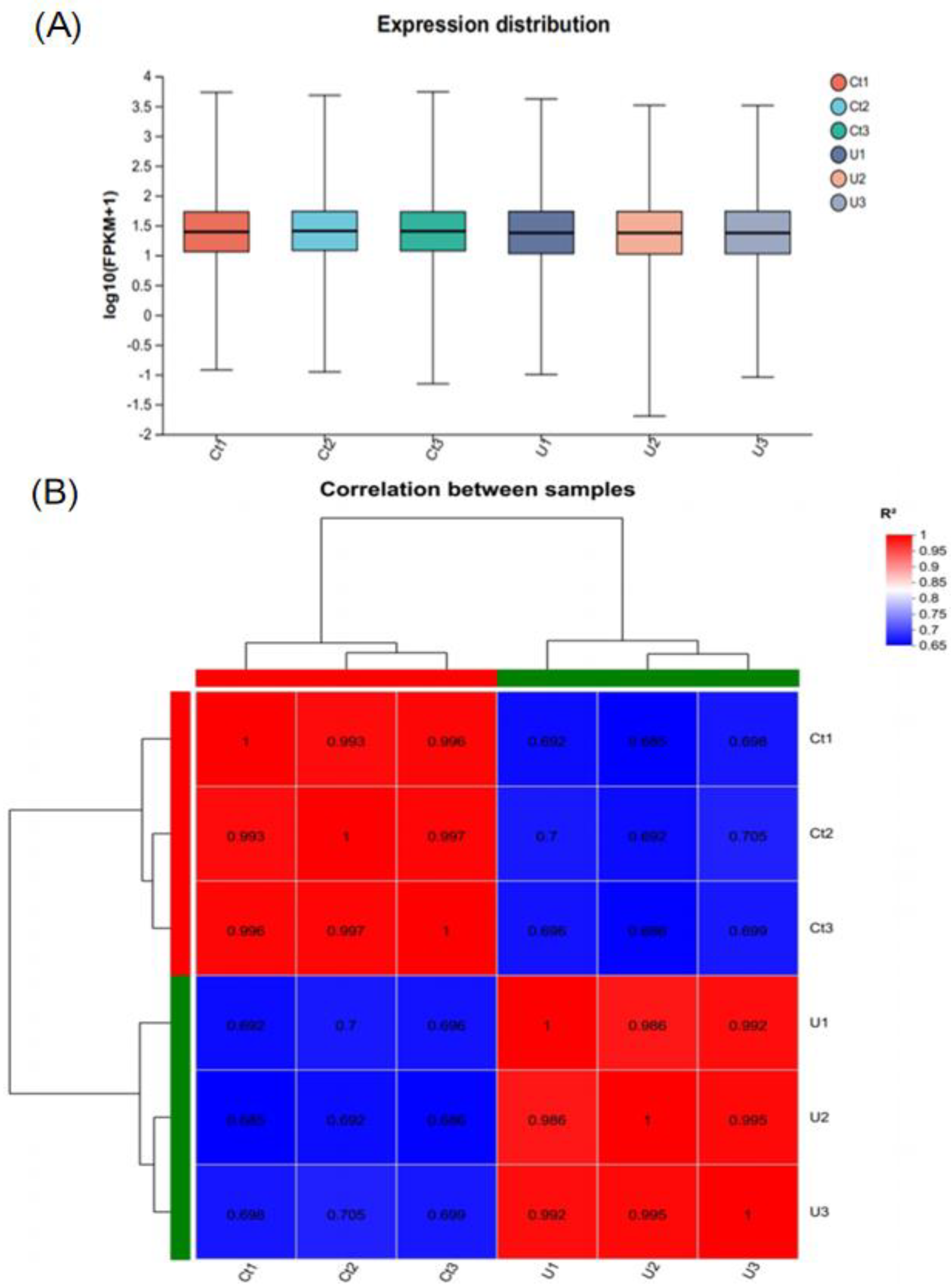
3.2.1. Transcriptome Data of N. oceanica Cruentum from Illumina Sequencing
3.2.2. Sample Correlation AnalysisInter-Sample Venn Analysis and Differentially Gene Expression Analysis by the Volcano Plot
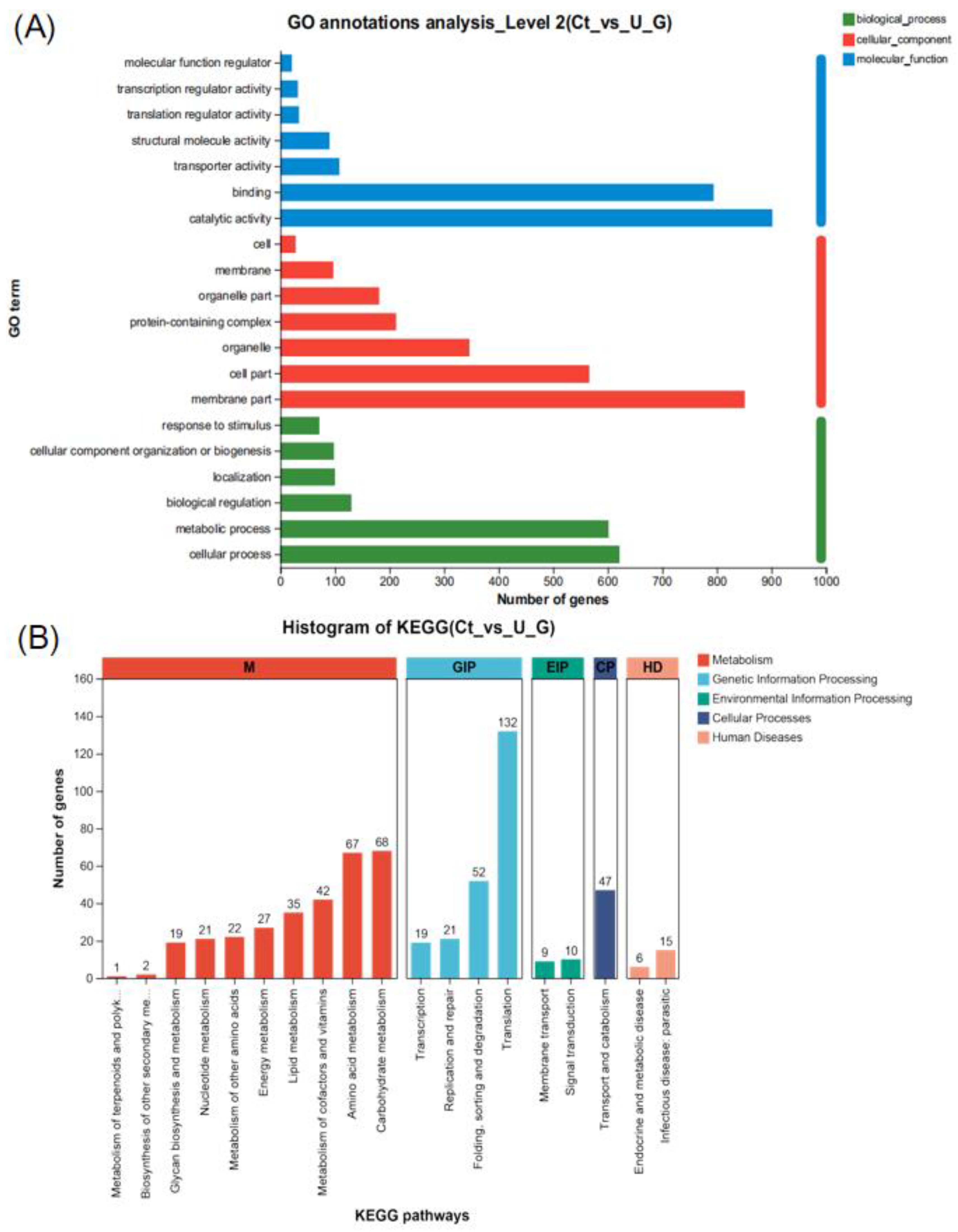
3.2.3. Functional Enrichment of Differential Expressed Gene by GO and KEGG
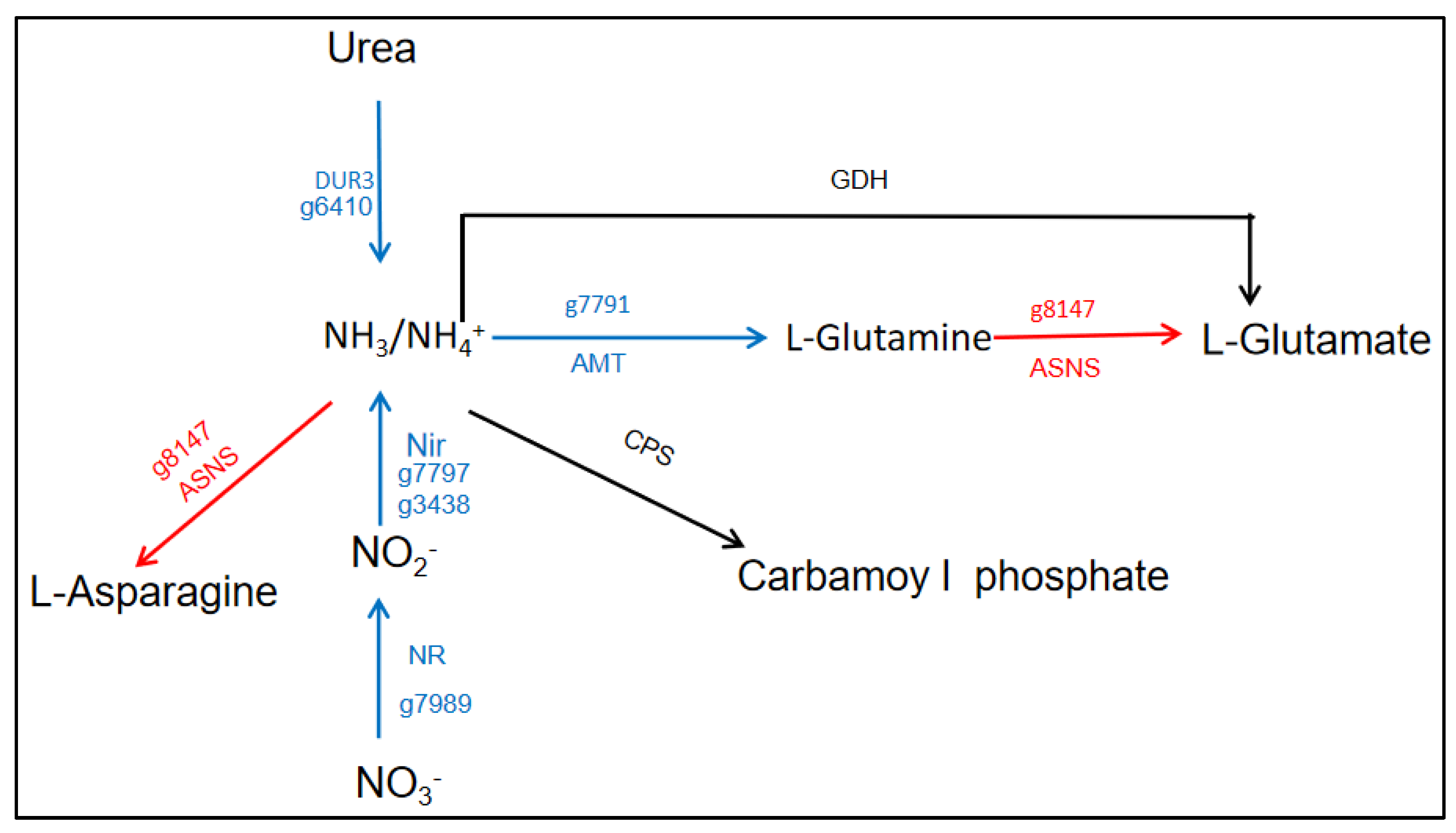
3.3. Nitrogen Metabolism Affected by Urea Addition in N. oceanica
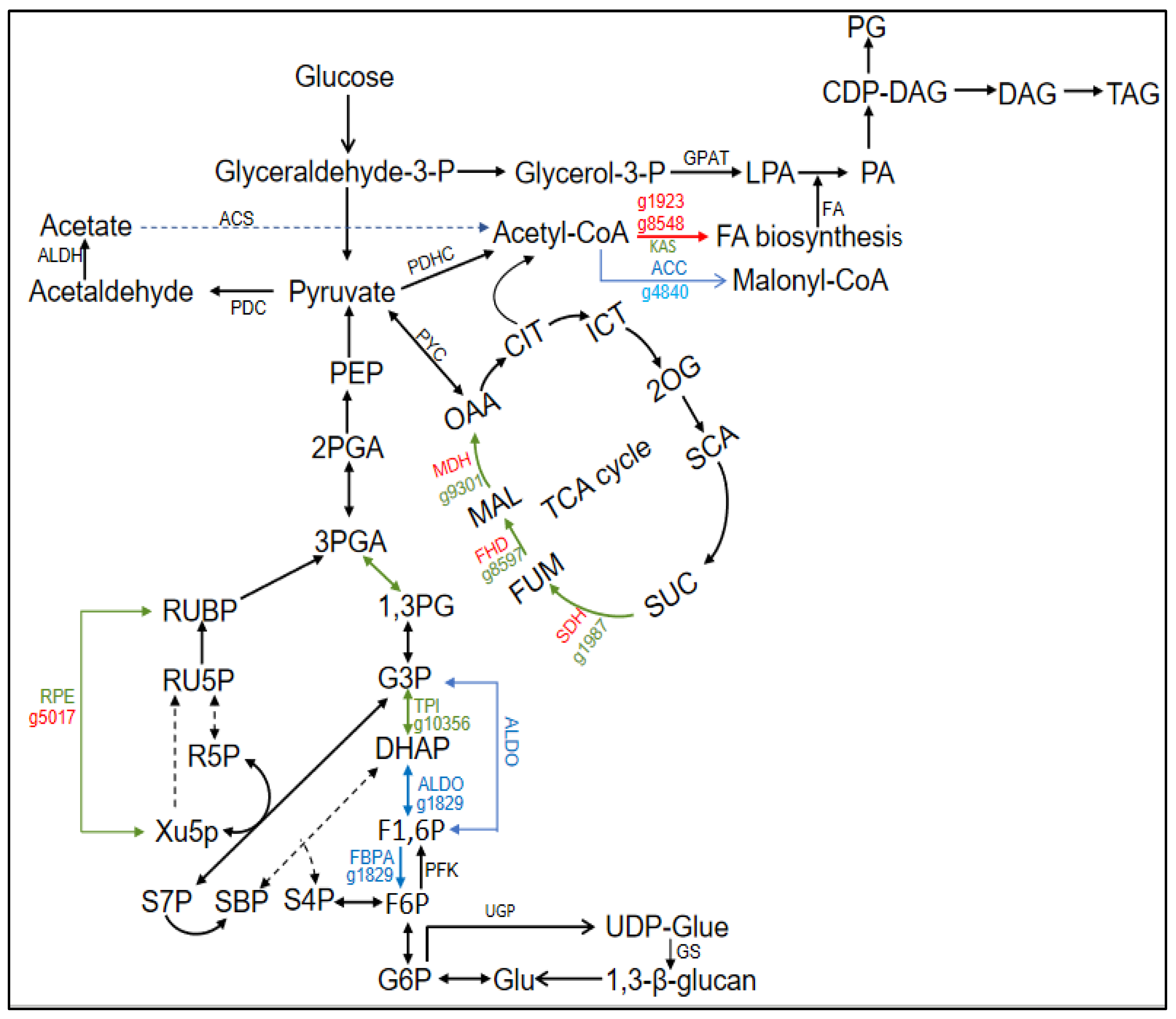
3.4. Change of Photosynthesis in Response to Urea Addition
3.5. Change of Carbon Fixation and Central Carbon Metabolism in Response to Urea Addition
3.6. Lipid Metabolism Affected by Urea Addition
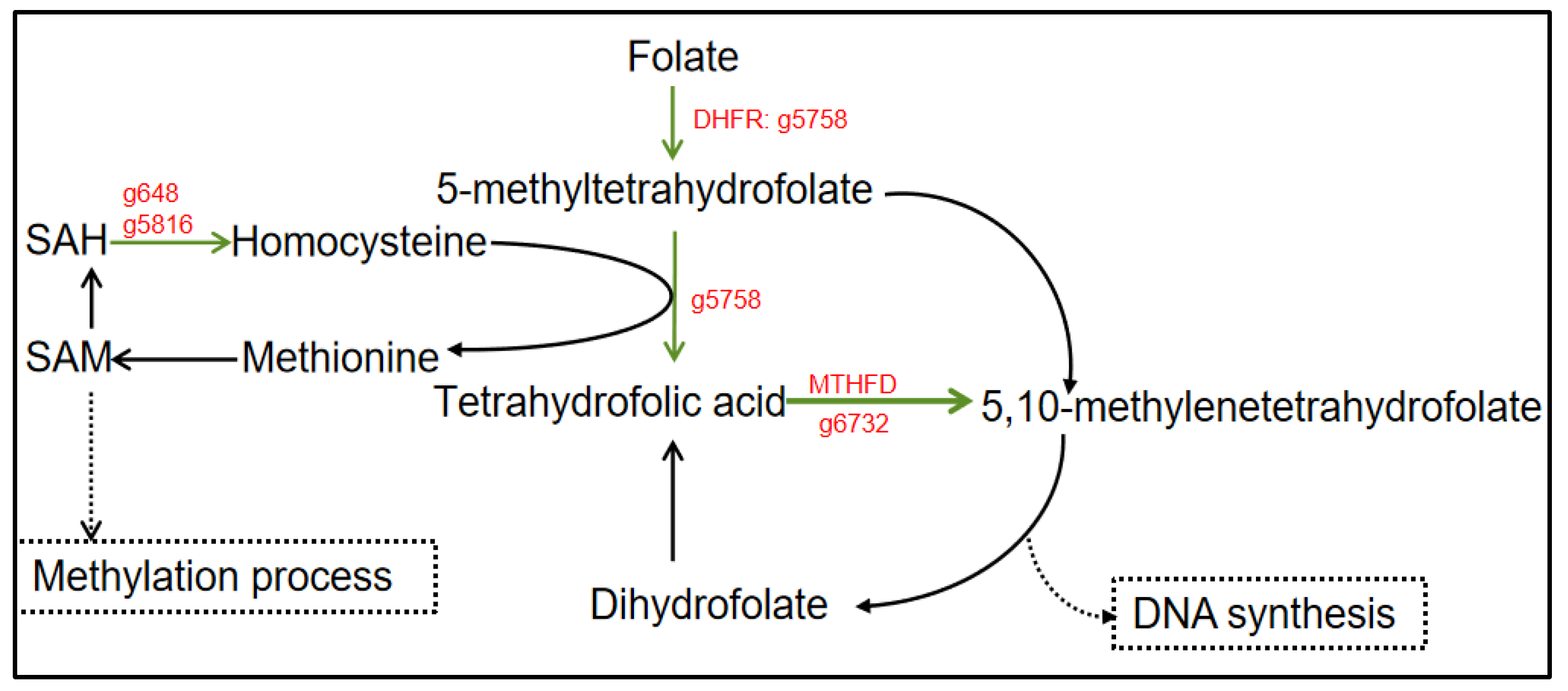
3.7. Folate Metabolism Affected by Urea Addition
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Rosa, R. M.; Machado, M.; Vaz, M.; Lopes-Santos, R.; Nascimento, A. G. D.; Araújo, W. L.; Nunes-Nesi, A., Urea as a source of nitrogen and carbon leads to increased photosynthesis rates in Chlamydomonas reinhardtii under mixotrophy. J Biotechnol 2023, 367, 20-30. [CrossRef]
