Submitted:
10 January 2024
Posted:
11 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
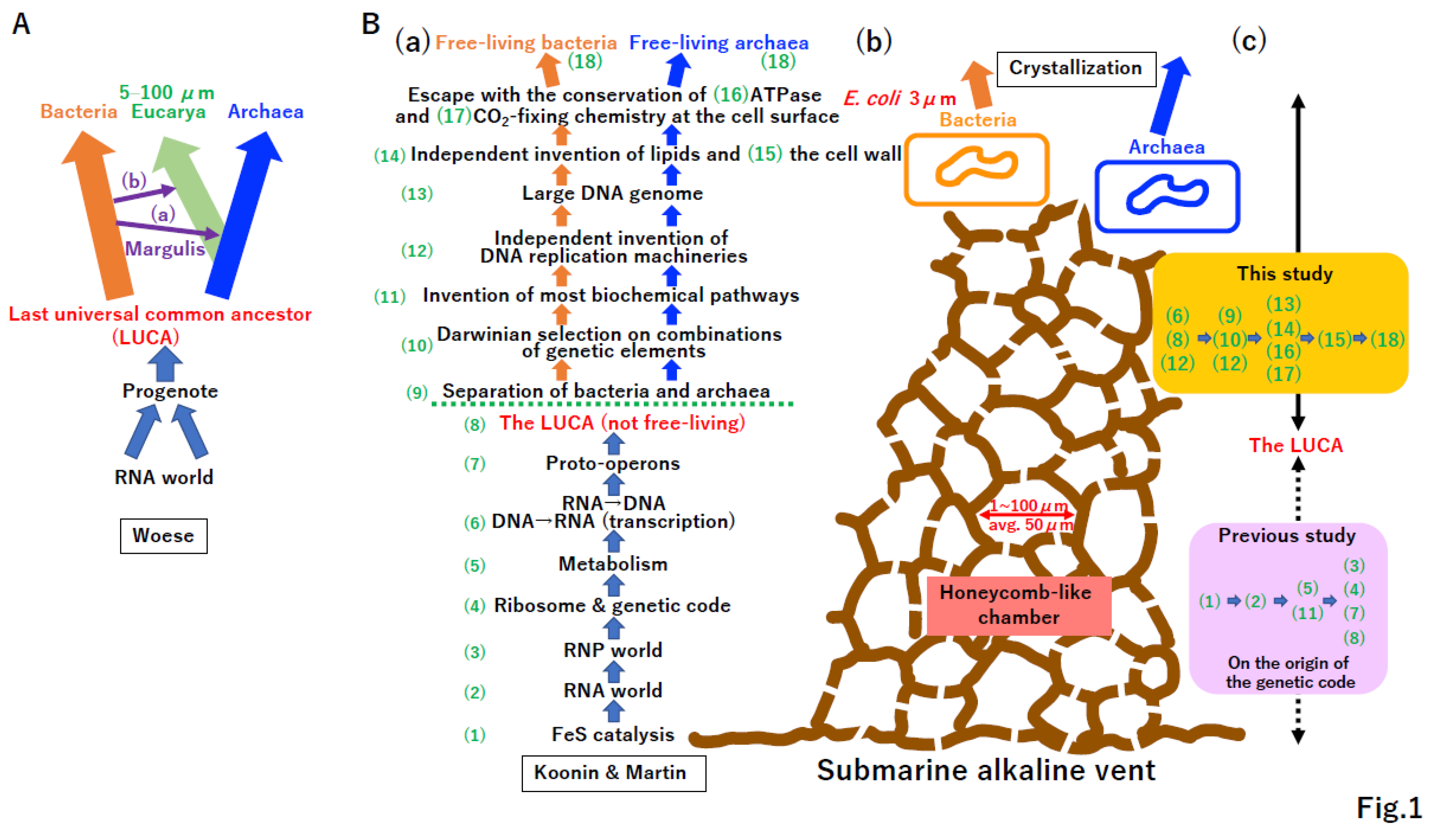
2. The LUCA
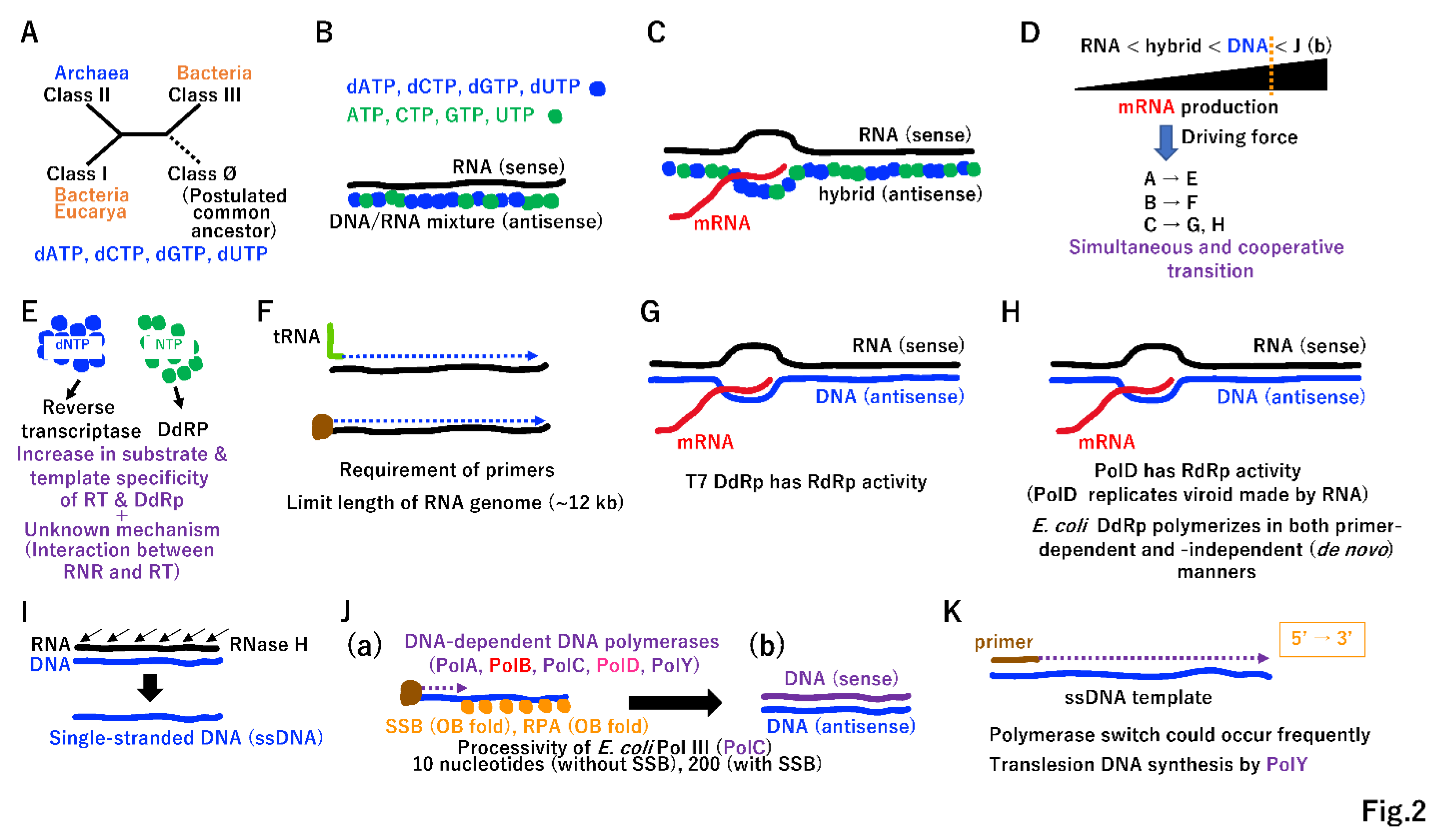
2.1. Double-stranded U-DNA on a single operon
2.2. Discrimination of sense and antisense DNA
2.3. Strand displacement and unidirectional replication
3. End of the LUCA
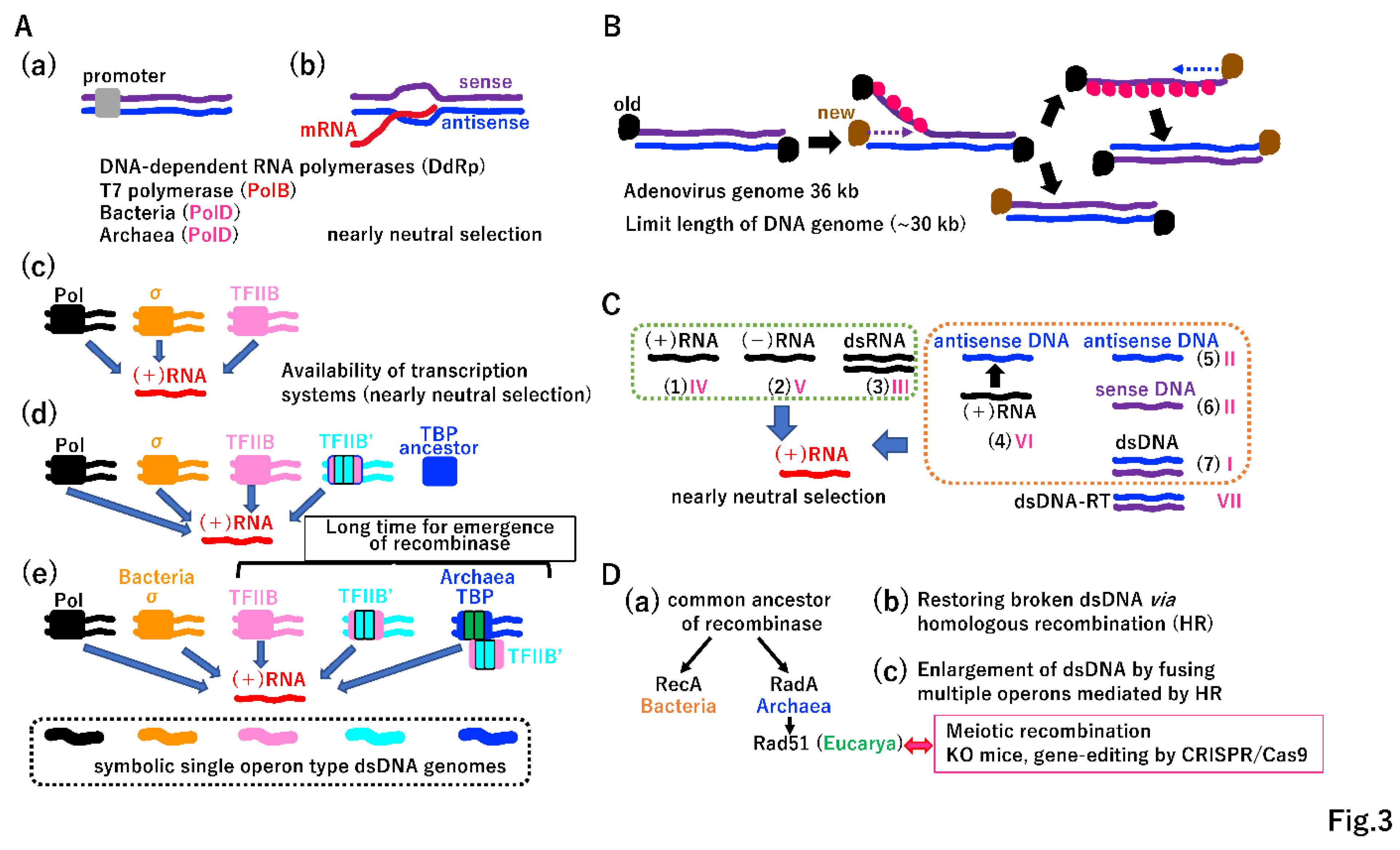
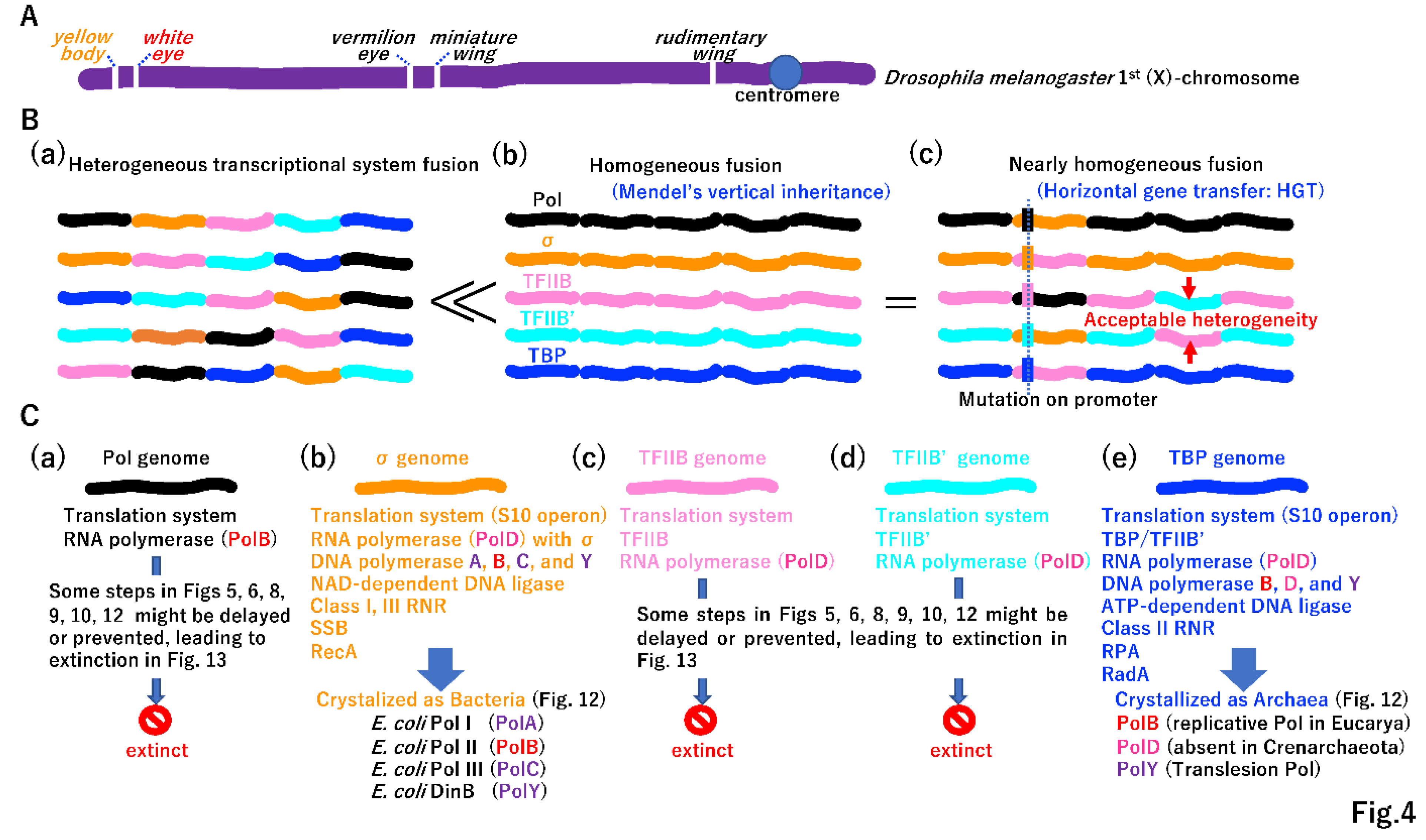
3.1. Beginning of genetic linkage and vertical inheritance
3.2. Beginning of horizontal gene transfer (HGT)
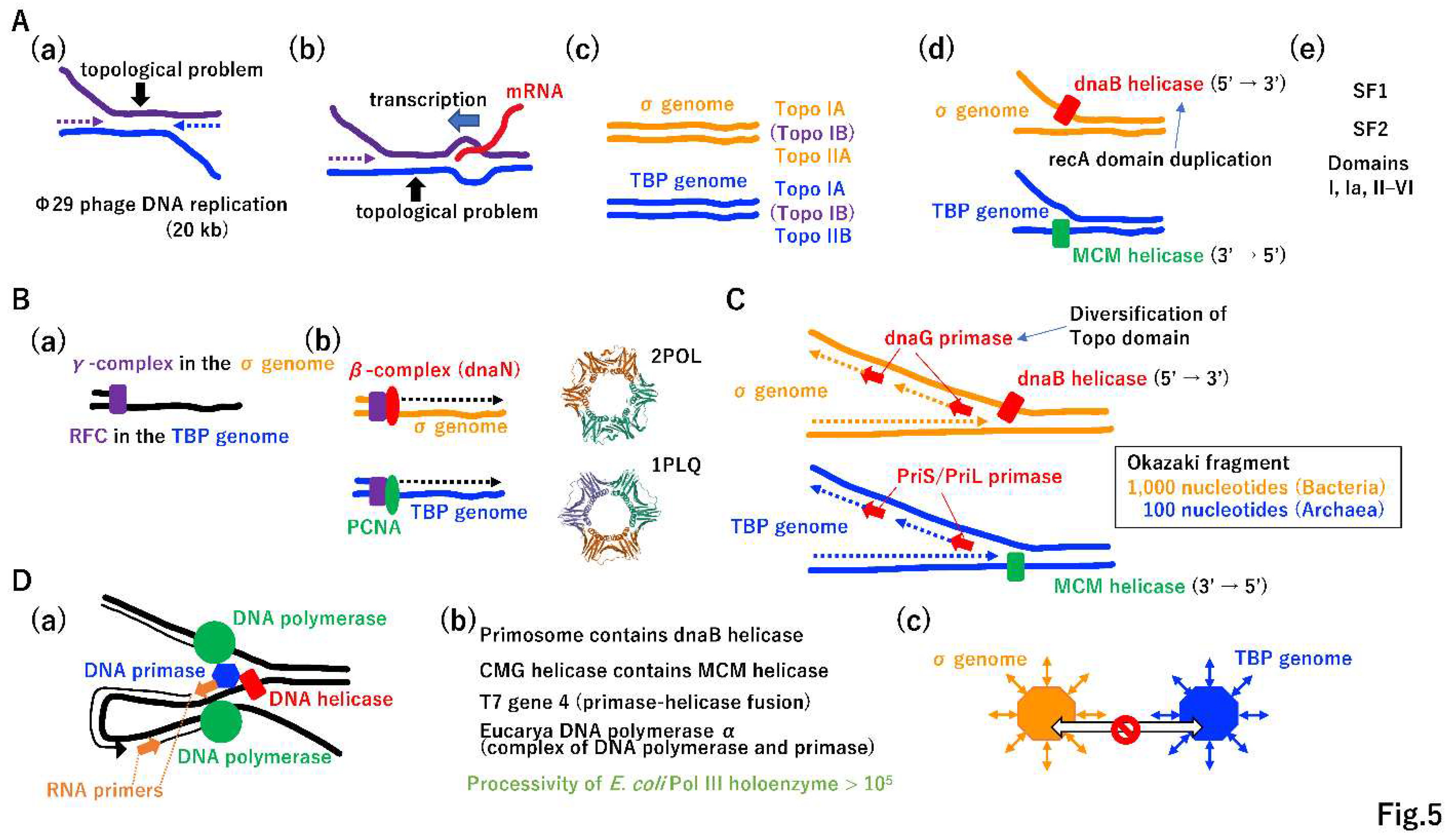
3.3. Evolution of two different DNA replication machineries
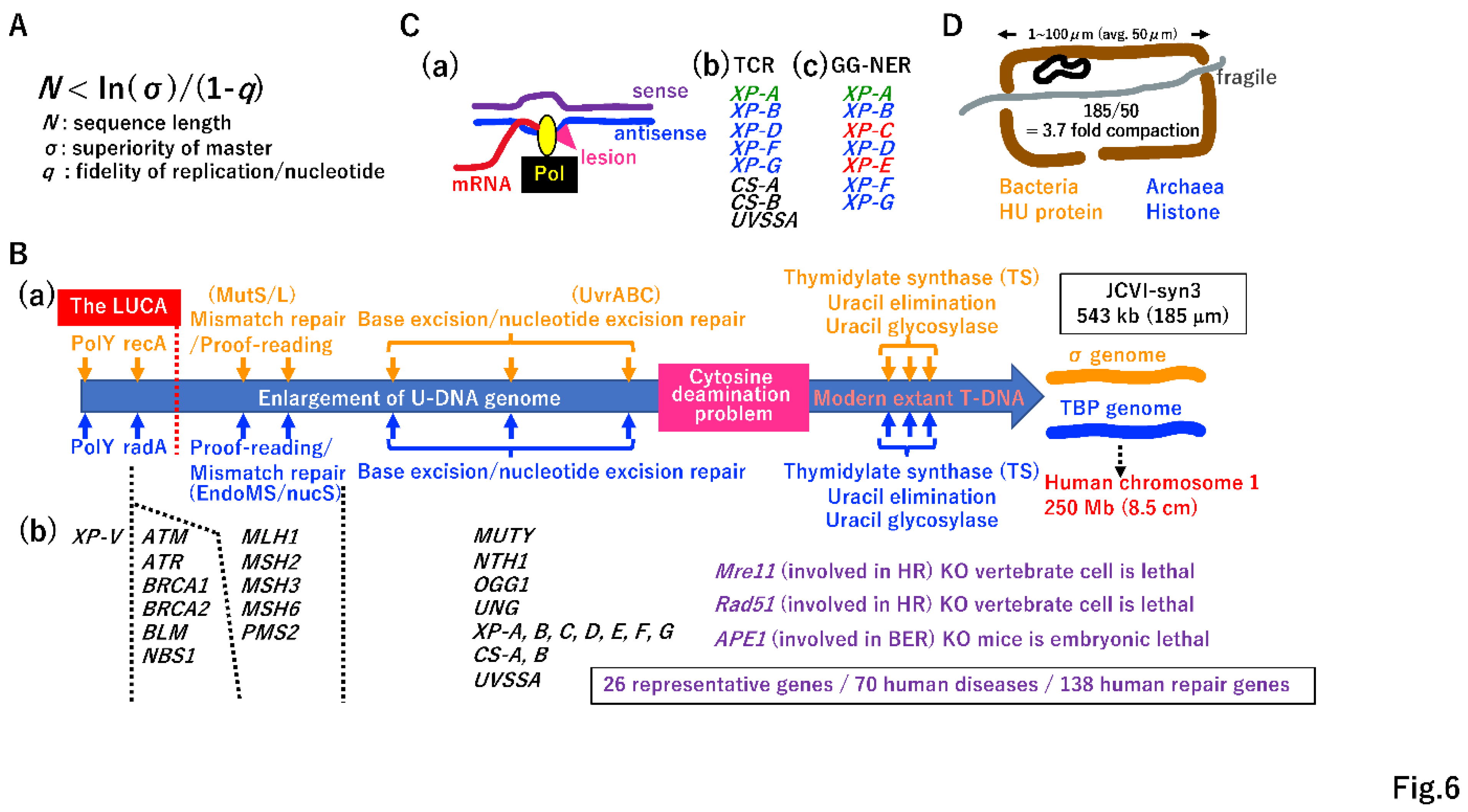
3.4. Necessity of DNA repair for the increase in DNA genome size
3.5. Transition from U-DNA to T-DNA
3.6. Transcription-coupled NER
3.7. Necessity of DNA compaction
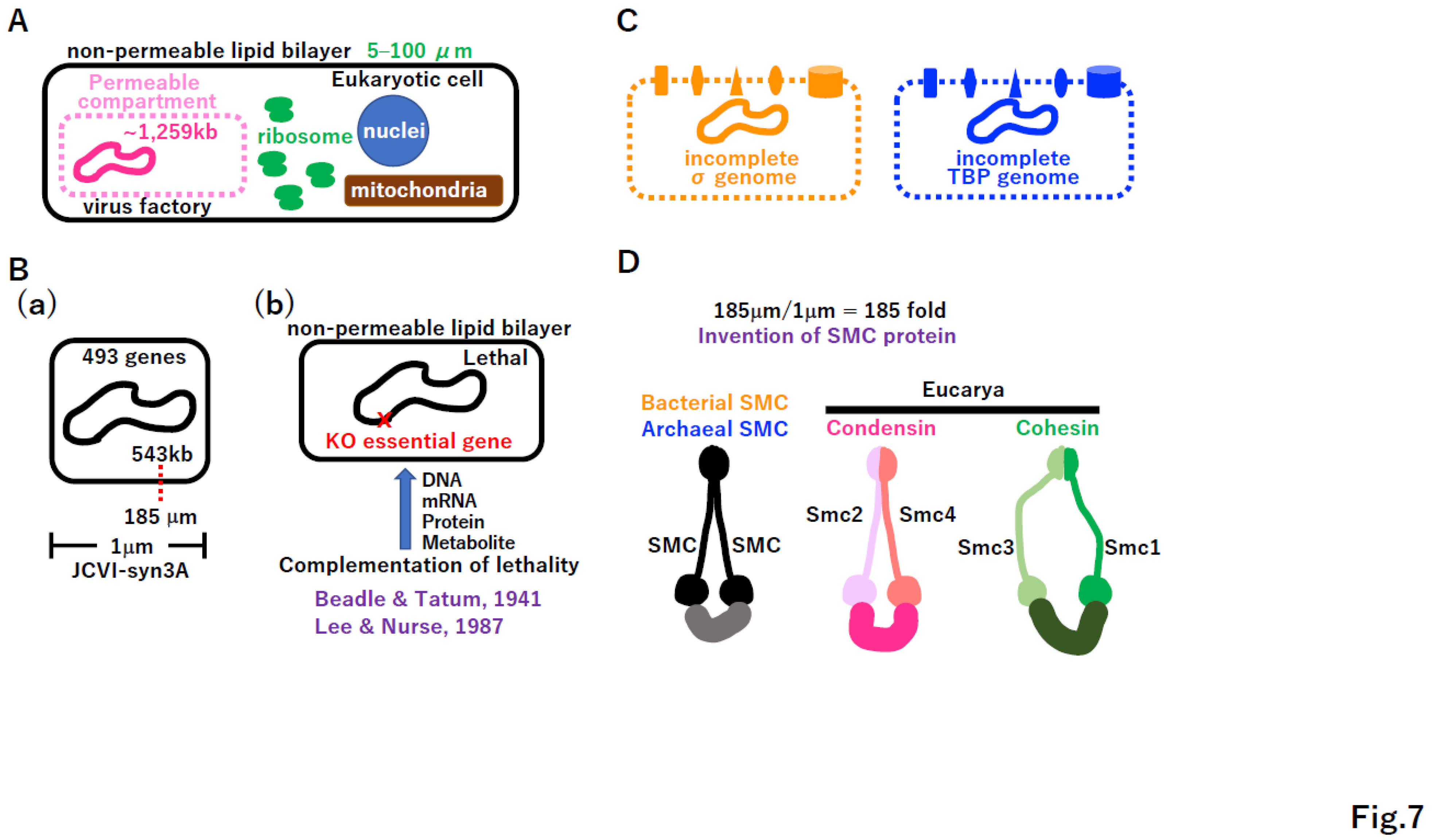
4. Toward proto-cells
4.1. A lack of essential genes in lethal in extant cells
4.2. Coacervate formation triggering lipid divide
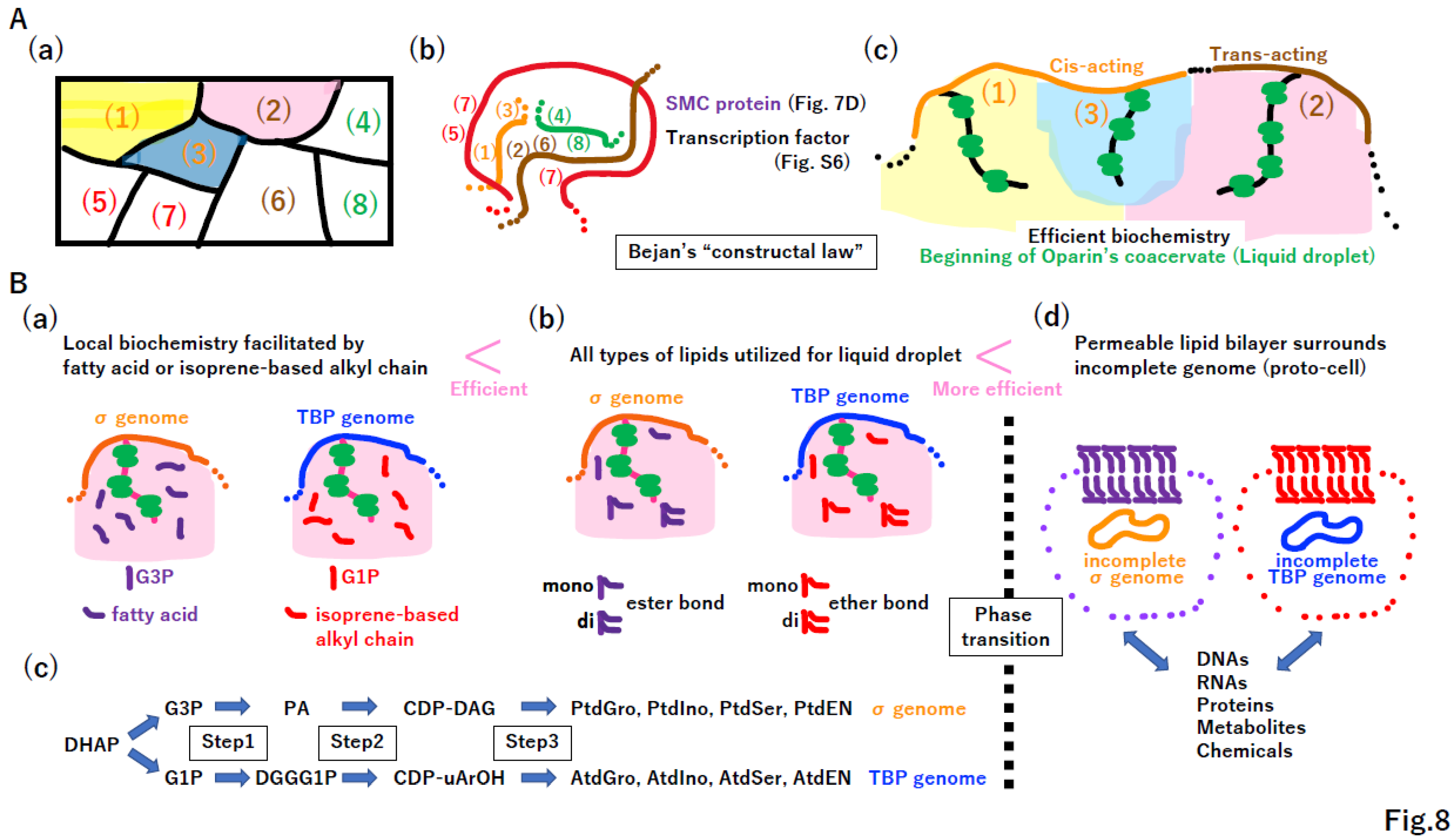
