Submitted:
11 January 2024
Posted:
12 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Soil Samples, Strains and Plasmids
2.1.2. Culture Media
2.1.3. Buffer Solutions
2.2. Methods
2.2.1. Screening of Keratinase-Producing Strains
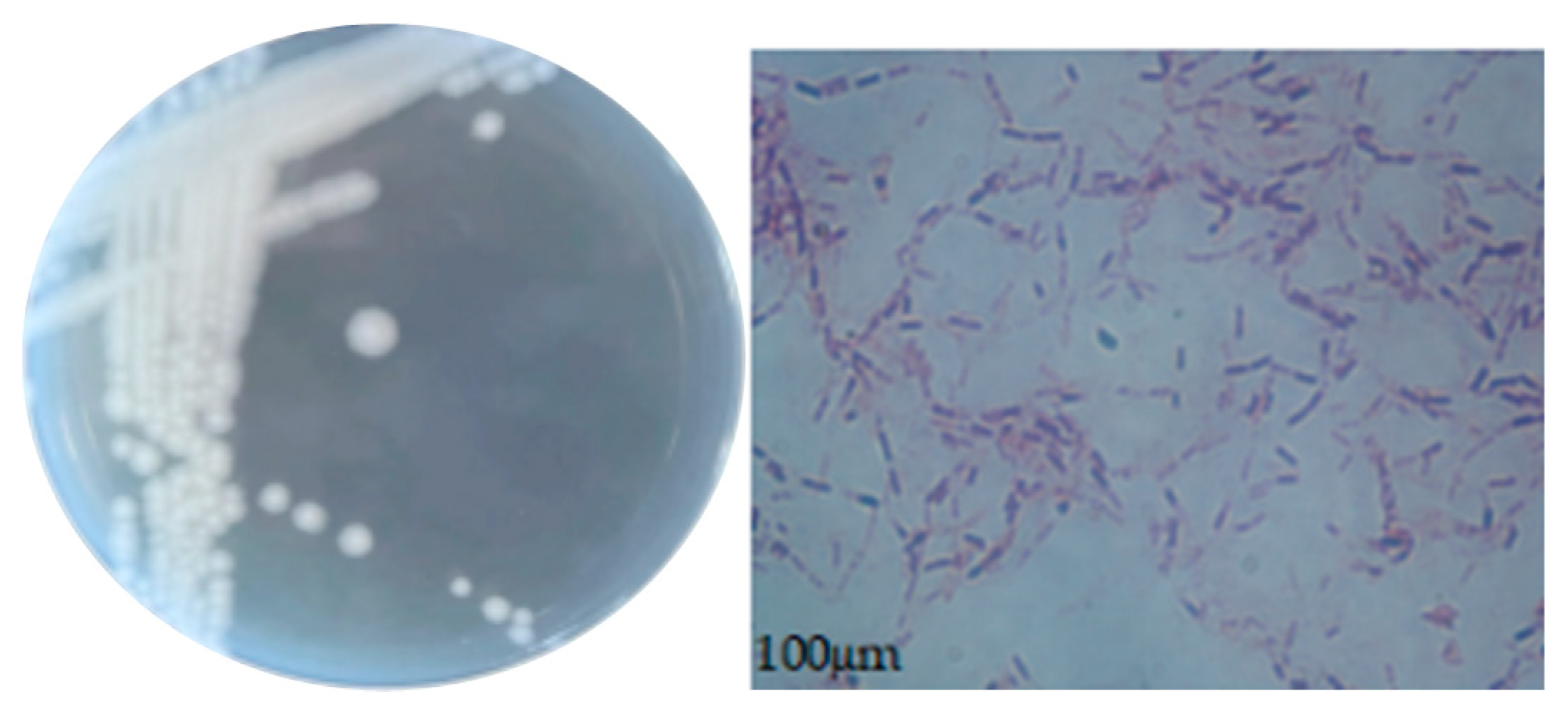
2.2.2. Morphological Observations of Colonies and Bacteria
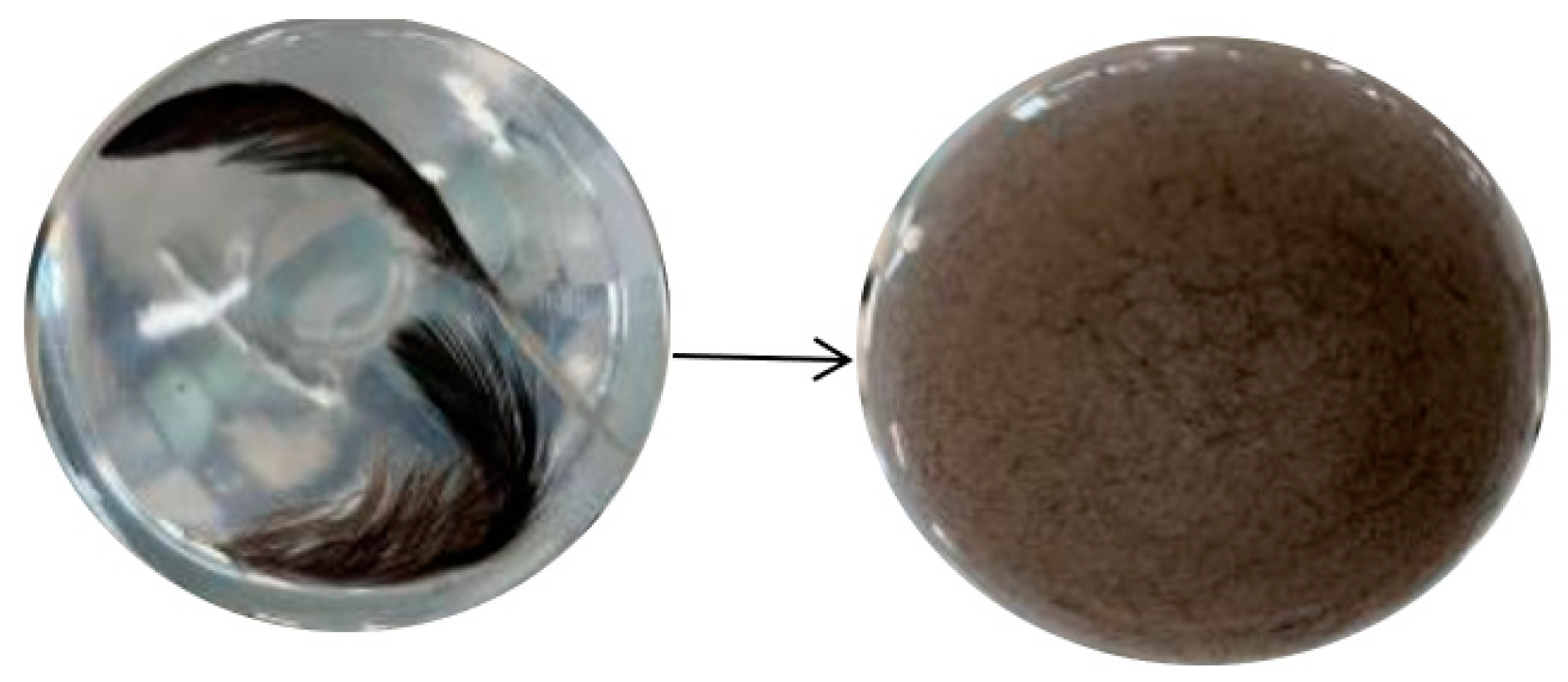
2.2.3. Determination of Keratinase Activity
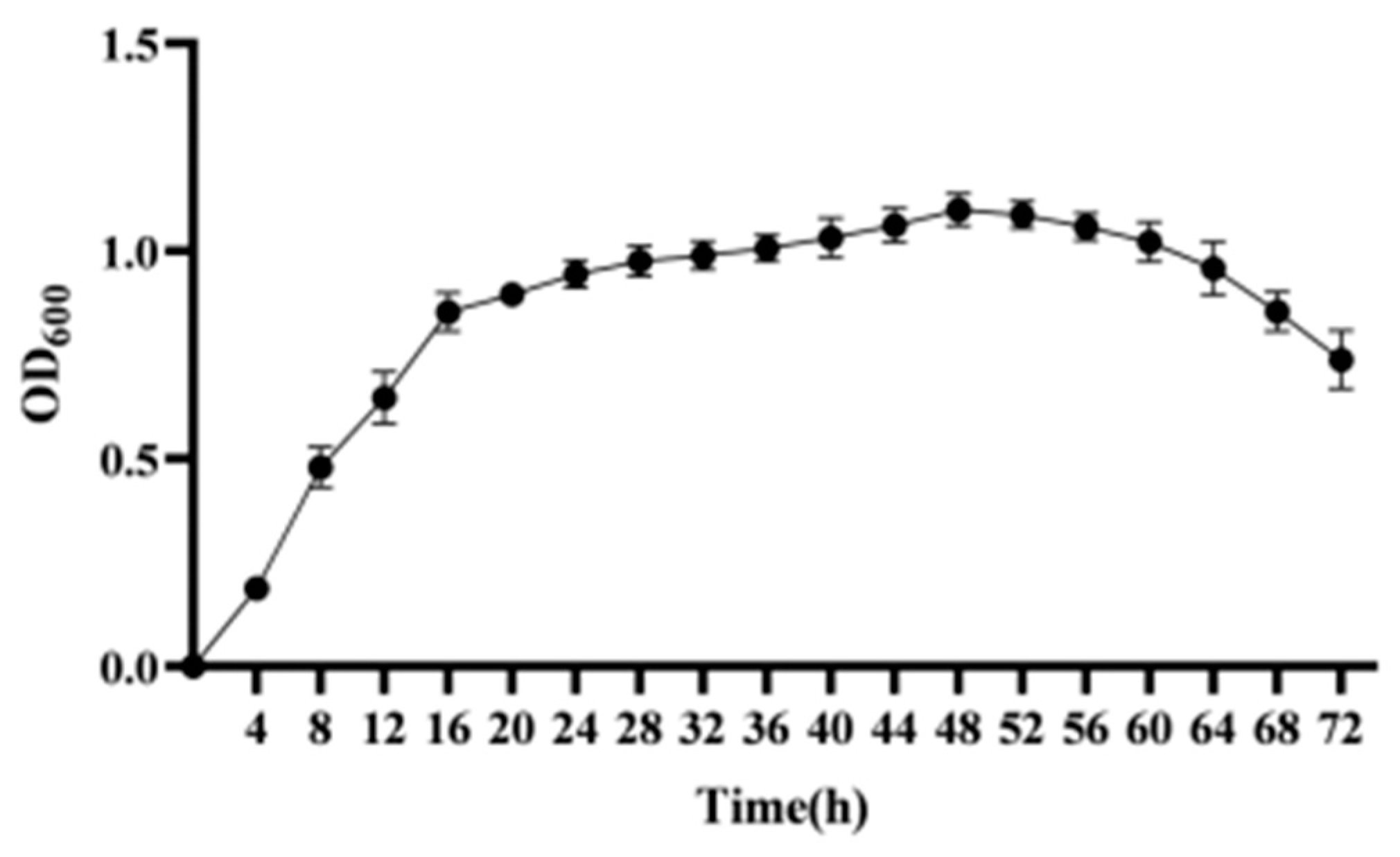