- Sun, M.; Li, S.; Yu, H.; Gong, Q.; Zhang, B.; Liu, G.; Xiao, Y.; Peng, F., Effects of Valine and Urea on Carbon and Nitrogen Accumulation and Lignin Content in Peach Trees. Plants (Basel) 2023, 12 (8). [CrossRef]
- Kumbhar, A. N.; He, M.; Rajper, A. R.; Memon, K. A.; Rizwan, M.; Nagi, M.; Woldemicael, A. G.; Li, D.; Wang, C.; Wang, C., The Use of Urea and Kelp Waste Extract is A Promising Strategy for Maximizing the Biomass Productivity and Lipid Content in Chlorella sorokiniana. Plants (Basel) 2020, 9 (4). [CrossRef]
- Kuo, C. M.; Yang, Y. C.; Zhang, W. X.; Wu, J. X.; Chen, Y. T.; Lin, C. H.; Lin, M. W.; Lin, C. S., A Low-Cost Fertilizer Medium Supplemented with Urea for the Lutein Production of Chlorella sp. and the Ability of the Lutein to Protect Cells against Blue Light Irradiation. Bioengineering (Basel) 2023, 10 (5). [CrossRef]
- Lai, Y. J., Omega-3 fatty acid obtained from Nannochloropsis oceanica cultures grown under low urea protect against Abeta-induced neural damage. J Food Sci Technol 2015, 52 (5), 2982-9. [CrossRef]
- Abougrara, A. M., Effects of Different Levels of Urea as Nitrogen Source on Chemical Composition of Marine Microalgae Nannochloropsis oculata. Al-Mukhtar Journal of Sciences 2021, 36 (1), 1-11. [CrossRef]
