Submitted:
29 December 2023
Posted:
29 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Patient Characteristics
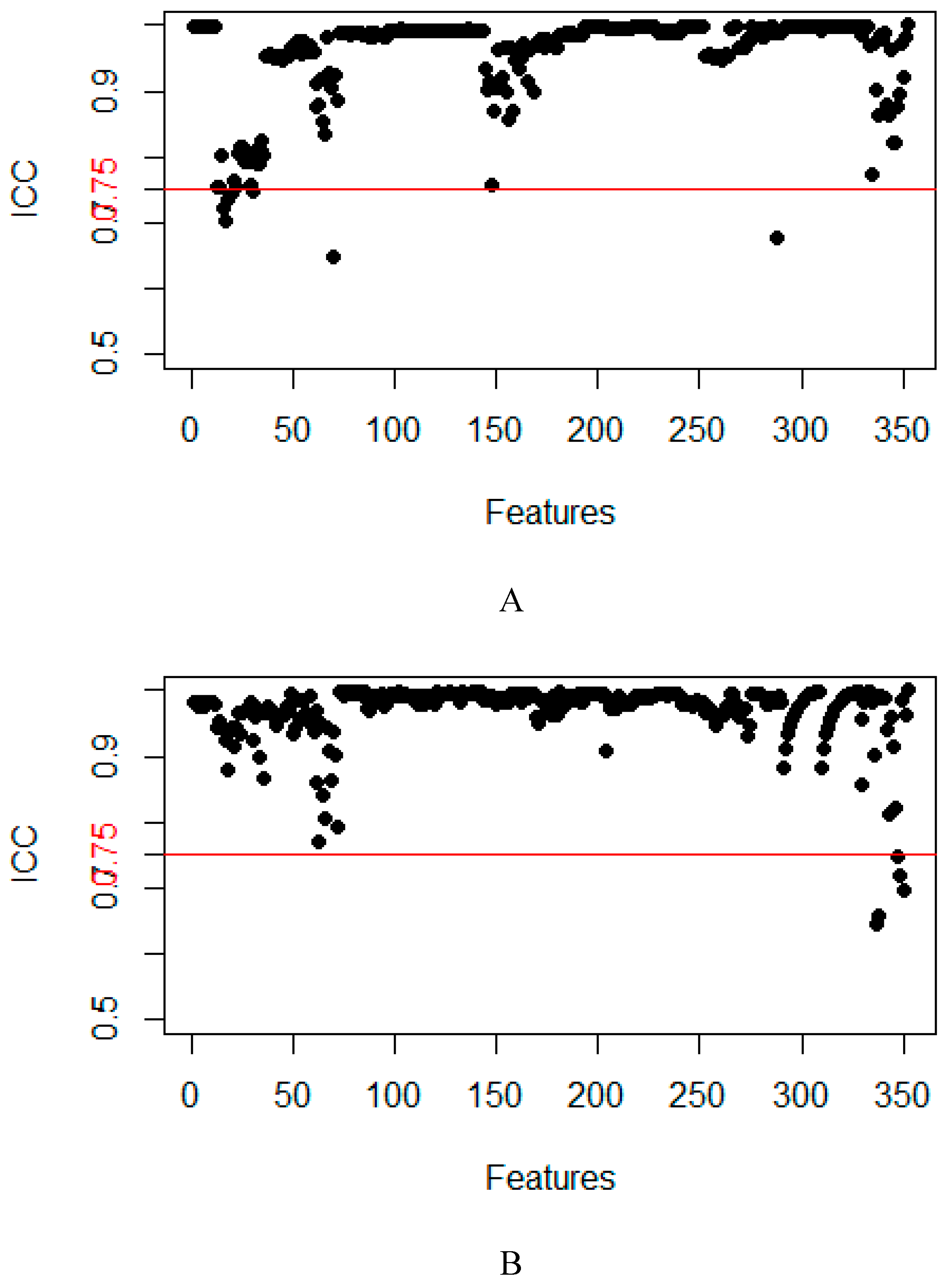
2.2. Feature Extraction and Selection