2.2.4. Determination of Strain Growth Curves
2.2.5. Physiological and Biochemical Experimental Identification
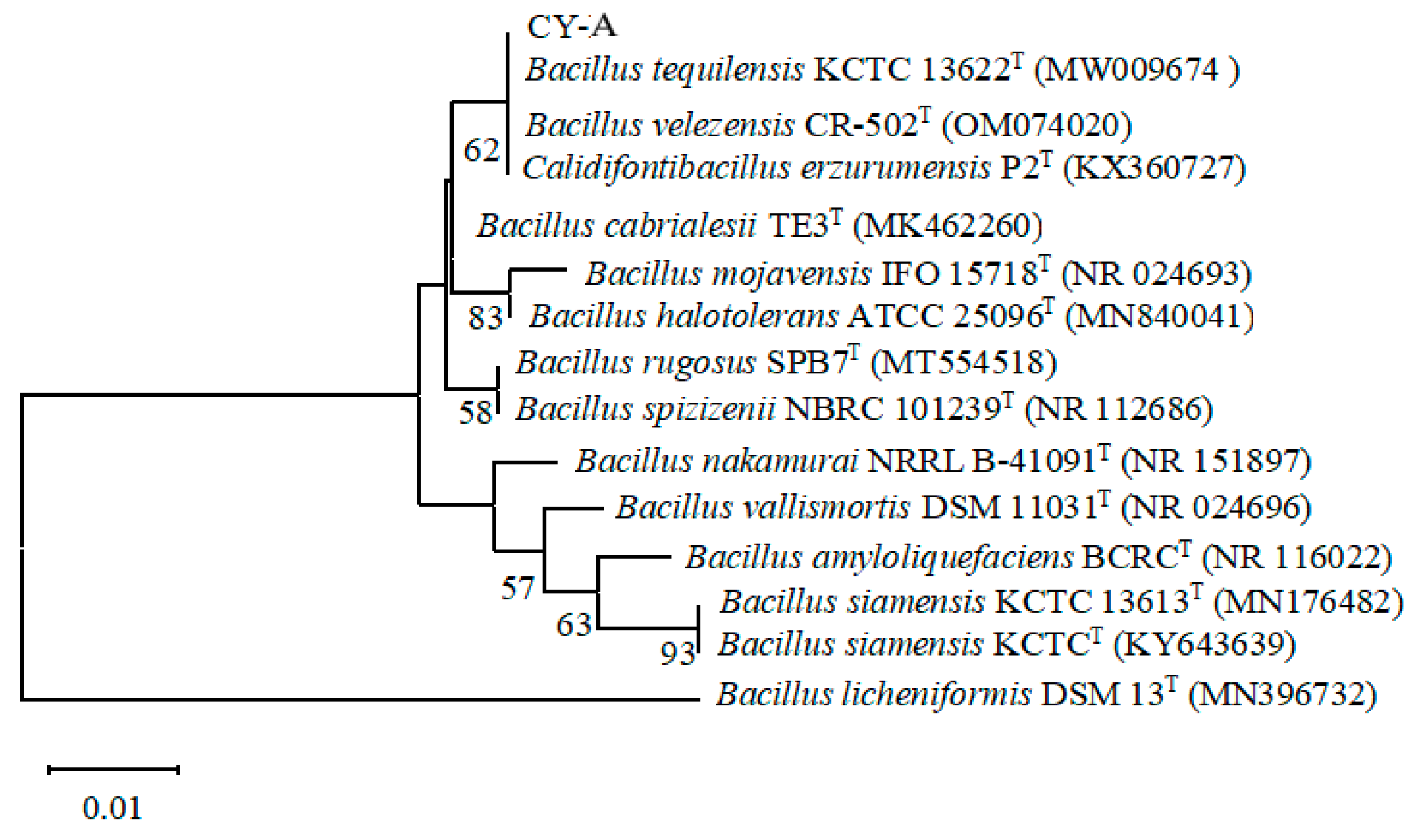
2.2.6. 16S rRNA Sequence Determination and Phylogenetic Tree Construction
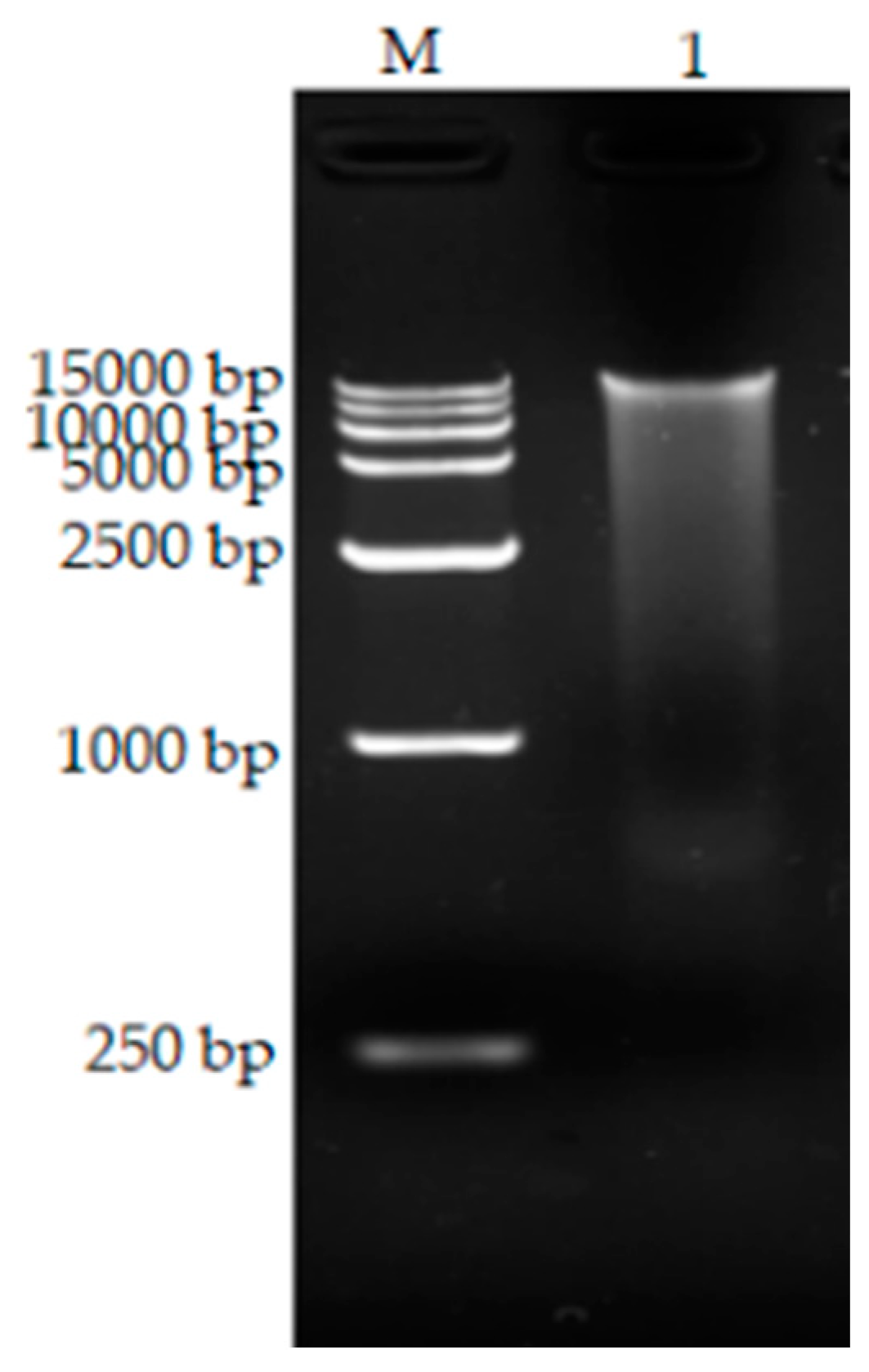
2.2.7. Extraction of Total DNA from Bacillus Tequilensis CY-A
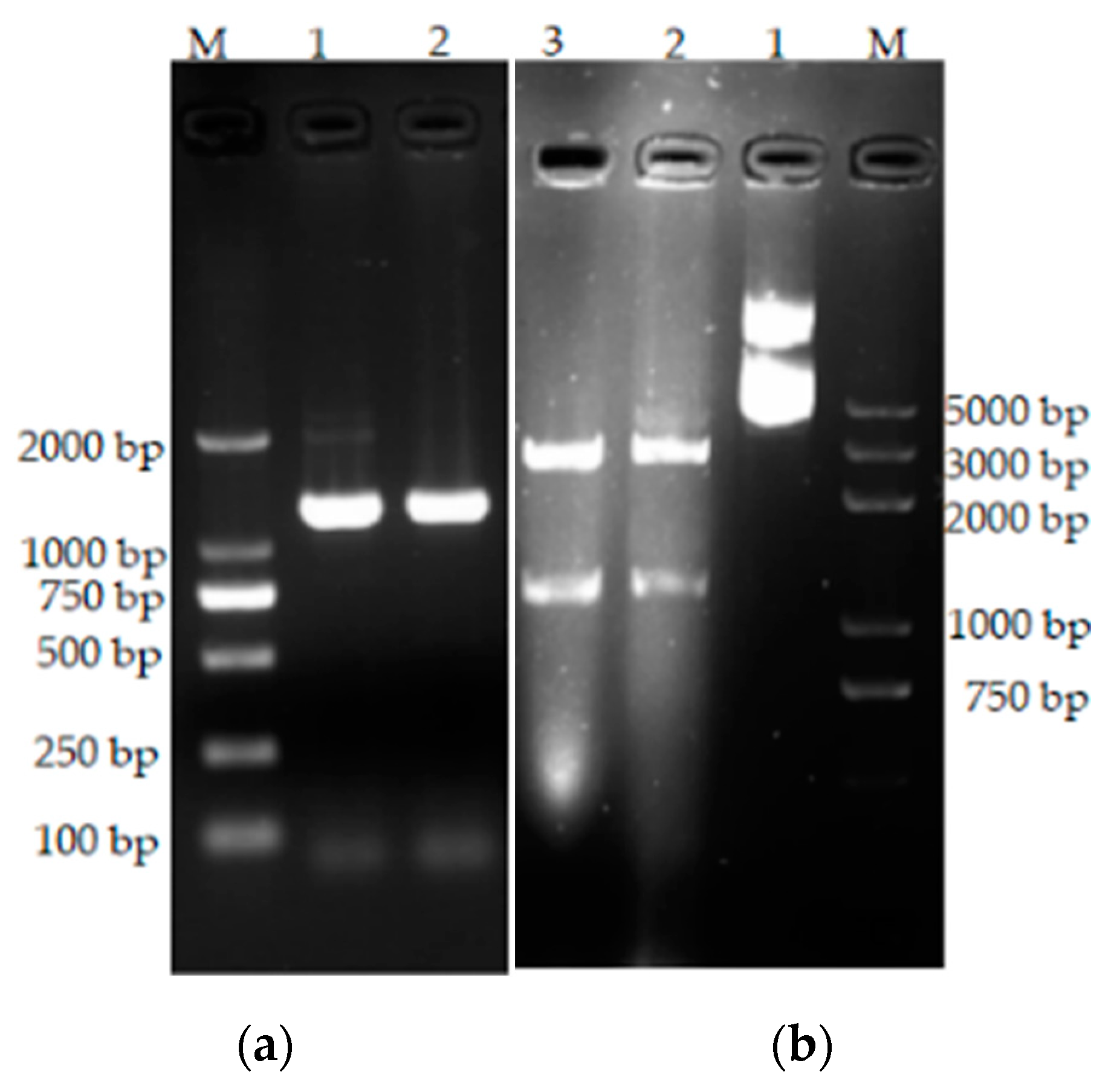
2.2.8. Primer Design and bta Gene Amplification