- Majid, M.; Salimeh, S., Characterization of the Growth, Total Lipid and Fatty Acid Profiles in Microalga, Nannochloropsis oceanica under Different Nitrogen Sources. Microbiol. Biotechnol. Lett 2019, 47 (1), 11-19.
- Dong, H. P.; Williams, E.; Wang, D. Z.; Xie, Z. X.; Hsia, R. C.; Jenck, A.; Halden, R.; Li, J.; Chen, F.; Place, A. R., Responses of Nannochloropsis oceanica IMET1 to Long-Term Nitrogen Starvation and Recovery. Plant Physiol 2013, 162 (2), 1110-26. [CrossRef]
- Kang, N. K.; Jeon, S.; Kwon, S.; Koh, H. G.; Shin, S. E.; Lee, B.; Choi, G. G.; Yang, J. W.; Jeong, B. R.; Chang, Y. K., Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol Biofuels 2015, 8, 200. [CrossRef]
- Li, J.; Han, D.; Wang, D.; Ning, K.; Jia, J.; Wei, L.; Jing, X.; Huang, S.; Chen, J.; Li, Y.; Hu, Q.; Xu, J., Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae. Plant Cell 2014, 26 (4), 1645-1665. [CrossRef]
- Haas, B. J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P. D.; Bowden, J.; Couger, M. B.; Eccles, D.; Li, B.; Lieber, M.; MacManes, M. D.; Ott, M.; Orvis, J.; Pochet, N.; Strozzi, F.; Weeks, N.; Westerman, R.; William, T.; Dewey, C. N.; Henschel, R.; LeDuc, R. D.; Friedman, N.; Regev, A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 