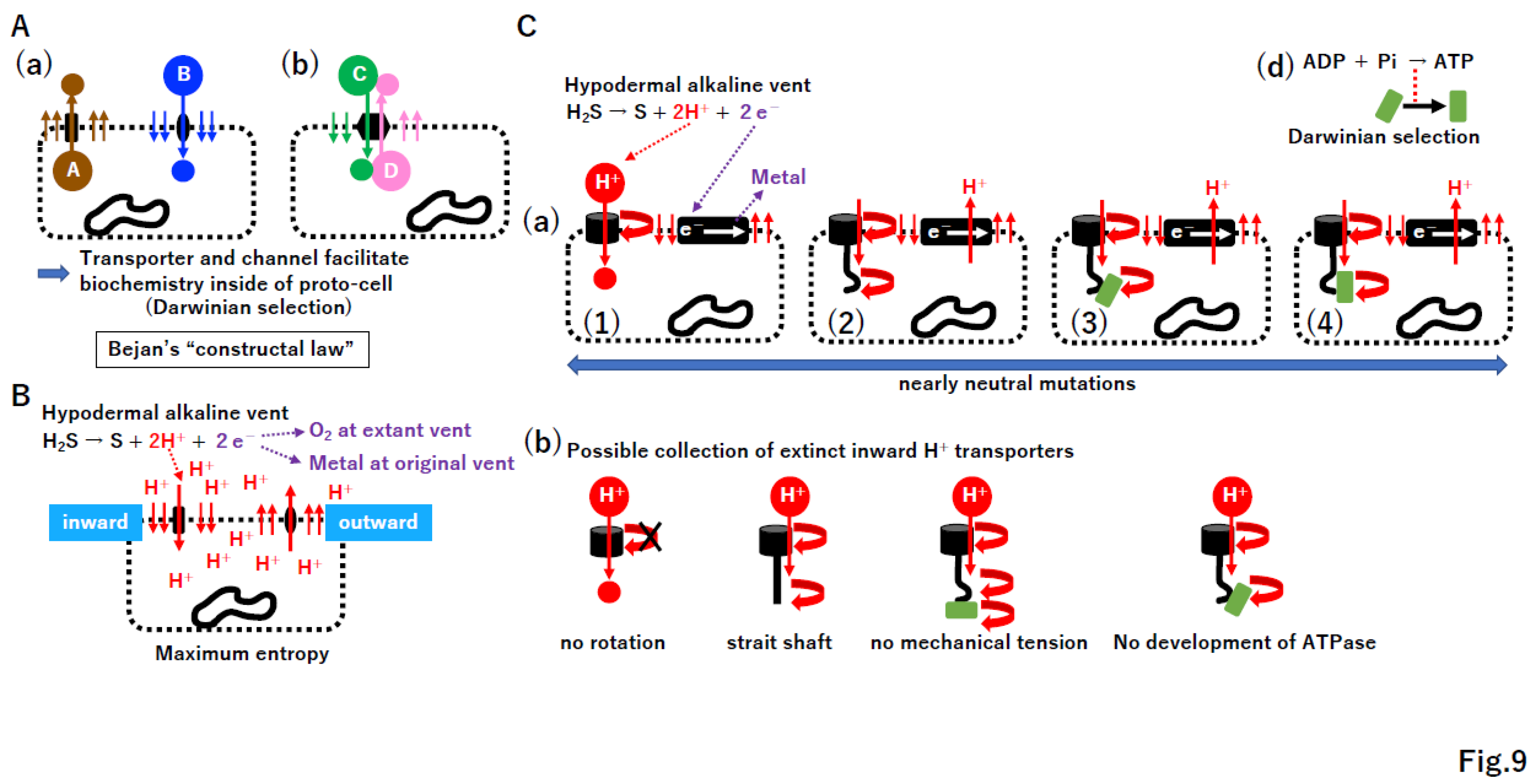
4.3. Bejan’s “constructal law” in transporter and channel creation
4.4. ATPase and the electron transport system
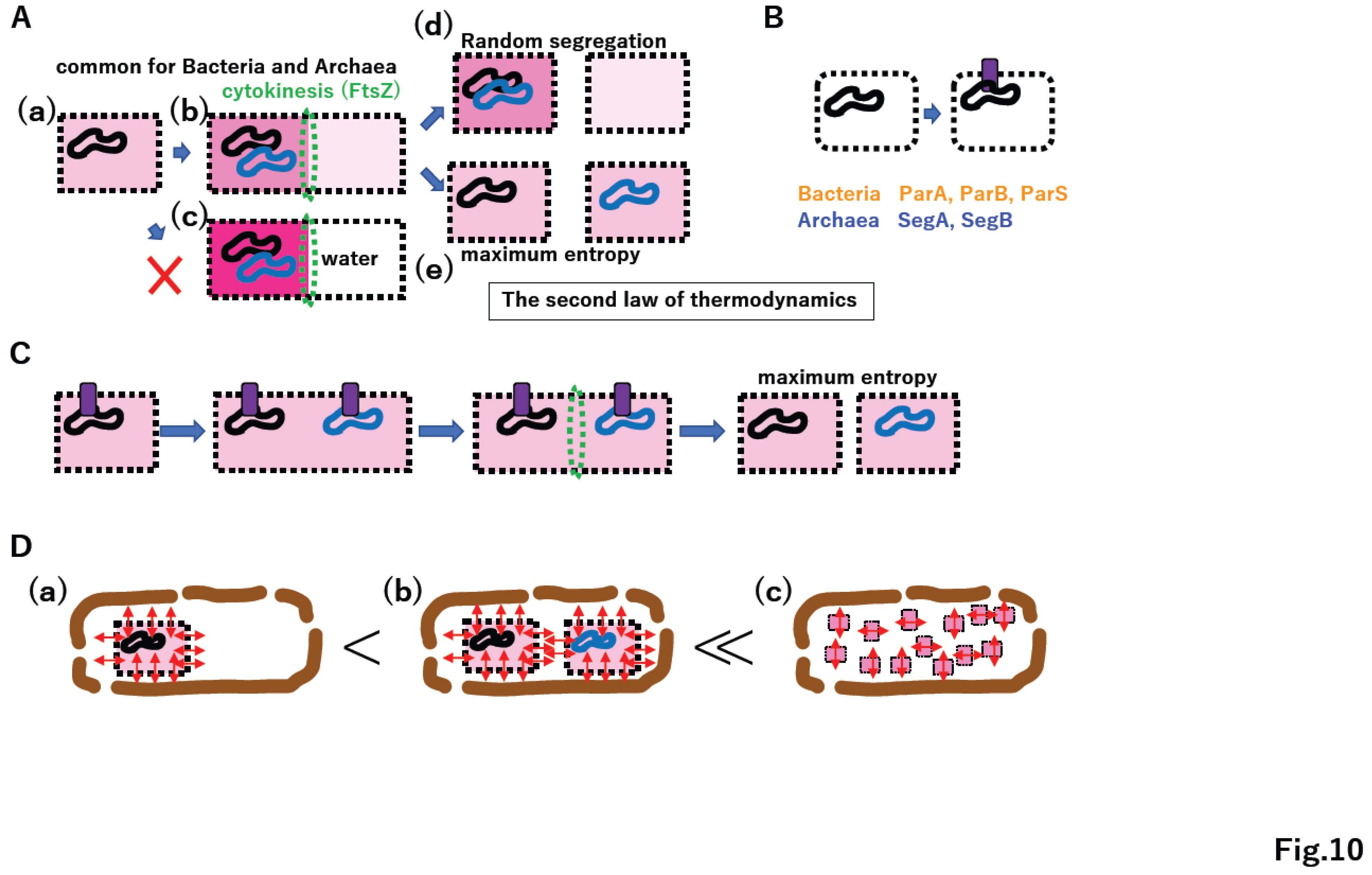
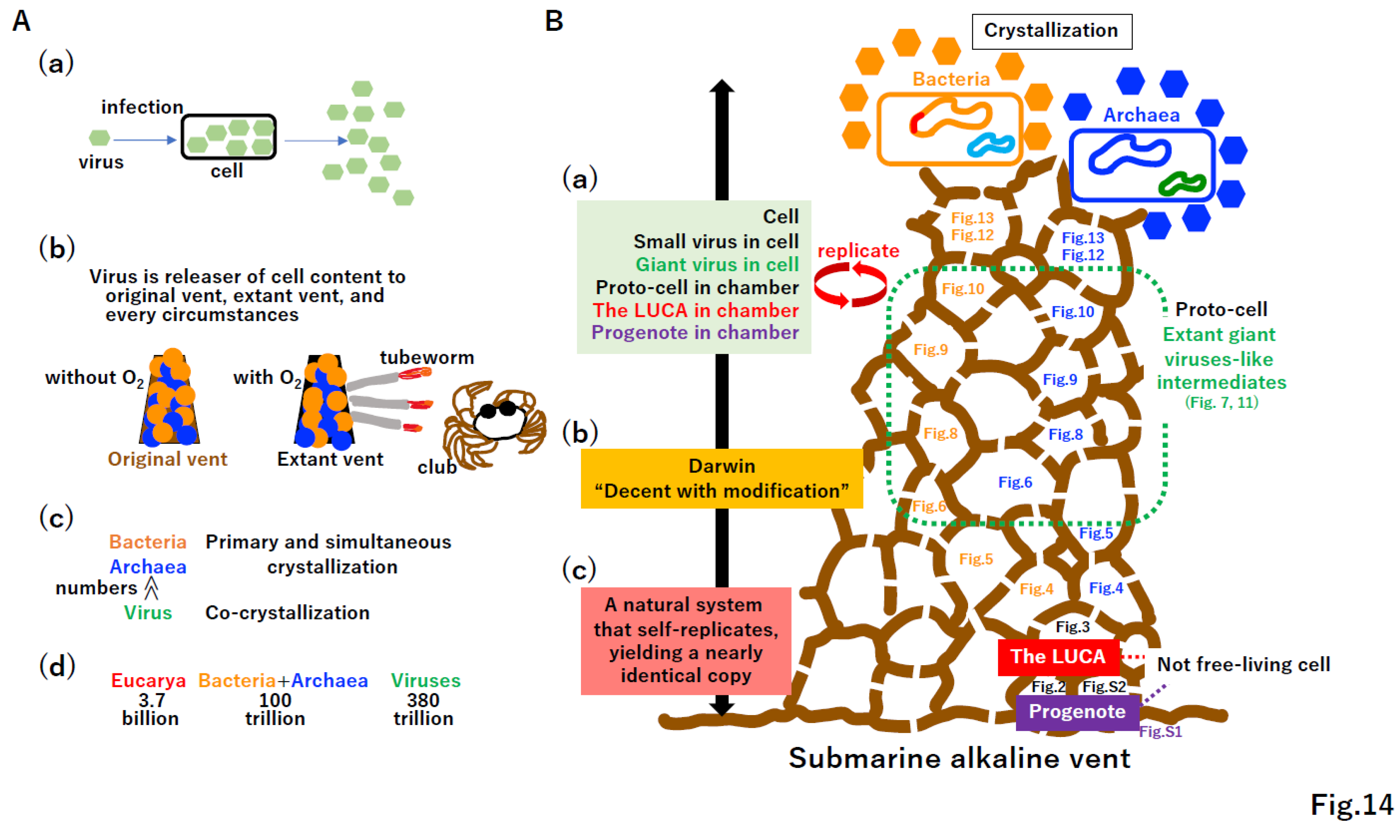
4.5. Omnis cellula a cellula
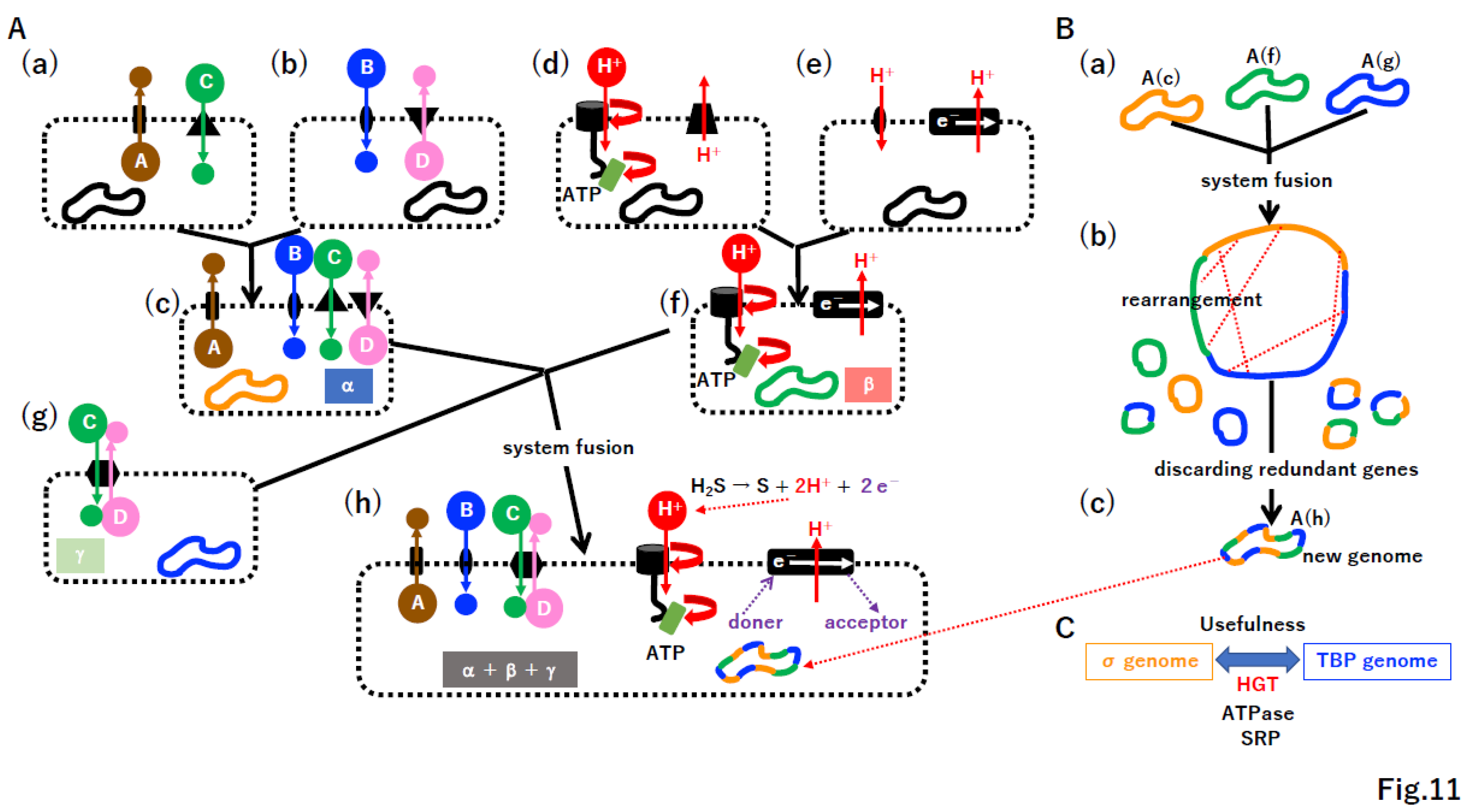
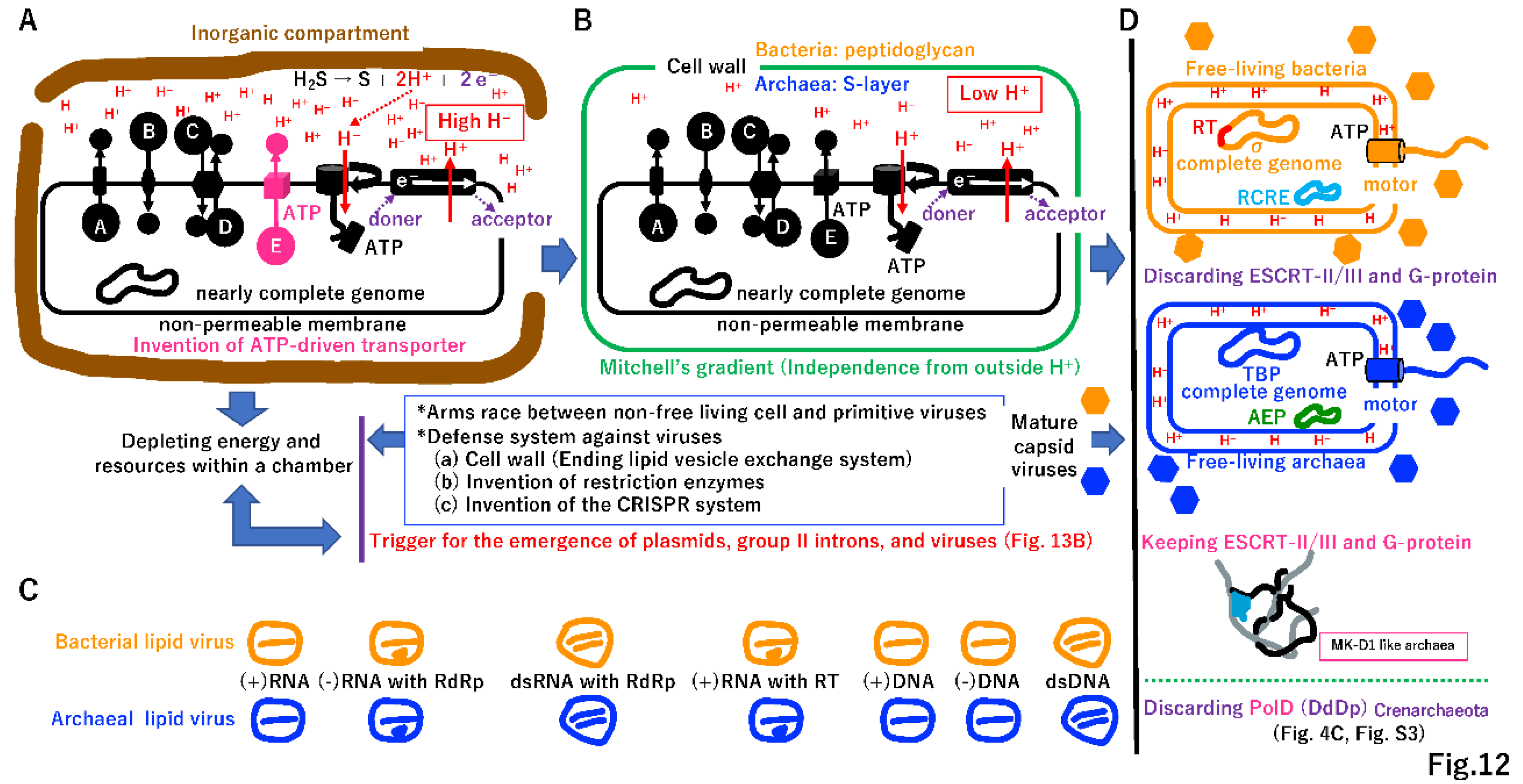
4.6. Punctuated equilibrium by the fusion of different systems
4.7. Selfish cells trigger the emergence of selfish genes
5. Independent time
5.1. Independent cell wall formation
5.2. Why is the genetic code universal?
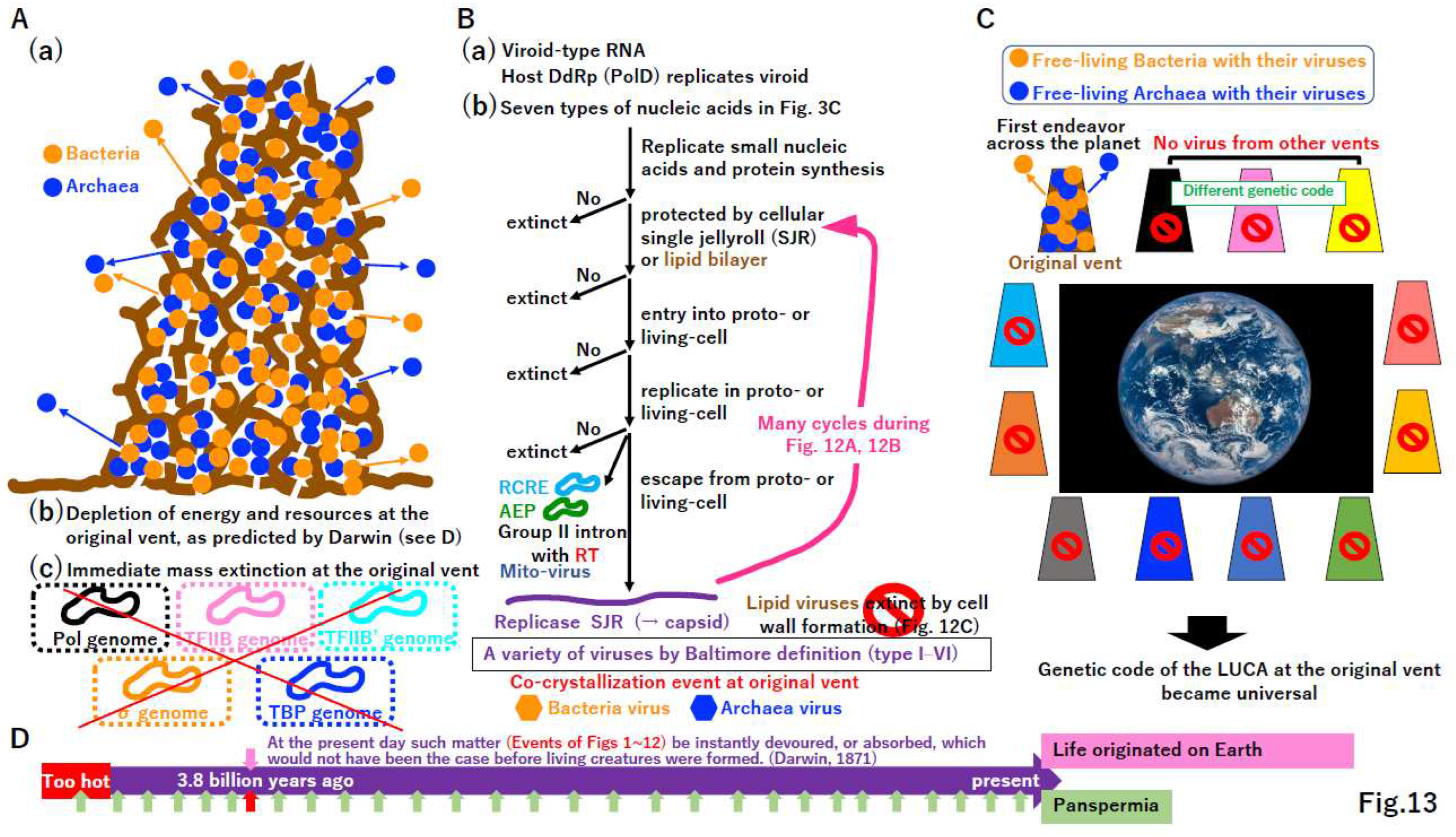
6. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balch, W. E.; Magrum, L. J.; Fox, G. E.; Wolfe, R. S.; Woese, C. R. An ancient divergence among the bacteria. J. Mol. Evol. 1977, 9, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Woese, C. R. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA. 2000, 97, 8392–8396. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L. Symbiosis and evolution. Sci. Am. 1971, 225, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Origin of life: RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Woese, C. R. The universal ancestor. Proc. Natl. Acad. Sci. USA. 1998, 95, 6854–6859. [Google Scholar] [CrossRef] [PubMed]
- Woese, C. R. On the evolution of cells. Proc. Natl. Acad. Sci. USA. 2002, 99, 8742–8747. [Google Scholar] [CrossRef]
- Koonin, E. V.; Martin, W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005, 21, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Branscomb, E.; Russell, M. J. Why the submarine alkaline vent is the most reasonable explanation for the emergence of life. BioEssays 2019, 41, e1800208. [Google Scholar] [CrossRef]
- Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003, 1, 127–136. [Google Scholar] [CrossRef]
- Tuller, T.; Birin, H.; Gophna, U.; Kupiec. M.; Ruppin, E. Reconstructing ancestral gene content by coevolution. Genome Res. 2010, 20, 122–132. [Google Scholar] [CrossRef]
- Seki, M. On the origin of the genetic code. Genes Genet. Syst. 2023, 98, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Bejan, A.; Lorente, S. The constructal law and the evolution of design in nature. Phys. Life Rev. 2011, 8, 209–240. [Google Scholar] [CrossRef] [PubMed]
- Burnim, A. A.; Spence, M. A.; Xu, D.; Jackson, C. J.; Ando, N. Comprehensive phylogenetic analysis of the ribonucleotide reductase family reveals an ancestral clade. Elife 2022, 11, e79790. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E. V.; Krupovic, M.; Ishino, S.; Ishino, Y. The replication machinery of LUCA: common origin of DNA replication and transcription. BMC Biol. 2020, 18, 61. [Google Scholar] [CrossRef] [PubMed]
- Ilina, T. V.; Brosenitsch, T.; Sluis-Cremer, N.; Ishima, R. Retroviral RNase H: Structure, mechanism, and inhibition. Enzymes 2021, 50, 227–247. [Google Scholar] [PubMed]
- Taib, N.; Gribaldo, S.; MacNeill, S. A. Single-stranded DNA-binding proteins in the archaea. Methods Mol. Biol. 2021, 2281, 23–47. [Google Scholar] [PubMed]
- Kornberg, A.; Baker, T. DNA Replication, Second Edition. Freeman, New York. 1992.