2.2.9. Sequence Analysis
- (1)
- ProtParm website (https://web.expasy.org/protparam/, accessed on) to analyze their physicochemical properties including relative molecular mass, amino acid composition, isoelectric point (pI), extinction coefficient, half-life, instability coefficient, and total average hydrophilicity;
- (2)
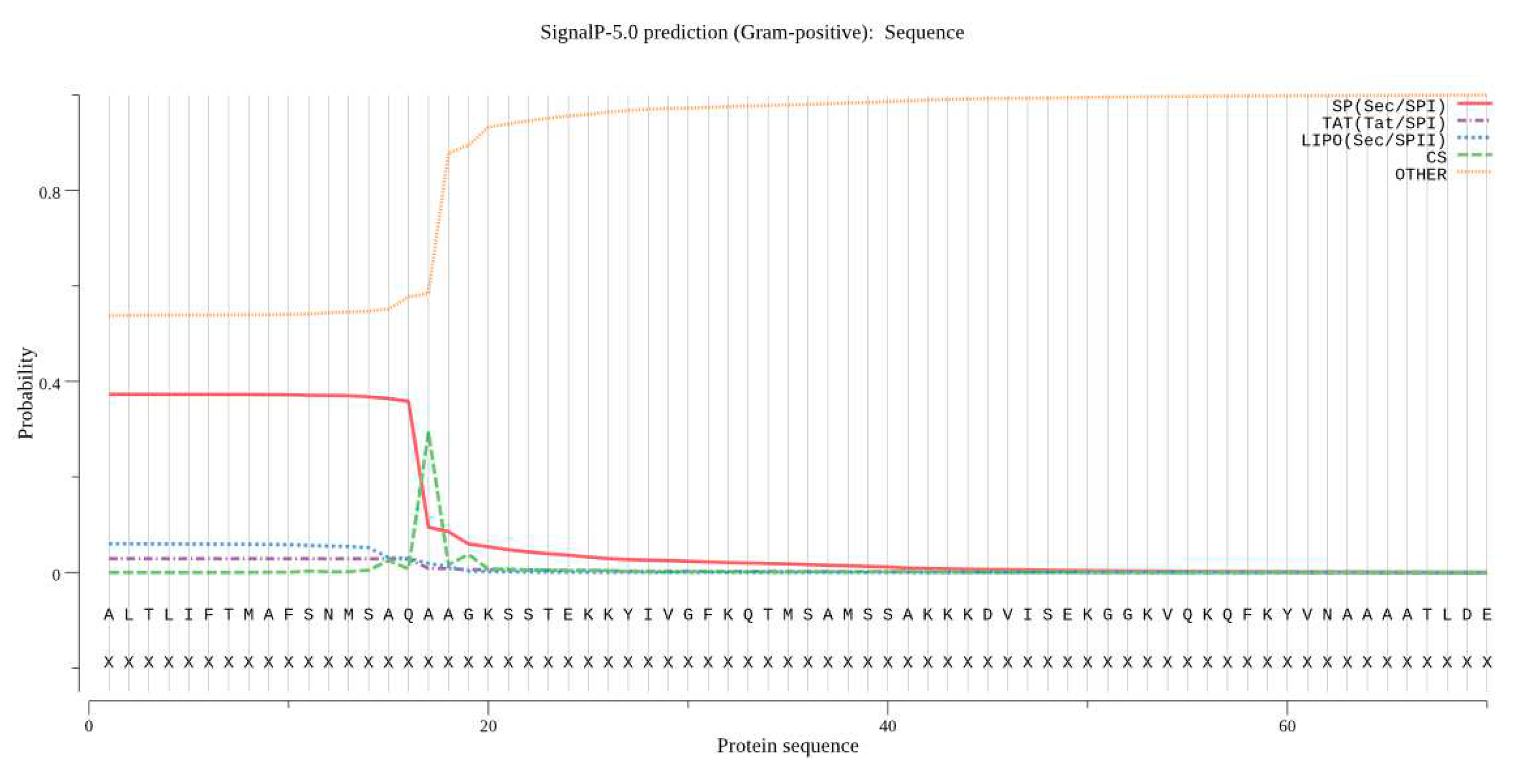
- SignalP (http://www.cbs.dtu.dk/services/SignalP/index.php, accessed on) to predict signal peptides;