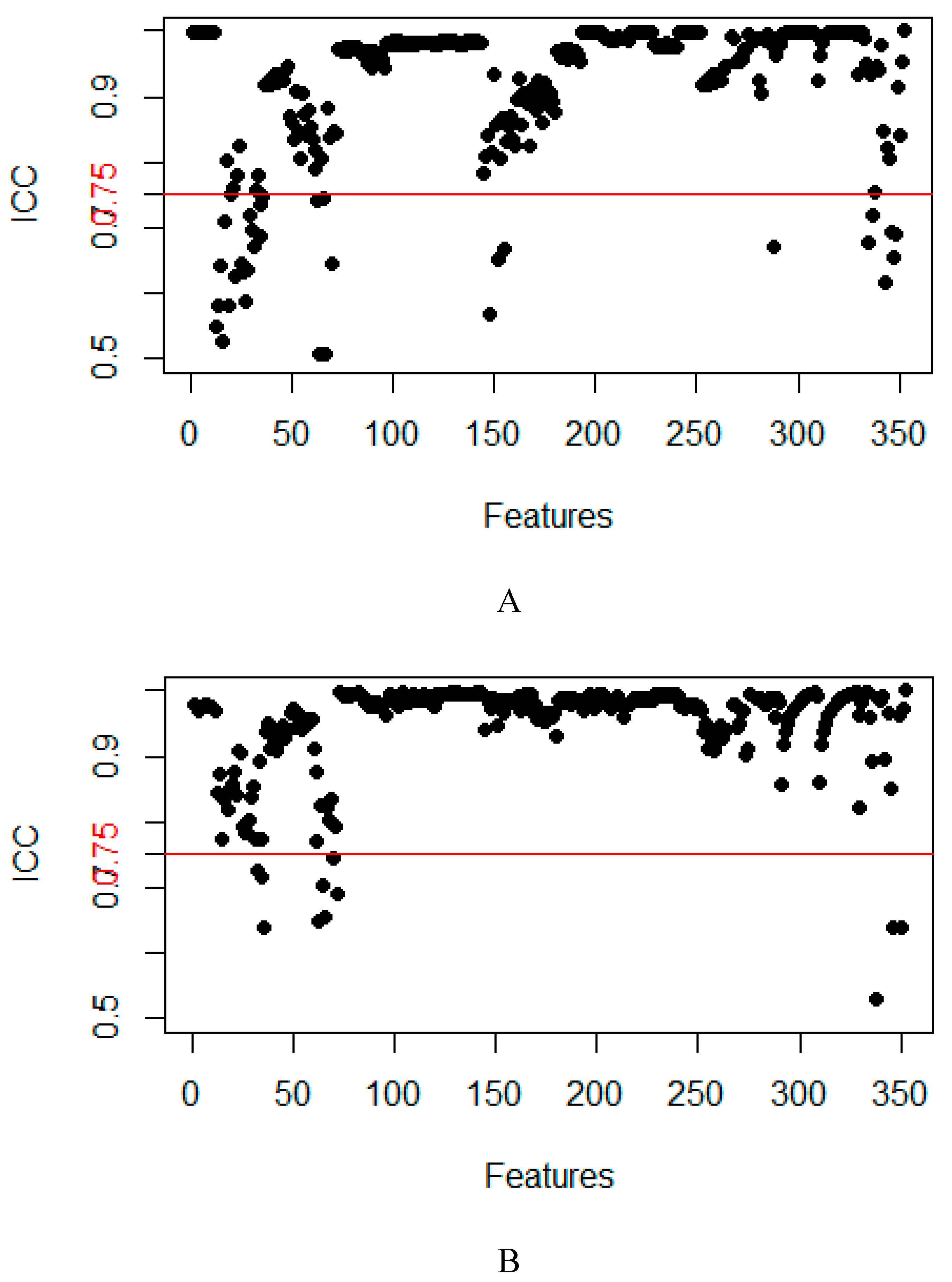
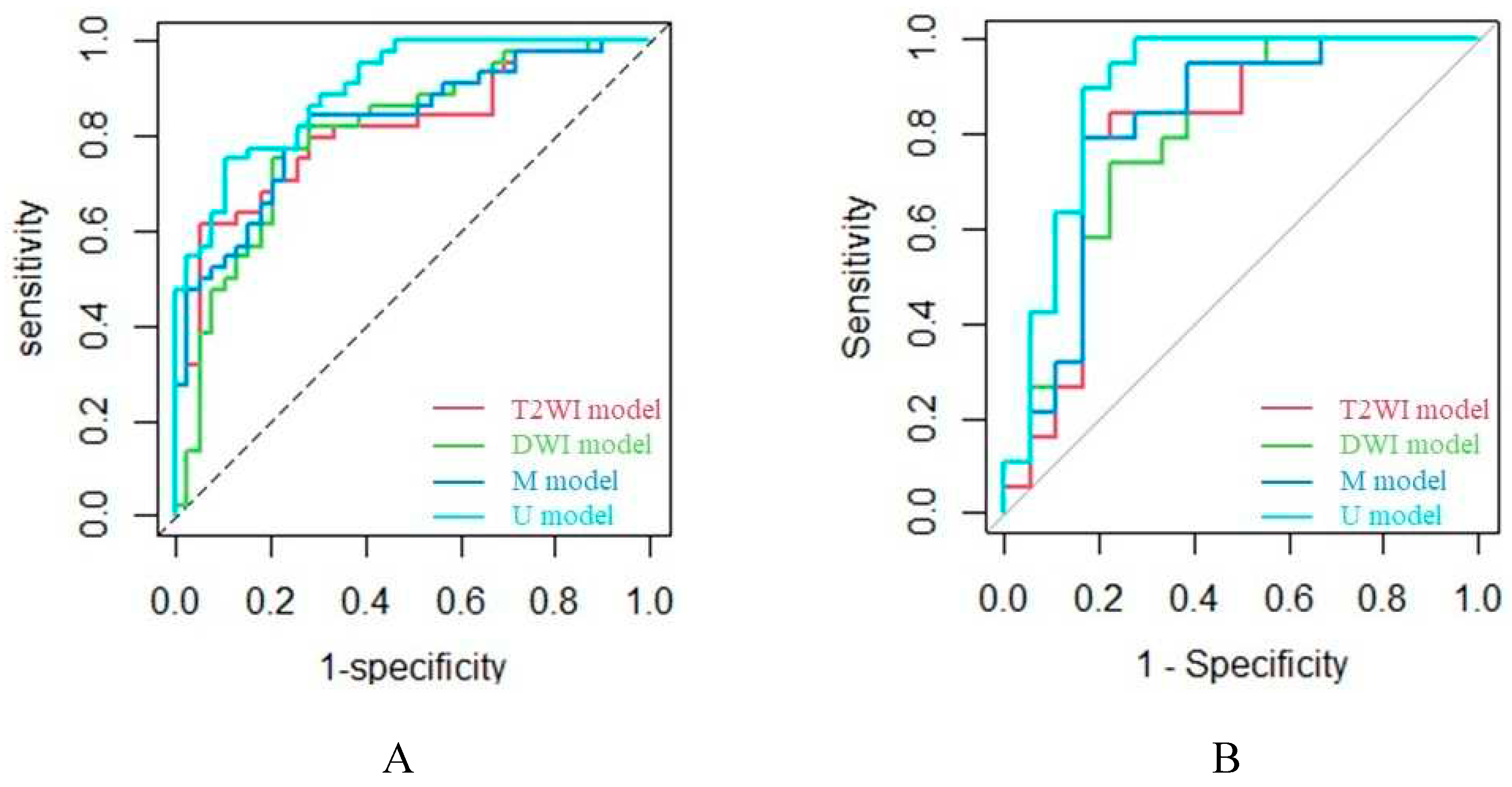
2.3. Model Evaluation