2013, 8 (8), 1494-512. [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J. M.; Terol, J.; Talon, M.; Robles, M., Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. BIOINFORMATICS 2005, 21 (18), 3674-6. [CrossRef]
- Goto, M. K. a. S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 2000, 28 (1), 27-30. [CrossRef]
- Raghavan, V.; Kraft, L.; Mesny, F.; Rigerte, L., A simple guide to de novo transcriptome assembly and annotation. Briefings in Bioinformatics 2022, 23 (2). [CrossRef]
- Soneson, C.; Love, M. I.; Robinson, M. D., Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M. W.; Gaffney, D. J.; Elo, L. L.; Zhang, X.; Mortazavi, A., A survey of best practices for RNA-seq data analysis. Genome Biology 2016, 17 (1), 13. [CrossRef]
- Li, B.; Dewey, C. N., RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [CrossRef]
- Robinson, M. D.; McCarthy, D. J.; Smyth, G. K., edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. BIOINFORMATICS 2010, 26 (1), 139-40. [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C. Y.; Wei, L., KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011, 39 (Web Server issue), W316-22. [CrossRef]
- Zhao, S.; Ye, Z.; Stanton, R., Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. Rna 2020, 26 (8), 903-909. [CrossRef]
- Vera Alvarez, R.; Pongor, L. S.; Mariño-Ramírez, L.; Landsman, D., TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 2019, 35 (11), 1960-1962. [CrossRef]
- Mohapatra, S.; Weisshaar, J. C., Modified Pearson correlation coefficient for two-color imaging in spherocylindrical cells. BMC Bioinformatics 2018, 19 (1), 428. [CrossRef]
- Salbitani, G.; Carfagna, S., Ammonium Utilization in Microalgae: A Sustainable Method for Wastewater Treatment. Sustainability 2021, 13 (2), 956. [CrossRef]
- Calabrese, S.; Pérez-Tienda, J.; Ellerbeck, M.; Arnould, C.; Chatagnier, O.; Boller, T.; Schüßler, A.; Brachmann, A.; Wipf, D.; Ferrol, N.; Courty, P.-E., GintAMT3 – a Low-Affinity Ammonium Transporter of the Arbuscular Mycorrhizal Rhizophagus irregularis. Frontiers in Plant Science 2016, 7. [CrossRef]
- Lanquar, V.; Loqué, D.; Hörmann, F.; Yuan, L.; Bohner, A.; Engelsberger, W. R.; Lalonde, S.; Schulze, W. X.; von Wirén, N.; Frommer, W. B., Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 2009, 21 (11), 3610-22.