- Burton, S. P.; Burton, Z. F. The σ enigma: bacterial σ factors, archaeal TFB and eukaryotic TFIIB are homologs. Transcription 2014, 5, e967599. [Google Scholar] [CrossRef]
- Adachi, N.; Senda, T.; Horikoshi, M. Uncovering ancient transcription systems with a novel evolutionary indicator. Sci. Rep. 2016, 6, 27922. [Google Scholar] [CrossRef]
- Lei, L.; Burton, Z. F. Early evolution of transcription systems and divergence of Archaea and Bacteria. Front. Mol. Biosci. 2021, 8, 651134. [Google Scholar] [CrossRef]
- Ohta, T. Simulation studies on the evolution of amino acid sequences. J. Mol. Evol. 1976, 8, 1–12. [Google Scholar] [CrossRef]
- Ikeda, J. E.; Enomoto, T.; Hurwitz, J. Adenoviral protein-primed initiation of DNA chains in vitro. Proc. Natl. Acad. Sci. USA. 1982, 79, 2442–2446. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. Expression of animal virus genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, G.; Wagner, G. K.; Bowater, R. P. Biochemical and structural characterisation of DNA ligases from bacteria and archaea. Biosci. Rep. 2016, 36, e00391. [Google Scholar] [CrossRef] [PubMed]
- Lobkovsky, A. E.; Wolf, Y. I.; Koonin, E. V. Evolvability of an optimal recombination rate. Genome Biol. Evol. 2015, 8, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I. R.; Bomblies, K. Evolution and plasticity of genome-wide meiotic recombination rates. Annu. Rev. Genet. 2021, 55, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K. R.; Capecchi, M. R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Doudna, J. A. Biotechnology: Rewriting a genome. Nature 2013, 495, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T. H.; Sturtevant, A. H.; Bridge, C. B. The evidence for the linear order of the genes. Proc. Natl. Acad. Sci. USA. 1920, 6, 162–164. [Google Scholar] [CrossRef]
- Leipe, D. D.; Aravind, L.; Koonin, E. V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999, 27, 3389–3401. [Google Scholar] [CrossRef]
- Mendel, G. Experiments in plant hybridization. Royal Horticultural Society of London, translation (1938), Harvard University Press, Cambridge, MA. 1866.