- (3)
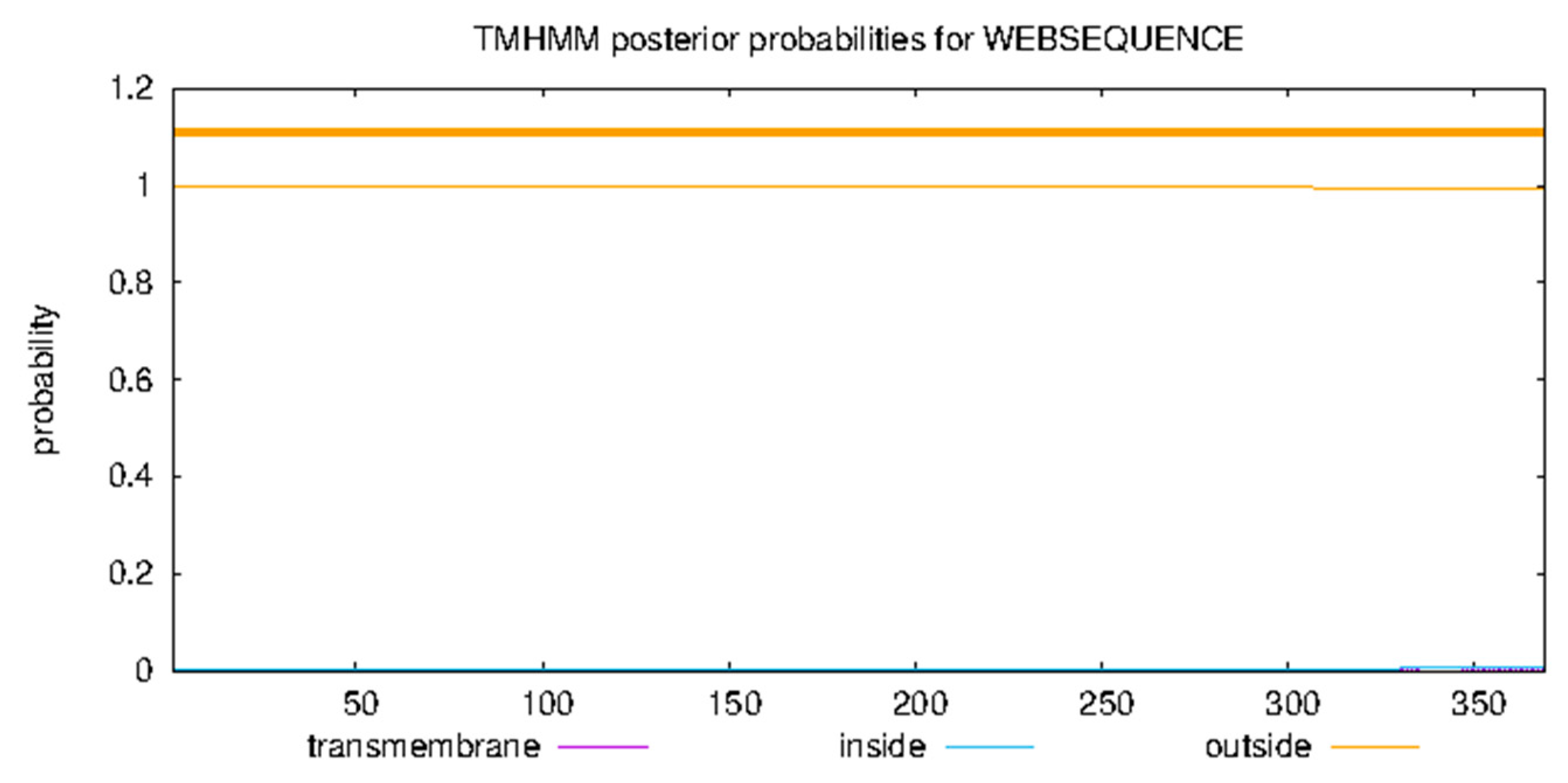
- The TMHMM program (http://www.cbs.dtu.dk/services/TMHMM, accessed on) to analyze protein transmembrane regions;
- (4)
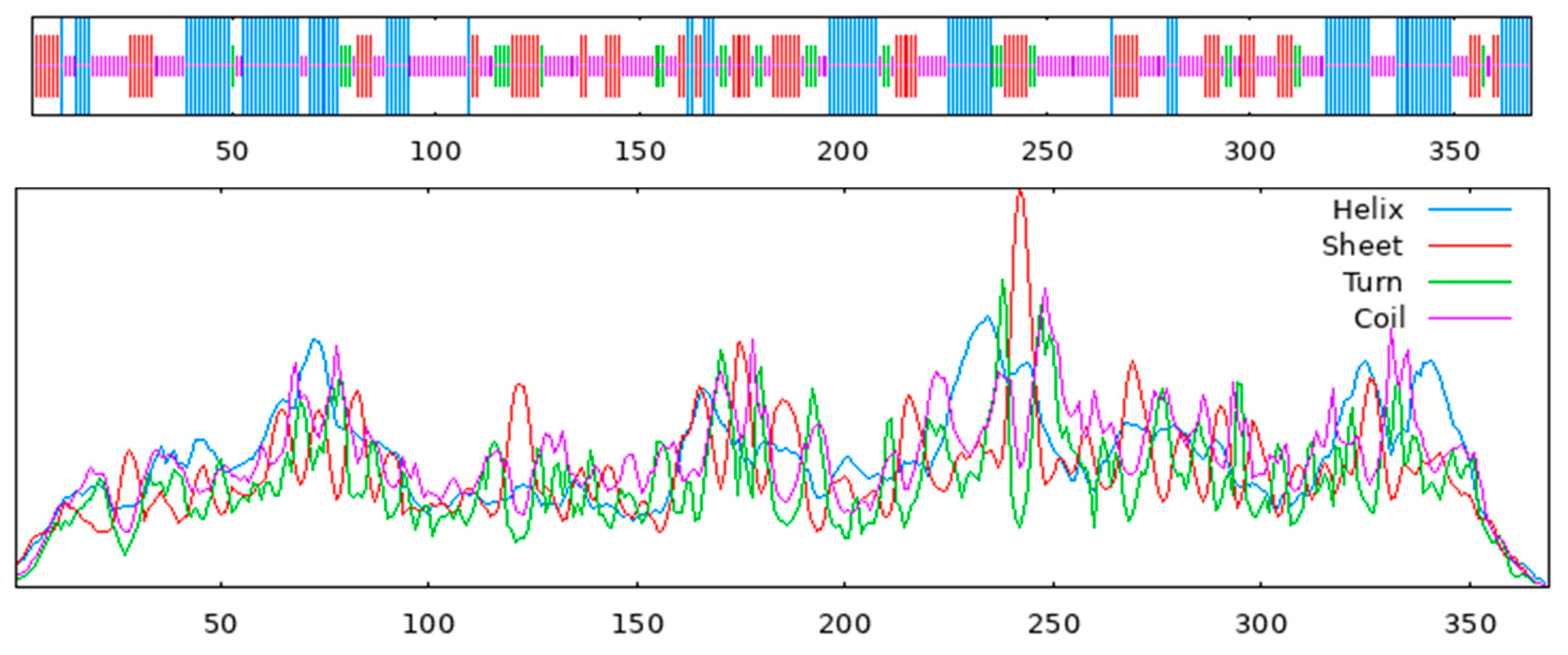
- SOPMA(http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on) to analyze secondary structures;
- (5)
- SWISS-MODELS (https://www.swissmodel.expasy.org/, accessed on) to predict tertiary structures.