- Hao, D.; Li, X.; Kong, W.; Chen, R.; Liu, J.; Guo, H.; Zhou, J., Phosphorylation regulation of nitrogen, phosphorus, and potassium uptake systems in plants. The Crop Journal 2023, 11 (4), 1034-1047. [CrossRef]
- Loqué, D.; Lalonde, S.; Looger, L. L.; von Wirén, N.; Frommer, W. B., A cytosolic trans-activation domain essential for ammonium uptake. Nature 446 (7132), 195-8. [CrossRef]
- Neuhäuser, B.; Dynowski, M.; Mayer, M.; Ludewig, U., Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 2007, 143 (4), 1651-9. [CrossRef]
- Miller, E. F.; Maier, R. J., Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J Bacteriol 2014, 196 (17), 3074-81. [CrossRef]
- Pechkovskaya, S. A.; Knyazev, N. A.; Matantseva, O. V.; Emelyanov, A. K.; Telesh, I. V.; Skarlato, S. O.; Filatova, N. A., Dur3 and nrt2 genes in the bloom-forming dinoflagellate Prorocentrum minimum: Transcriptional responses to available nitrogen sources. Chemosphere 2020, 241, 125083. [CrossRef]
- Veaudor, T.; Cassier-Chauvat, C.; Chauvat, F., Genomics of Urea Transport and Catabolism in Cyanobacteria: Biotechnological Implications. Frontiers in Microbiology 2019, 10. [CrossRef]
- Su, Z.; Olman, V.; Mao, F.; Xu, Y., Comparative genomics analysis of NtcA regulons in cyanobacteria: regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res 2005, 33 (16), 5156-71. [CrossRef]
- Beckers, G.; Bendt, A. K.; Krämer, R.; Burkovski, A., Molecular Identification of the Urea Uptake System and Transcriptional Analysis of Urea Transporter- and Urease-Encoding Genes in Corynebacterium glutamicum. Journal of Bacteriology 2004, 186, 7645 - 7652. [CrossRef]
- Hodin, J.; Lind, C.; Marmagne, A.; Espagne, C.; Bianchi, M. W.; De Angeli, A.; Abou-Choucha, F.; Bourge, M.; Chardon, F.; Thomine, S.; Filleur, S., Proton exchange by the vacuolar nitrate transporter CLCa is required for plant growth and nitrogen use efficiency. Plant Cell 2023, 35 (1), 318-335. [CrossRef]
- Mérigout, P.; Lelandais, M.; Bitton, F.; Renou, J. P.; Briand, X.; Meyer, C.; Daniel-Vedele, F., Physiological and transcriptomic aspects of urea uptake and assimilation in Arabidopsis plants. Plant Physiol 2008, 147 (3), 1225-38. [CrossRef]
- Gutierrez, J.; Kwan, T.; Zimmerman, J.; Peccia, J., Ammonia inhibition in oleaginous microalgae. Algal Research 2016, 19, 123-127. [CrossRef]
- Takeuchi, T.; Benning, C., Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas. Biotechnology for Biofuels 2019, 12 (1), 292. [CrossRef]
- Ort, D. R.; Zhu, X.; Melis, A., Optimizing Antenna Size to Maximize Photosynthetic Efficiency. Plant Physiology 2010, 155 (1), 79-85. [CrossRef]
- Kirst, H.; Formighieri, C.; Melis, A., Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2014, 1837 (10), 1653-1664. [CrossRef]
- Nakajima, Y.; Itayama, T., Analysis of photosynthetic productivity of microalgal mass cultures. Journal of Applied Phycology 2003, 15 (6), 497-505. [CrossRef]
- Chadee, A.; Alber, N. A.; Dahal, K.; Vanlerberghe, G. C., The Complementary Roles of Chloroplast Cyclic Electron Transport and Mitochondrial Alternative Oxidase to Ensure Photosynthetic Performance. Front Plant Sci 2021, 12, 748204. [CrossRef]
- Wei, L.; Shen, C.; El Hajjami, M.; You, W.; Wang, Q.; Zhang, P.; Ji, Y.; Hu, H.; Hu, Q.; Poetsch, A.; Xu, J., Knockdown of carbonate anhydrase elevates Nannochloropsis productivity at high CO2 level. Metab Eng 2019, 54, 96-108. [CrossRef]
- Wei, L.; El Hajjami, M.; Shen, C.; You, W.; Lu, Y.; Li, J.; Jing, X.; Hu, Q.; Zhou, W.; Poetsch, A.; Xu, J., Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol Biofuels 2019, 12, 168. [CrossRef]