- Liu, L. F.; Wang, J. C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987, 84, 7024–7027. [Google Scholar] [CrossRef]
- Mirkin, E. V.; Mirkin, S. M. Mechanisms of transcription-replication collisions in bacteria. Mol. Cell. Biol. 2005, 25, 888–895. [Google Scholar] [CrossRef]
- Pomerantz, R. T.; O’Donnell, M. What happens when replication and transcription complexes collide? Cell Cycle 2010, 9, 2537–2543. [Google Scholar] [CrossRef]
- St Germain, C.; Zhao, H.; Barlow, J. H. Transcription-replication collisions–A series of unfortunate events. Biomolecules 2021, 11, 1249. [Google Scholar] [CrossRef]
- Watson, J. D.; Crick, F. H. C. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Delbrück, M. On the replication of deoxyribonucleic acid (DNA). Proc. Natl. Acad. Sci. USA. 1954, 40, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. C. DNA topoisomerases. Annu. Rev. Biochem. 1985, 54, 665–697. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Gribaldo, S.; Gadelle, D.; Serre, M. C. Origin and evolution of DNA topoisomerases. Biochimie 2007, 89, 427–446. [Google Scholar] [CrossRef] [PubMed]
- LeBowitz, J. H.; McMacken, R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986, 261, 4738–4748. [Google Scholar] [CrossRef]
- Ishimi, Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997, 272, 24508–24513. [Google Scholar] [CrossRef]
- Leipe, D. D.; Aravind, L.; Grishin, N. V.; Koonin, E. V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000, 10, 5–16. [Google Scholar]
- Gorbalenya, A. E.; Koonin, E. V.; Donchenko, A. P.; Blinov, V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989, 17, 4713–4730. [Google Scholar] [CrossRef]
- Maki, S.; Kornberg, A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J. Biol. Chem. 1988, 263, 6555–6560. [Google Scholar] [CrossRef] [PubMed]
- Tsurimoto, T.; Stillman, B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 1989, 9, 609–619. [Google Scholar]
- Krishna, T. S.; Kong, X. P.; Gary, S.; Burgers, P. M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E. V. The origins of cellular life. Antonie Van Leeuwenhoek. 2014, 106, 27–41. [Google Scholar] [CrossRef]
- Bocquier, A. A.; Liu, L.; Cann, I. K.; Komori, K.; Kohda, D.; Ishino, Y. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr. Biol. 2011, 11, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Leipe, D. D.; Koonin, E. V. Toprim-a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998, 26, 4205–4213. [Google Scholar] [CrossRef]
- Sugino, A.; Hirose, S.; Okazaki, R. RNA-linked nascent DNA fragments in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1972, 69, 1863–1867. [Google Scholar] [CrossRef]
- Matsunaga, F.; Norais, C.; Forterre, P.; Myllykallio, H. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003, 4, 154–158. [Google Scholar] [CrossRef]
- Samson, R. Y.; Bell, S. D. Archaeal chromosome biology. J. Mol. Microbiol. Biotechnol. 2014, 24, 420–427. [Google Scholar] [CrossRef]
- McInerney, P.; Johnson, A.; Katz, F.; O’Donnell, M. Characterization of a triple DNA polymerase replisome. Mol. Cell 2007, 27, 527–538. [Google Scholar] [CrossRef]
- Masai, H.; Nomura, N.; Kubota, Y.; Arai, K. Roles of phi X174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand syntheses of DNA replication. J. Biol. Chem. 1990, 265, 15124–15133. [Google Scholar] [CrossRef]
- Tanaka, S.; Araki, H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010371. [Google Scholar] [CrossRef]
- Takayama, Y.; Kamimura, Y.; Okawa, M.; Muramatsu, S.; Sugino, A.; Araki, H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003, 17, 1153–1165. [Google Scholar] [CrossRef]
- Hamdan, S. M, Richardson CC. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009, 78, 205–243. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Enomoto, T.; Masutani, C.; Hanaoka, F.; Yamada, M.; Ui, M. DNA primase-DNA polymerase α assembly from mouse FM3A cells. Purification of constituting enzymes, reconstitution, and analysis of RNA priming as coupled to DNA synthesis. J. Biol. Chem. 1989, 264, 10065–10071. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F. On regulation of DNA replication in Bacteria. Cold Spring Harbor Symposia on Quantttative Biology 1963, 28, 329. [Google Scholar] [CrossRef]
- Ohmori, H.; Friedberg, E. C.; Fuchs, R. P.; Goodman, M. F.; Hanaoka, F.; Hinkle, D.; Kunkel, T. A.; Lawrence, C. W.; Livneh, Z.; Nohmi, T.; Prakash, L.; Prakash, S.; Todo, T.; Walker, G. C.; Wang, Z.; Woodgate, R. M. The Y-family of DNA polymerases. Mol. Cell 2001, 8, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Lu, A. L.; Clark, S.; Modrich, P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl. Acad. Sci. USA. 1983, 80, 4639–4643. [Google Scholar] [CrossRef]
- Nakae, S.; Hijikata, A.; Tsuji, T.; Yonezawa, K.; Kouyama, K. I.; Mayanagi, K.; Ishino, S.; Ishino, Y.; Shirai, T. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 2016, 24, 1960–1971. [Google Scholar] [CrossRef]
- Cebrián-Sastre, E.; Martín-Blecua, I.; Gullón, S.; Blázquez, J.; Castañeda-García, A. Control of genome stability by EndoMS/NucS-mediated non-canonical mismatch repair. Cells 2021, 10, 1314. [Google Scholar] [CrossRef]
- Maki, H.; Sekiguchi, M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 1992, 355, 273–275. [Google Scholar] [CrossRef]
- Tajiri, T.; Maki, H.; Sekiguchi, M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 1995, 336, 257–267. [Google Scholar] [CrossRef]
- Takao, M.; Kanno, S.; Shiromoto, T.; Hasegawa, R.; Ide, H.; Ikeda, S.; Sarker, A. H.; Seki, S.; Xing, J. Z.; Le, X. C.; Weinfeld, M.; Kobayashi, K.; Miyazaki, J.; Muijtjens, M.; Hoeijmakers, J. H.; van der Horst, G.; Yasui, A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002, 21, 3486–3493. [Google Scholar] [CrossRef]
- White, M. F.; Allers, T. DNA repair in the archaea - an emerging picture. FEMS Microbiol. Rev. 2018, 42, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C. J.; Santangelo, T. J. Archaeal DNA repair mechanisms. Biomolecules 2020, 10, 1472. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arnaiz, P.; Dattani, A.; Smith, V.; Allers, T. Haloferax volcanii - a model archaeon for studying DNA replication and repair. Open Biol. 2020, 10, 200293. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Rupp, W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 1983, 33, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Scheijen, E. E. M.; Wilson, D. M. 3rd. Genome integrity and neurological disease. Int. J. Mol. Sci. 2022, 23, 4142. [Google Scholar] [CrossRef]
- Masutani, C.; Kusumoto, R.; Yamada, A.; Dohmae, N.; Yokoi, M.; Yuasa, M.; Araki, M.; Iwai, S.; Takio, K.; Hanaoka, F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase . Nature 1999, 399, 700–704. [Google Scholar] [CrossRef]
- Matsuura, S.; Tauchi, H.; Nakamura, A.; Kondo, N.; Sakamoto, S.; Endo, S.; Smeets, D.; Solder, B.; Belohradsky, B. H.; Der Kaloustian, V. M.; Oshimura, M.; Isomura, M.; Nakamura, Y.; Komatsu, K. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 1998, 19, 179–181. [Google Scholar] [CrossRef]
- Tanaka, K.; Miura, N.; Satokata, I.; Miyamoto, I.; Yoshida, M. C.; Satoh, Y.; Kondo, S.; Yasui, A.; Okayama, H.; Okada, Y. Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature 1990, 348, 73–76. [Google Scholar] [CrossRef]
- Lindahl, T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA. 1974, 71, 3649–3653. [Google Scholar] [CrossRef]
- Madhani, H. D.; Bohr, V. A.; Hanawalt, P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell 1986, 45, 417–423. [Google Scholar] [CrossRef]
- Zhang, X.; Horibata, K.; Saijo, M.; Ishigami, C.; Ukai, A.; Kanno, S.; Tahara, H.; Neilan, E. G.; Honma, M.; Nohmi, T.; Yasui, A.; Tanaka, K. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat. Genet. 2012, 44, 593–597. [Google Scholar] [CrossRef]
- Kim, J.; Li, C. L.; Chen, X.; Cui, Y.; Golebiowski, F. M.; Wang, H.; Hanaoka, F.; Sugasawa, K.; Yang, W. Lesion recognition by XPC, TFIIH and XPA in DNA excision repair. Nature 2023, 617, 170–175. [Google Scholar] [CrossRef]
- Hutchison, C. A. 3rd; Chuang, R. Y.; Noskov, V. N.; Assad-Garcia, N.; Deerinck, T. J.; Ellisman, M. H.; Gill, J.; Kannan, K.; Karas, B. J.; Ma, L.; Pelletier, J. F.; Qi, Z. Q.; Richter, R. A.; Strychalski, E. A.; Sun, L.; Suzuki, Y.; Tsvetanova, B.; Wise, K. S.; Smith, H. O.; Glass, J. I.; Merryman, C.; Gibson, D. G.; Venter, J. C. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [PubMed]
- Macvanin, M.; Adhya, S. Architectural organization in E. coli nucleoid. Biochim. Biophys. Acta. 2012, 1819, 830–835. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Bhattacharyya, S.; Dyer, P. N.; White, A. E.; Sandman, K.; Burkhart, B. W. Byrne, K. R.; Lee, T.; Ahn, N. G.; Santangelo, T. J.; Reeve, J. N.; Luger, K. Structure of histone-based chromatin in Archaea. Science 2017, 357, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E. V.; Yutin, N. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 2019, 103, 167–202. [Google Scholar] [PubMed]
- Queiroz, V. F.; Rodrigues, R. A. L.; de Miranda Boratto, P. V.; La Scola, B.; Andreani, J.; Abrahão, J. S. Amoebae: Hiding in plain sight: unappreciated hosts for the very large viruses. Annu. Rev. Virol. 2022, 9, 79–98. [Google Scholar] [CrossRef]
- Beadle, G. W.; Tatum, E. L. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA. 1941, 27, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. G.; Nurse, P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 1987, 327, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Paula, S.; Volkov, A. G.; Van Hoek, A. N.; Haines, T. H.; Deamer, D. W. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 1996, 70, 339–348. [Google Scholar] [CrossRef]
- Deamer, D. W. Origins of life: How leaky were primitive cells? Nature 2008, 454, 37–38. [Google Scholar] [CrossRef]
- Niki, H.; Jaffé, A.; Imamura, R.; Ogura, T.; Hiraga, S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991, 10, 183–193. [Google Scholar] [CrossRef]
- Hirano, T.; Mitchison, T. J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 1994, 79, 449–458. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Lengronne, A.; Katou, Y.; Mori, S.; Yokobayashi, S.; Kelly, G. P.; Itoh, T.; Watanabe, Y.; Shirahige, K.; Uhlmann, F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 2004, 430, 573–578. [Google Scholar] [CrossRef]
- Oparin, A. I. The origin of life. McMillan, New York, USA. 1938.