2.2.10. Expression of the bta Gene in Escherichia coli
2.2.11. Ni2+ Chelation Chromatographic Purification of Recombinant Keratinase Bta
- (1)
- Inclusion of body fragmentation and purification
- (2)
- Dialysis of inclusion bodies for dialysis reversibility and enzyme activity measurement
3. Results
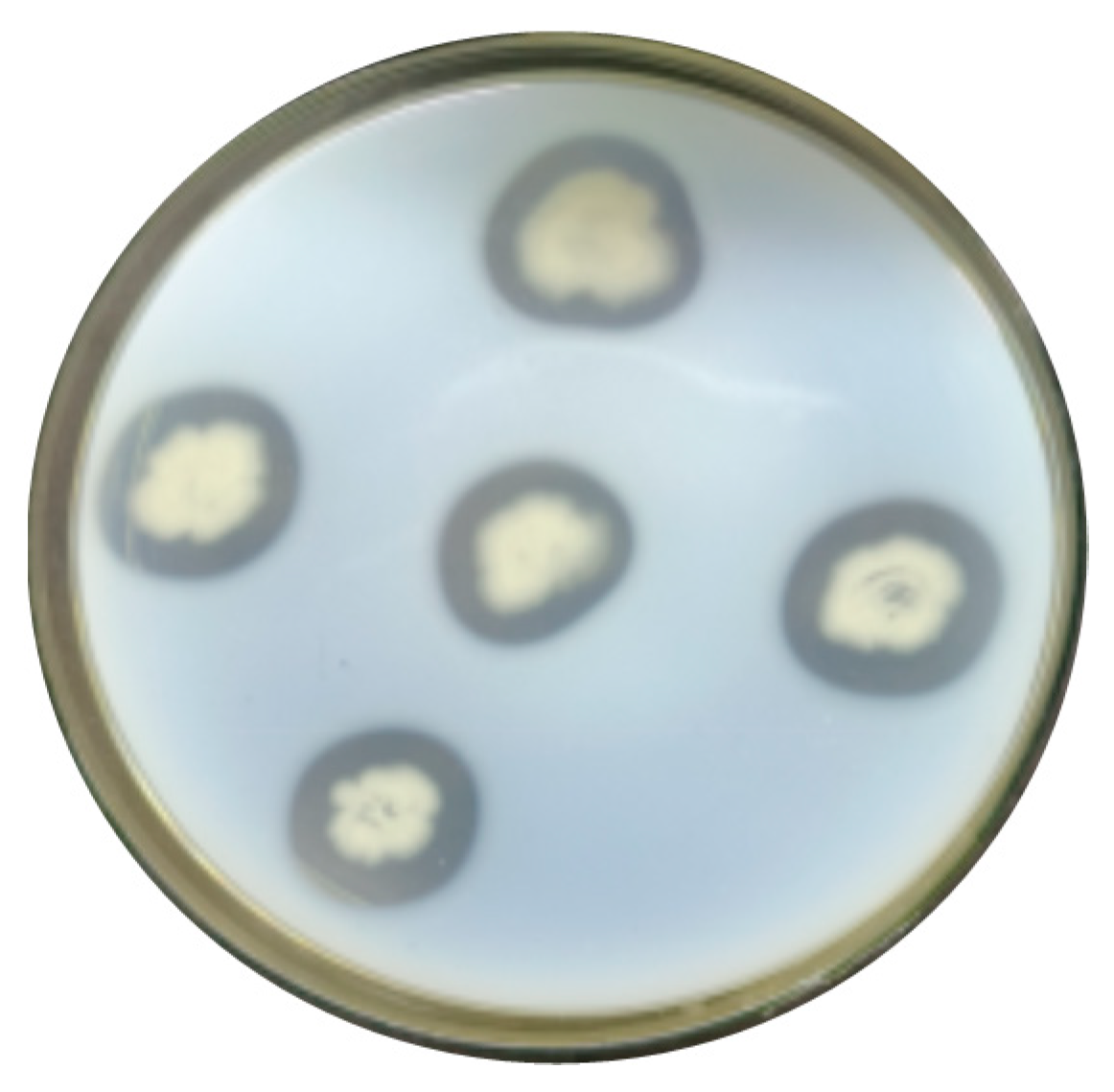
3.1. Screening of Strains
3.2. Re-Screening of Strains and Determination of Enzyme Viability
3.3. Growth Curve of Strain CY-A
3.4. Strain Identification
3.4.1. Morphological Characteristics
3.4.2. Physiological and Biochemical Characteristics
3.4.3. 16S rRNA Sequence Determination and Phylogenetic Tree Construction
3.5. Extraction of Total DNA
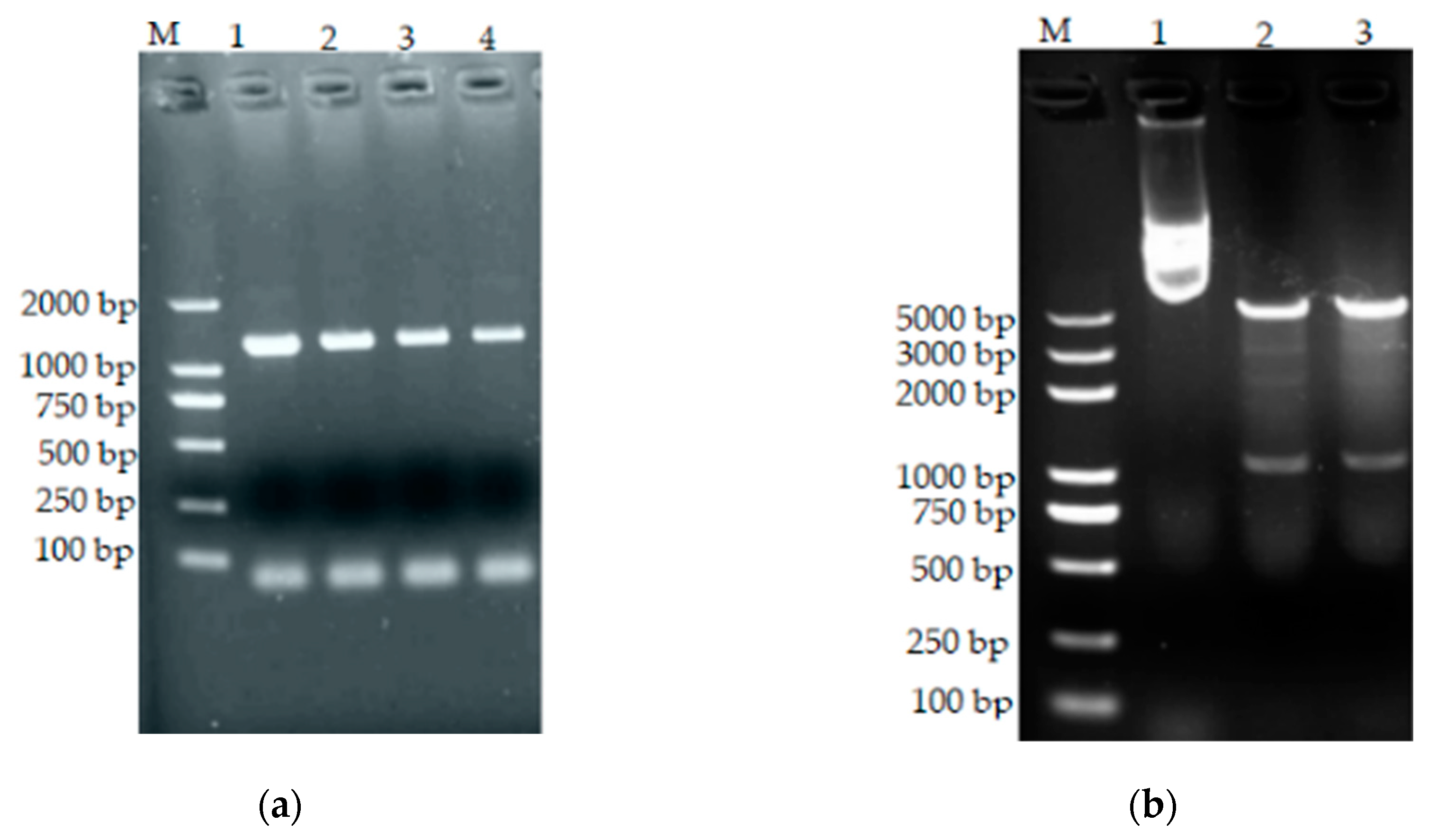
3.6. Cloning of the bta Gene
3.7. Sequence Analysis
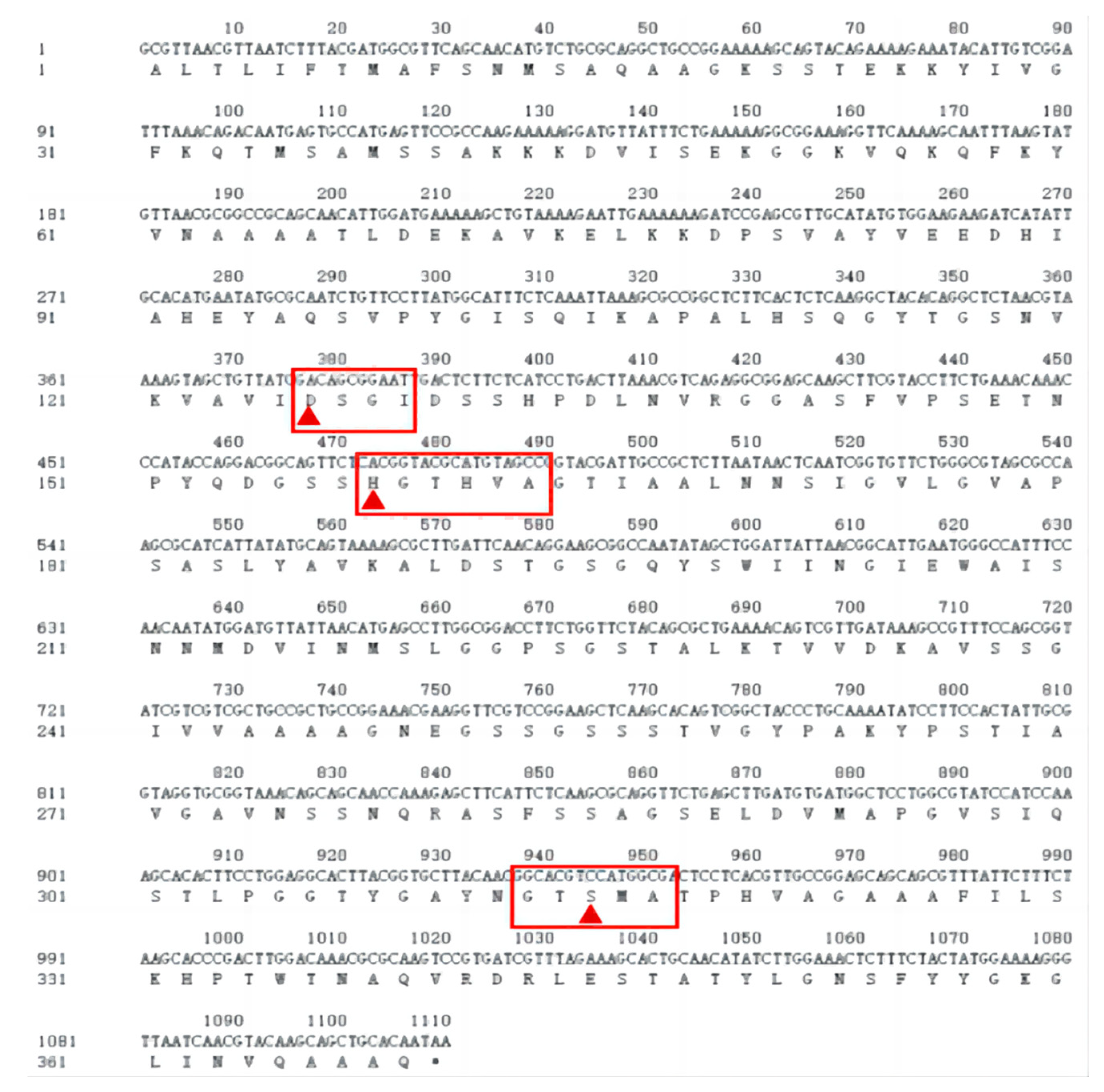

3.8. Predicting Basic Protein Physicochemical Properties
3.9. Structural Analysis
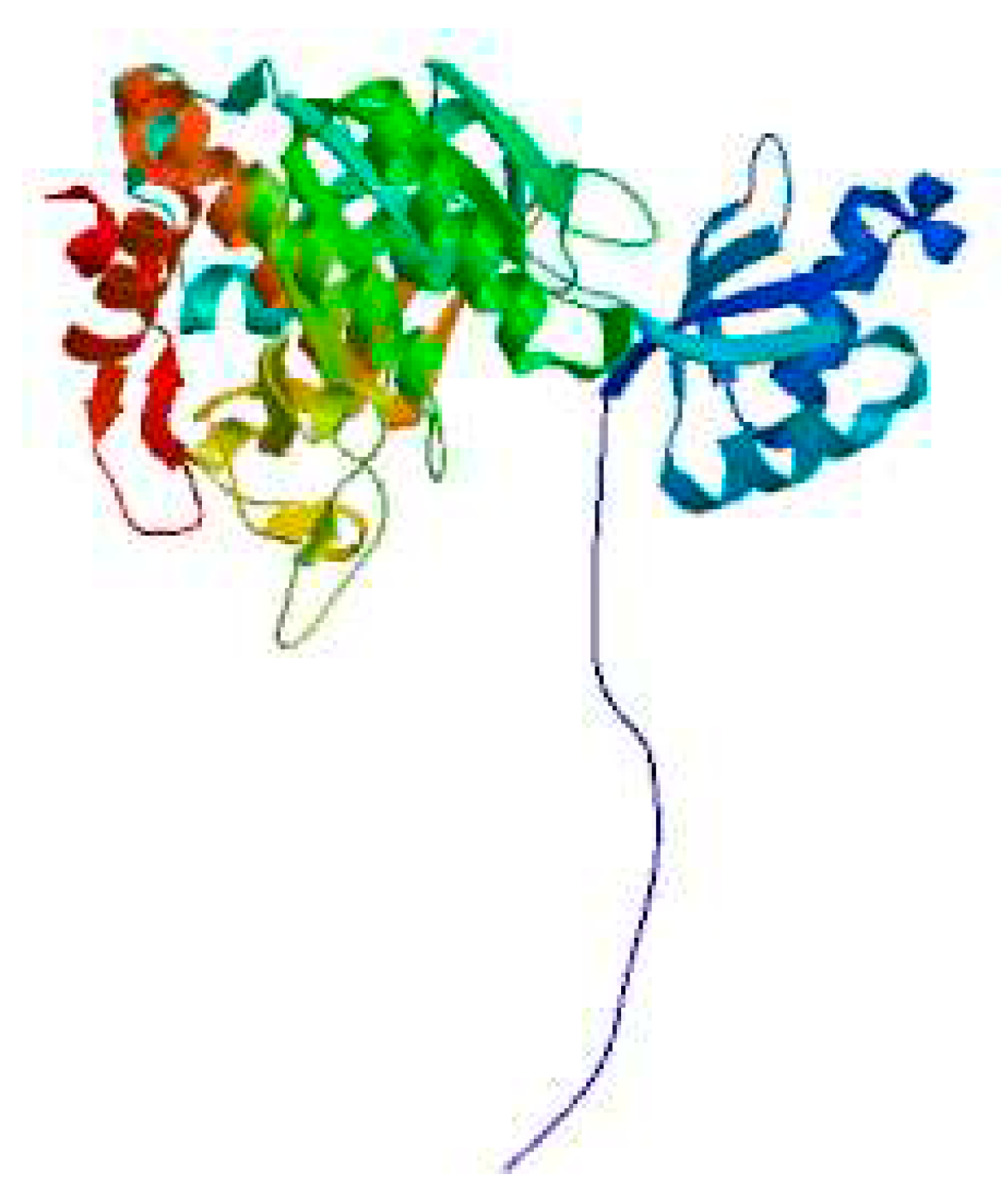
3.10. Construction of Prokaryotic Expression Vectors
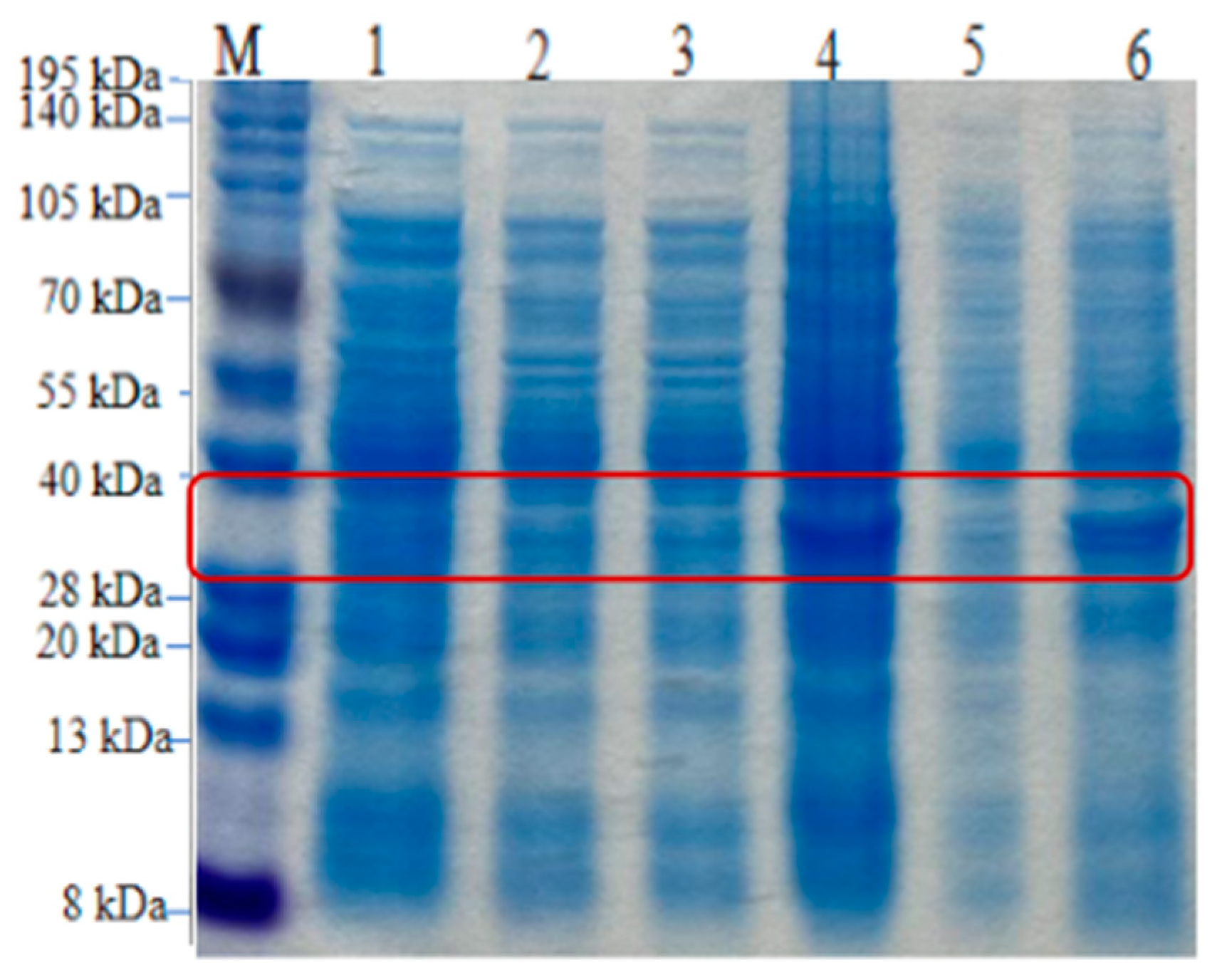
3.11. Induced Expression of Recombinant Proteins
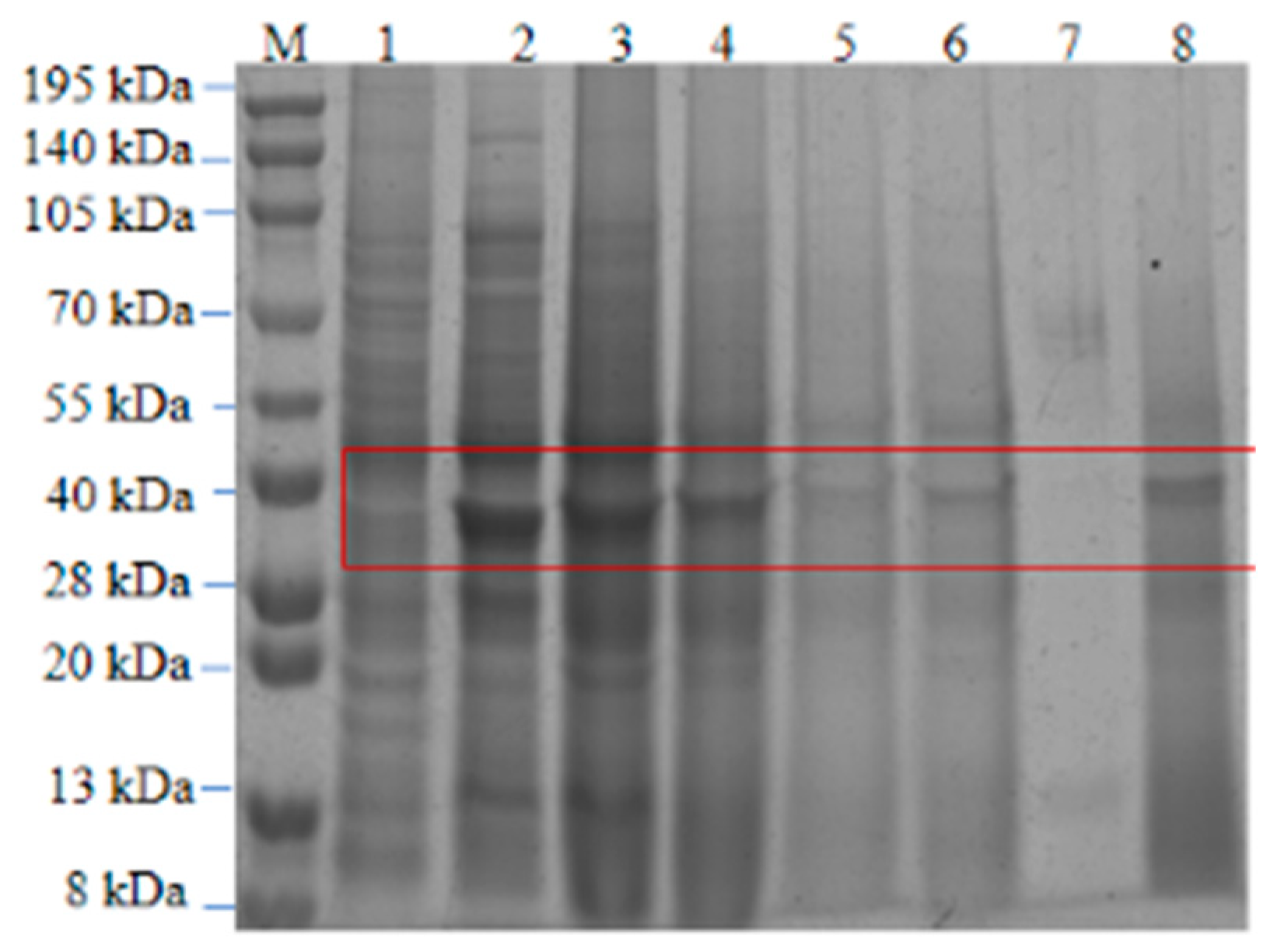
3.12. Purification of Recombinant Keratin
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gozali, C.; Suryanto, D.; Hartanto, A. Partial Purification and Characterization of Keratinase Produced by Keratinolytic Fungi, Earliella Scabrosa A2 and Aspergillus Flavus A11; IOP Publishing Ltd.: Bristol, UK, 2023. [Google Scholar]
- Bhari, R.; Kaur, M.; Singh, R.S.; Pandey, A.; Larroche, C. Bioconversion of chicken feathers by Bacillus aerius NSMk2: A potential approach in poultry waste management. Bioresour. Technol. Rep. 2018, 3, 224–230. [Google Scholar] [CrossRef]
- Khumalo, M.; Sithole, B.; Tesfaye, T. Valorisation of waste chicken feathers: Optimisation of keratin extraction from waste chicken feathers by sodium bisulphite, sodium dodecyl sulphate and urea. J. Environ. Manag. 2020, 262, 110329.1–110329.7. [Google Scholar] [CrossRef]
- Yahaya, R.S.R.; Normi, Y.M.; Lai Yee, P.; Ahmad, S.A.; Abdullah, J.O.; Sabri, S. Molecular strategies to increase keratinase production in heterologous expression systems for industrial applications. Appl. Microbiol. Biotechnol. 2021, 5, 3955–3969. [Google Scholar] [CrossRef]
- Fang, Z.; Yong, Y.C.; Zhang, J.; Du, G.; Chen, J. Keratinolytic protease: A green biocatalyst for the leather industry. Appl. Microbiol. Biotechnol. 2017, 101, 7771–7779. [Google Scholar] [CrossRef]
- Mamangkey, J.; Suryanto, D.; Munir, E.; Mustopa, A. Isolation, Molecular Identification and Verification of Gene Encoding Bacterial Keratinase from Crocodile (Crocodylus porosus) Feces. In Proceedings of the 4th International Conference on Biological Sciences and Biotechnology, Kuala Lumpur, Malaysia, 21–22 February 2019. [Google Scholar]