- Krausfeldt, L. E.; Farmer, A. T.; Castro Gonzalez, H. F.; Zepernick, B. N.; Campagna, S. R.; Wilhelm, S. W., Urea Is Both a Carbon and Nitrogen Source for Microcystis aeruginosa: Tracking 13C Incorporation at Bloom pH Conditions. Frontiers in Microbiology 2019, 10. [CrossRef]
- Matsuda, Y.; Hopkinson, B. M.; Nakajima, K.; Dupont, C. L.; Tsuji, Y., Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism. Philos Trans R Soc Lond B Biol Sci 2017, 372 (1728). [CrossRef]
- Becker, P.; Naughton, F.; Brotherton, D.; Pacheco-Gomez, R.; Beckstein, O.; Cameron, A. D., Mechanism of substrate binding and transport in BASS transporters. Elife 2023, 12. [CrossRef]
- Arnold, P. K.; Finley, L. W. S., Regulation and function of the mammalian tricarboxylic acid cycle. J Biol Chem 2023, 299 (2), 102838. [CrossRef]
- Zarrinmehr, M. J.; Farhadian, O.; Heyrati, F. P.; Keramat, J.; Koutra, E.; Kornaros, M.; Daneshvar, E., Effect of nitrogen concentration on the growth rate and biochemical composition of the microalga, Isochrysis galbana. The Egyptian Journal of Aquatic Research 2020, 46 (2), 153-158. [CrossRef]
- Choi, I.; Son, H.; Baek, J. H., Tricarboxylic Acid (TCA) Cycle Intermediates: Regulators of Immune Responses. Life (Basel) 2021, 11 (1). [CrossRef]
- Rosa, R. M.; Machado, M.; Vaz, M. G. M. V.; Lopes-Santos, R.; Nascimento, A. G. d.; Araújo, W. L.; Nunes-Nesi, A., Urea as a source of nitrogen and carbon leads to increased photosynthesis rates in Chlamydomonas reinhardtii under mixotrophy. Journal of Biotechnology 2023, 367, 20-30. [CrossRef]
- Ollis, D. L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S. M.; Harel, M.; Remington, S. J.; Silman, I.; Schrag, J.; et al., The alpha/beta hydrolase fold. Protein Eng 1992, 5 (3), 197-211.
- Nardini, M.; Dijkstra, B. W., Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 1999, 9 (6), 732-7. [CrossRef]
- Edwards, M.; Mohiuddin, S. S., Biochemistry, Lipolysis. In StatPearls, StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC.: Treasure Island (FL) ineligible companies. Disclosure: Shamim Mohiuddin declares no relevant financial relationships with ineligible companies., 2023.
- Bhuyar, P.; Sundararaju, S.; Rahim, M. H. A.; Maniam, G. P.; Govindan, N., Enhanced productivity of lipid extraction by urea stress conditions on marine microalgae Coelastrum sp. for improved biodiesel production. Bioresource Technology Reports 2021, 15, 100696. [CrossRef]
- Popko, J.; Herrfurth, C.; Feussner, K.; Ischebeck, T.; Iven, T.; Haslam, R.; Hamilton, M.; Sayanova, O.; Napier, J.; Khozin-Goldberg, I.; Feussner, I., Metabolome Analysis Reveals Betaine Lipids as Major Source for Triglyceride Formation, and the Accumulation of Sedoheptulose during Nitrogen-Starvation of Phaeodactylum tricornutum. PLoS One 2016, 11 (10), e0164673. [CrossRef]
- Gorelova, V.; Bastien, O.; De Clerck, O.; Lespinats, S.; Rébeillé, F.; Van Der Straeten, D., Evolution of folate biosynthesis and metabolism across algae and land plant lineages. Scientific Reports 2019, 9 (1), 5731. [CrossRef]
- Raghubeer, S.; Matsha, T. E., Methylenetetrahydrofolate (MTHFR), the One-Carbon Cycle, and Cardiovascular Risks. Nutrients 2021, 13 (12). [CrossRef]
- Geryk, J.; Krsička, D.; Vlčková, M.; Havlovicová, M.; Macek, M., Jr.; Kremlíková Pourová, R., The Key Role of Purine Metabolism in the Folate-Dependent Phenotype of Autism Spectrum Disorders: An In Silico Analysis. Metabolites 2020, 10 (5). [CrossRef]