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef]
- Deng, N. N. Complex coacervates as artificial membraneless organelles and protocells. Biomicrofluidics 2020, 14, 051301. [Google Scholar] [CrossRef]
- Matsuo, M.; Kurihara, K. Proliferating coacervate droplets as the missing link between chemistry and biology in the origins of life. Nat. Commun. 2021, 12, 5487. [Google Scholar] [CrossRef]
- Matsuo, M.; Toyota, T.; Suzuki, K.; Sugawara, T. Evolution of proliferative model protocells highly responsive to the environment. Life (Basel) 2022, 12, 1635. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Mann, S. Membranized coacervate microdroplets: from versatile protocell models to cytomimetic materials. Acc. Chem. Res. 2023, 56, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y. From promiscuity to the lipid divide: on the evolution of distinct membranes in Archaea and Bacteria. J. Mol. Evol. 2014, 78, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Bejan, A. Constructal-theory network of conducting paths for cooling a heat generating volume. Int. J. Heat Mass Transfer. 1997, 40, 799–816. [Google Scholar] [CrossRef]
- Nakamura, R.; Takashima, T.; Kato, S.; Takai, K.; Yamamoto, M.; Hashimoto, K. Electrical current generation across a black smoker chimney. Angew. Chem. Int. Ed. Engl. 2010, 49, 7692–7694. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Nakamura, R.; Oguri, K.; Kawagucci, S.; Suzuki, K.; Hashimoto, K.; Takai, K. Generation of electricity and illumination by an environmental fuel cell in deep-sea hydrothermal vents. Angew. Chem. Int. Ed. Engl. 2013, 52, 10758–10761. [Google Scholar] [CrossRef] [PubMed]
- Martin, W. F.; Sousa, F. L.; Lane. N. Evolution. Energy at life’s origin. Science 2014, 344, 1092–1093. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Kawaichi, S.; Nakagawa, H.; Hashimoto, K.; Nakamura, R. From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe(II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front. Microbiol. 2015, 6, 994. [Google Scholar] [CrossRef]
- McGlynn, S. E.; Chadwick, G. L.; Kempes, C. P.; Orphan, V. J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 2015, 526, 531–535. [Google Scholar] [CrossRef]
- Lane, N.; Allen, J. F.; Martin, W. How did LUCA make a living? Chemiosmosis in the origin of life. Bioessays 2010, 32, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019, 43, 273–303. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cvirkaite-Krupovic, V.; Commere, P. H.; Yang, Y.; Zhou, F.; Forterre, P.; Shen, Y.; Krupovic, M. Archaeal extracellular vesicles are produced in an ESCRT-dependent manner and promote gene transfer and nutrient cycling in extreme environments. ISME J. 2021, 15, 2892–2905. [Google Scholar] [CrossRef]
- Pende, N.; Sogues, A.; Megrian, D.; Sartori-Rupp, A.; England, P.; Palabikyan, H.; Rittmann, S. K. R.; Graña, M.; Wehenkel, A. M.; Alzari, P. M.; Gribaldo, S. SepF is the FtsZ anchor in archaea, with features of an ancestral cell division system. Nat. Commun. 2021, 12, 3214. [Google Scholar] [CrossRef]
- Santana-Molina, C.; Del Saz-Navarro, D.; Devos, D. P. Early origin and evolution of the FtsZ/tubulin protein family. Front. Microbiol. 2023, 13, 1100249. [Google Scholar] [CrossRef]
- Badrinarayanan, A.; Le, T. B.; Laub, M. T. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015, 31, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D. Rudolf Virchow, the founder of cellular pathology. Rom. J. Morphol. Embryol. 2019, 60, 1381–1382. [Google Scholar]
- Gould, S. J. Punctuated equilibrium in fact and theory. J. Social Biol. Struct. 1989, 12, 117–136. [Google Scholar] [CrossRef]
- Mulkidjanian, A. Y.; Makarova, K. S.; Galperin, M. Y.; Koonin, E. V. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat. Rev. Microbiol. 2007, 5, 892–899. [Google Scholar] [CrossRef]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E. V.; Raoult, D. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef] [PubMed]
- van Wolferen, M.; Pulschen, A. A.; Baum, B.; Gribaldo, S.; Albers, S. V. The cell biology of archaea. Nat. Microbiol. 2022, 7, 1744–1755. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Dolja, V. V.; Koonin, E. V. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef]
- Miyata, M.; Robinson, R. C.; Uyeda, T. Q. P.; Fukumori, Y.; Fukushima, S. I.; Haruta, S.; Homma, M.; Inaba, K.; Ito, M.; Kaito, C.; Kato, K.; Kenri, T.; Kinosita, Y.; Kojima, S.; Minamino, T.; Mori, H.; Nakamura, S.; Nakane, D.; Nakayama, K.; Nishiyama, M.; Shibata, S.; Shimabukuro, K.; Tamakoshi, M.; Taoka, A.; Tashiro, Y.; Tulum, I.; Wada, H.; Wakabayashi, K. I. Tree of motility - A proposed history of motility systems in the tree of life. Genes Cells 2020, 25, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Matzke, N. J.; Lin, A.; Stone, M.; Baker, M. A. B. Flagellar export apparatus and ATP synthetase: homology evidenced by synteny predating the Last Universal Common Ancestor. Bioessays 2021, 43, e2100004. [Google Scholar] [CrossRef]
- Daimon, K.; Ishino, S.; Imai, N.; Nagumo, S.; Yamagami, T.; Matsukawa, H.; Ishino, Y. Two family B DNA polymerases from Aeropyrum pernix, based on revised translational frames. Front. Mol. Biosci. 2018, 5, 37. [Google Scholar] [CrossRef]
- Imachi, H.; Nobu, M. K.; Nakahara, N.; Morono, Y.; Ogawara, M.; Takaki, Y.; Takano, Y.; Uematsu, K.; Ikuta, T.; Ito, M.; Matsui, Y.; Miyazaki, M.; Murata, K.; Saito, Y.; Sakai, S.; Song, C.; Tasumi, E.; Yamanaka, Y.; Yamaguchi, T.; Kamagata, Y.; Tamaki, H.; Takai, K. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020, 577, 519–525. [Google Scholar] [CrossRef]
- Arber, W. Restriction endonucleases. Angew. Chem. Int. Ed. Engl. 1978, 17, 73–79. [Google Scholar] [CrossRef]
- Wang, Y. Current view and perspectives in viroid replication. Curr. Opin. Virol. 2021, 47, 32–37. [Google Scholar] [CrossRef]
- Hillman, B. I.; Cai, G. The family narnaviridae: simplest of RNA viruses. Adv. Virus Res. 2013, 86, 149–176. [Google Scholar]
- Follmann, H.; Brownson, C. Darwin’s warm little pond revisited: from molecules to the origin of life. Naturwissenschaften 2009, 96, 1265–1292. [Google Scholar] [CrossRef]
- Crick, F. H. C.; Orgel, L. E. (1973) Directed panspermia. Icarus 1973, 19, 341–346. [Google Scholar] [CrossRef]
- Aydinoglu, A. U.; Taskin, Z. Origins of life research: a bibliometric approach. Orig. Life Evol. Biosph. 2018, 48, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Dolja, V. V.; Koonin, E. V. The LUCA and its complex virome. Nat. Rev. Microbiol. 2020, 18, 661–670. [Google Scholar] [CrossRef]
- Call, L.; Nayfach, S.; Kyrpides, N. C. Illuminating the virosphere through global metagenomics. Annu. Rev. Biomed. Data. Sci. 2021, 4, 369–391. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Bushman, F. D. The human virome: assembly, composition and host interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Forterre, P. Viruses in the 21st century: from the curiosity-driven discovery of giant viruses to new concepts and definition of life. Clin. Infect Dis. 2017, 65 (suppl 1), S74–S79. [Google Scholar] [CrossRef] [PubMed]
- Harris, H. M. B.; Hillm, C. A place for viruses on the tree of life. Front. Microbiol. 2021, 11, 604048. [Google Scholar] [CrossRef]
- Kornberg, A. Never a dull enzyme. Annu. Rev. Biochem. 1989, 58, 1–30. [Google Scholar] [CrossRef]
- Penny, D. Darwin’s theory of descent with modification, versus the biblical tree of life. PLoS Biol. 2011, 9, e1001096. [Google Scholar] [CrossRef]
- Mak, J.; Kleiman, L. Primer tRNAs for reverse transcription. J. Virol. 1997, 71, 8087–8095. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. A Tyr residue in the reverse transcriptase domain can mimic the protein-priming Tyr residue in the terminal protein domain of a hepadnavirus P protein. J. Virol. 2011, 85, 7742–7753. [Google Scholar] [CrossRef]
- Jain, N.; Blauch, L. R.; Szymanski, M. R.; Das, R.; Tang, S. K. Y.; Yin, Y. W.; Fire, A. Z. Transcription polymerase-catalyzed emergence of novel RNA replicons. Science 2020, 368, eaay0688. [Google Scholar] [CrossRef]
- Skalenko, K. S.; Li, L.; Zhang, Y.; Vvedenskaya, I. O.; Winkelman, J. T.; Cope, A. L. Taylor, D. M.; Shah, P.; Ebright, R. H.; Kinney, J. B.; Zhang, Y.; Nickels, B. E. Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2021, 118, e2106388118. [Google Scholar] [CrossRef]
- Stasiak, A.; Di Capua, E. The helicity of DNA in complexes with recA protein. Nature 1982, 299, 185–186. [Google Scholar] [CrossRef]
- Shinohara, A.; Ogawa, H.; Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 1992, 69, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Haruta, N.; Kurokawa, Y.; Murayama, Y.; Akamatsu, Y.; Unzai, S.; Tsutsui, Y.; Iwasaki, H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006, 13, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Wolf, U.; Atkin, N. B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, E.; Sasaki, M. S.; Buerstedde, J. M.; Bezzubova, O.; Shinohara, A.; Ogawa, H.; Takata, M.; Yamaguchi-Iwai. Y.; Takeda, S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998, 17, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Iwai, Y.; Sonoda, E.; Sasaki, M. S.; Morrison, C.; Haraguchi, T.; Hiraoka, Y.; Yamashita, Y. M.; Yagi, T.; Takata, M.; Price, C.; Kakazu, N.; Takeda, S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999, 18, 6619–6629. [Google Scholar] [CrossRef] [PubMed]
- Izumi, T,; Brown, D. B.; Naidu, C. V.; Bhakat, K. K.; Macinnes, M. A.; Saito, H.; Chen, D. J.; Mitra, S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. USA. 2005, 102, 5739–5743. [Google Scholar] [CrossRef] [PubMed]
- Neri, U.; Wolf, Y. I.; Roux, S.; Camargo, A. P.; Lee, B.; Kazlauskas, D.; Chen, I. M.; Ivanova, N.; Zeigler Allen, L.; Paez-Espino, D.; Bryant, D. A.; Bhaya, D.; RNA Virus Discovery Consortium; Krupovic, M. ; Dolja, V. V.; Kyrpides, N. C.; Koonin, E. V.; Gophna, U. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell 2022, 185, 4023–4037. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