- Gupta, R.; Ramnani, P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006, 70, 21–33. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature—A new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef]
- Sharma, I.; Kango, N. Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. Int. J. Biol. Macromol. 2021, 166, 1046–1056. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Periasamy, A.; Thangavel, L.; Tang, T.-H.; Chen, Y.; Hashim, U.; Ruslinda, R.; Arshad, M.K.M. Biotechnological Aspects and Perspective of Microbial Keratinase Production. J. Biomed. Biotechnol. 2015, 2015, 140726. [Google Scholar] [CrossRef]
- Vidmar, B.; Vodovnik, M. Microbial Keratinases: Enzymes with Promising Biotechnological Applications. Food Technol. Biotechnol. 2018, 56, 312–328. [Google Scholar] [CrossRef]
- Imania, G.; Aqsa, I.; Ali, H.; Arshad, J.; Faiza, J.; Muhammad, A.; Javed, I.Q. Microbial production and industrial applications of keratinases: An overview. Int. Microbiol. 2018, 21, 163–174. [Google Scholar]
- Cheng, S.W.; Hu, H.M.; Shen, S.W.; Takagi, H.; Asano, M.; Tsai, Y.-C. Production and characterization of keratinase of a feather-degrading Bacillus licheniformis PWD-1. Biosci. Biotechnol. Biochem. 1995, 59, 2239–2243. [Google Scholar] [CrossRef]
- Liang, B. Extraction, Cloning and Expression of Keratinase Gene kerB; Jilin University: Changchun, China, 2003. [Google Scholar]
- Radha, S.; Gunasekaran, P. Sustained expression of keratinase gene under PxylA and PamyL promoters in the recombinant Bacillus megaterium MS941. Bioresour. Technol. 2008, 99, 5528–5537. [Google Scholar] [CrossRef]
- Fakhfakh, N.; Kanoun, S.; Manni, L.; Nasri, M. Production and biochemical and molecular characterization of a keratinolytic serine protease from chicken feather-degrading Bacillus licheniformis RPk. Can. J. Microbiol. 2009, 55, 427. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Huang, Y.; Wei, Q.; Huang, H.; Guo, C. Cloning and expression of a thermostable keratinase gene from Thermoactinomyces sp. YT06 in Escherichia coli and characterization of purified recombinant enzymes. World J. Microbiol. Biotechnol. 2019, 35, 135. [Google Scholar] [CrossRef]
- Gurunathan, R.; Huang, B.; Ponnusamy, V.K.; Hwang, J.-S.; Dahms, H.-U. Novel recombinant keratin degrading subtilisin like serine alkaline protease from Bacillus cereus isolated from marine hydrothermal vent crabs. Sci. Rep. 2021, 11, 12007. [Google Scholar] [CrossRef]