- Rozen, R., Biochemistry and genetics of folate metabolism. Cerebrospinal Fluid Research 2010, 7 (1), S4. [CrossRef]
- Stover, P. J., One-carbon metabolism-genome interactions in folate-associated pathologies. J Nutr 2009, 139 (12), 2402-5. [CrossRef]
- Zheng, Y.; Cantley, L. C., Toward a better understanding of folate metabolism in health and disease. J Exp Med 2019, 216 (2), 253-266. [CrossRef]

| Gene ID | Gene name | Abbreviation | Fold change (U Vs Ct; fold) |
|
|---|---|---|---|---|
| g9735 | Kynureninase | KYNU | ↑177.17 | |
| g8147 | Asparagine synthase | ASNS | ↑63.69 | |
| g10184 | Delta-1-pyrroline-5-carboxylate synthetase | P5CS | ↑6.46 | |
| g4389 | Similar to dimethylanaline monooxygenase-like (predicted) | FMO | ↑4.23 | |
| g615 | Urate oxidase | UOX | ↑2.12 | |
| g8006 | Urease accessory protein | URE | ↓2.17 | |
| g6250 | Putative urate catabolism protein | UCP | ↓2.25 | |
| g7989 | Nitrate high affinity transporter | NR | ↓9.35 | |
| g6410 | Urea/Na+ high-affinity symporter | DUR3 | ↓13.33 | |
| g3438 | Ferredoxin nitrite reductase | NiR | ↓17.54 | |
| g7791 | Ammonium transporter | AMT | ↓21.7 | |
| g7797 | Nitroreductase-like protein | NTR | ↓28.57 |
| Gene ID | Gene name | Abbreviation | Fold change (U VsCt; fold) |
|---|---|---|---|
| g5017 | Ribulose-phosphate 3-epimerase | RPE | ↑20.97 |
| g9470 | Cytochrome b6-f complex iron-sulfur subunit | PetC | ↑2.42 |
| g876 | Light-dependent protochlorophyllide reductase | LPOR | ↑2.40 |
| g4344 | Iron-sulfur assembly-like protein | ISU | ↑15.12 |
| g240 | Light harvesting complex protein | LHC | ↓2.04 |
| g5628 | Light harvesting complex protein | LHC | ↓2.05 |
| g7977 | 3,8-divinyl protochlorophyllide a 8-vinyl reductase,putative chloroplast precursor | DVR | ↓2.12 |
| g4337 | Cytochrome c biogenesis protein,thiol reduction transmembrane region | CcdA | ↓2.14 |
| g5629 | Light harvesting complex protein | LHC | ↓2.18 |
| g3077 | Light harvesting complex protein | LHC | ↓2.00 |
| g6113 | Light harvesting complex protein | LHC | ↓2.32 |
| g9713 | Light harvesting complex protein | LHC | ↓2.46 |
| g903 | Light harvesting complex protein | LHC | ↓2.55 |
| g5529 | Ferredoxin(cyanobacterial type ferredoxin family) | Fd | ↓6.62 |
| Gene ID Gene name | Abbreviation | Fold change (U Vs Ct; fold) |
||
|---|---|---|---|---|
| Calvin Cycle | ||||
| g6144 | Phosphoglycerate kinase | PGK | ↑2.18 | |
| g10356 | Glyceraldehyde-3-phosphate dehydrogenase | GPDH | ↑2.47 | |
| g1829 | Fructose-1,6-bisphosphate aldolase | FBPA | ↓3.02 | |
| g8036 | Transketolase | TL | ↑2.07 | |
| g5017 | Ribulose-phosphate 3-epimerase | RPE | ↑20.97/3.41 | |
| CCM | ||||
| g2018 | Carbonic anhydrase | CA | ↑26.86/3.06 | |
| C4-like pathway | ||||
| g9301 | Malate dehydrogenase | MDH | ↑265.43 | |
| Degradation of 1,3-β glucan | ||||
| g5401 | Endoglucanase A | EG | ↓2.07 | |
| g4700 | Glucan 1,3-beta-glucosidase | GluB | ↑16.24 | |
| Glycolysis | ||||
| g1829 | Fructose-1,6-bisphosphate aldolase | ALDO | ↓3.02 | |
| g4700 | Triosephosphate isomerase | TPI | ↑2.47 | |
| g5401 | Phosphoglycerate kinase | PGK | ↑2.18 | |
| PDHC Bypass | ||||
| g2887 g956 |
Aldehyde dehydrogenase | ALDH | ↑2.71 | |
| ALDH | ↓2.36 | |||
| TCA cycle | ||||
| g1987 | Succinate dehydrogenase | SDH | ↑2.01 | |
| g8597 | Fumarate hydratase | FHD | ↑2.47 | |
| g9301 | Malate dehydrogenase | MDH | ↑265.43 | |
| Transporter | ||||
| g10029 | Sodium/hydrogen exchanger | NDH | ↓2.64 | |
| g1797 | ATP/ADP transporter | AAT | ↑4.62 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