- Yamamura, S.; Morita, Y.; Hasan, Q.; Rao, S.R.; Murakami, Y.; Yokoyama, K.; Tamiya, E. Characterization of a new keratin-degrading bacterium isolated from deer fur. J. Biosci. Bioeng. 2002, 93, 595–600. [Google Scholar] [CrossRef]
- Zhu, J.G. Manual of Identification of Common Clinical Bacteria; Beijing Medical University and Peking Union Medical College Joint Publishing House: Beijing, China, 1993. [Google Scholar]
- Xiao, J.; Yang, X.; Guo, H. Bioinformatics and expression analysis of the flavonoid-3′5′-hydroxylase (F3′5′H) gene of the colourful potato variety ‘Turning Heart U’. 2015(7):9.
- Mabrouk, M.E.M. Feather degradation by a new keratinolytic Streptomyces sp. MS-2. World J. Microbiol. Biotechnol. 2008, 24, 2331–2338. [Google Scholar] [CrossRef]
- Arokiyaraj, S.; Varghese, R.; Ahmed, B.A.; Duraipandiyan, V.; Al-Dhabi, N. Optimizing the fermentation conditions and enhanced production of keratinase from Bacillus cereus isolated from halophilic environment. Saudi J. Biol. Sci. 2019, 26, 378–381. [Google Scholar] [CrossRef]
- Sharma, I.; Kango, N. Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. Int. J. Biol. Macromol. 2021, 166, 1046–1056. [Google Scholar] [CrossRef]
- Mamangkey, J.; Suryanto, D.; Munir, E.; Mustopa, A. Isolation, Molecular Identification and Verification of Gene Encoding Bacterial Keratinase from Crocodile (Crocodylus porosus) Feces. In Proceedings of the 4th International Conference on Biological Sciences and Biotechnology, Kuala Lumpur, Malaysia, 21–22 February 2019. [Google Scholar]
- ZHANG Ni, ZHANG Hongyan, YANG Mengying, et al. Screening, characterisation and enzymatic properties of a marine-derived strain of highly efficient keratinase-producing bacteria[J]. Food and Fermentation Industry 2020, 46, 7.
- DONG Mengmeng, HAO Pengze, WANG Jialiu, et al. Isolation and identification of feather-degrading bacterium DHW-06 and study on enzyme-producing characteristics[J]. Journal of Northwest Agriculture and Forestry University: Natural Science Edition 2021, 49, 9.
- Hossain, M.S.; Azad, A.K.; Abu Sayem, S.M.; Mostafa, G.; Hoq, M. Production and partial characterization of feather-degrading keratinolytic serine protease from Bacillus licheniformis MZK-3. J. Biological. Sci. 2007, 7, 599–606. [Google Scholar]
- Manivasagan, P.; Sivakumar, K.; Gnanam, S.; Venkatesan, J.; Kim, S.-K. Production, Biochemical Characterization and Detergents Application of Keratinase from the Marine Actinobacterium Actinoalloteichus sp. MA-32. J. Surfactants Deterg. 2014, 17, 669–682. [Google Scholar] [CrossRef]
- Mwanza, E.P.; van der Westhuizen, W.A.; Boucher, C.E.; Charimba, G.; Hugo, C. Heterologous expression and characterisation of a keratinase produced by Chryseobacterium carnipullorum. Protein Expr. Purif. 2021, 186, 105926. [Google Scholar] [CrossRef]
- Hu, H.; Gao, J.; He, J.; Yu, B.; Zheng, P.; Huang, Z.; Mao, X.; Yu, J.; Han, G.; Chen, D. Codon Optimization Significantly Improves the Expression Level of a Keratinase Gene in Pichia pastoris. PLoS ONE 2013, 8, e58393. [Google Scholar] [CrossRef]
- Hu, H.; He, J.; Yu, B.; Zheng, P.; Huang, Z.; Mao, X.; Yu, J.; Han, G.; Chen, D. Expression of a keratinase (kerA) gene from Bacillus licheniformis in Escherichia coli and characterization of the recombinant enzymes. Biotechnol. Lett. 2013, 35, 239–244. [Google Scholar] [CrossRef]
- Porres, J.M.; Benito, M.J.; Lei, X.G. Functional expression of keratinase (kerA) gene from Bacillus licheniformis in Pichia pastoris. Biotechnol. Lett. 2002, 24, 631–636. [Google Scholar] [CrossRef]
- Wang, J.-J.; Rojanatavorn, K.; Shih, J.C.H. Increased production of Bacillus keratinase by chromosomal integration of multiple copies of the kerA gene. Biotechnol. Bioeng. 2004, 87, 459–464. [Google Scholar] [CrossRef]
- Lin, X.; Lee, C.G.; Casale, E.S.; Shih, J.C.H. Purification and Characterization of a Keratinase from a Feather-Degrading Bacillus licheniformis Strain. Appl. Environ. Microbiol. 1992, 58, 3271–3275. [Google Scholar] [CrossRef]
- Tork, S.; Aly, M.M.; Nawar, L. Biochemical and Molecular Characterization of a New Local Keratinase Producing Pseudomomanas sp. MS21. Asian J. Biotechnol. 2010, 2, 1–13. [Google Scholar] [CrossRef]
- Cai, C.; Chen, J.S.; Qi, J.J.; Yin, Y.; Zheng, X.-D. Purification and characterization of keratinase from a new Bacillus subtilis strain. J. Zhejiang Univ. Sci. B 2008, 9, 713–720. [Google Scholar] [CrossRef]
- Rao, C.S.; Sathish, T.; Ravichandra, P.; Prakasham, R. Characterization of thermo- and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem. 2009, 44, 262–268. [Google Scholar]
- ZHANG Qi, SUN Dan, YANG Wenbo. Purification process, physicochemical properties and application of keratinase produced by Bacillus sphaericus L_4[J]. Ion exchange and adsorption 2009, 25, 448–455.
| Strain | Keratinase Activity (U/mL) |
|---|---|
| CY-A | 28.64 |
| CY-C | 19.99 |
| CY-E | 25.04 |
| CY-F | 28.14 |
| CY-G | 25.42 |
| WQ4-3 | 16.70 |
| WO5-3 | 20.83 |
| QT4-1 | 27.23 |
| QT4-3 | 25.42 |
| Assay | Reaction Result |
|---|---|
| CY-A | |
| Glucose fermentation | + |
| Lactose fermentation | - |
| Sucrose fermentation | - |
| Ammonia production experiment | + |
| Malonic acid utilization | - |
| Decomposition of milk with litmus | + |
| Methyl red | + |
| Citrate decomposition | - |
| L-Arabinose fermentation | - |
| V-P experiment | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