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. MRI Acquisition
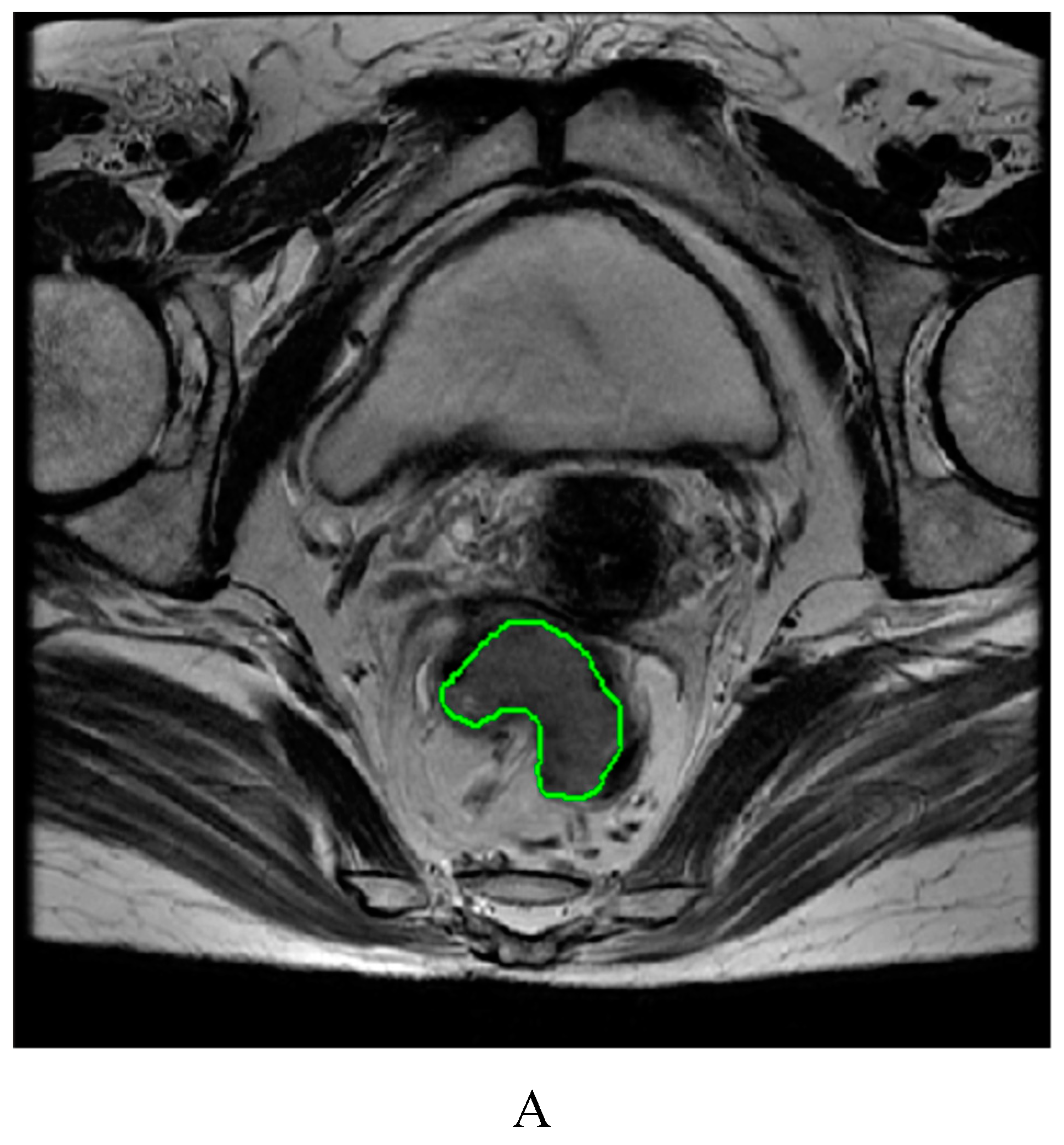
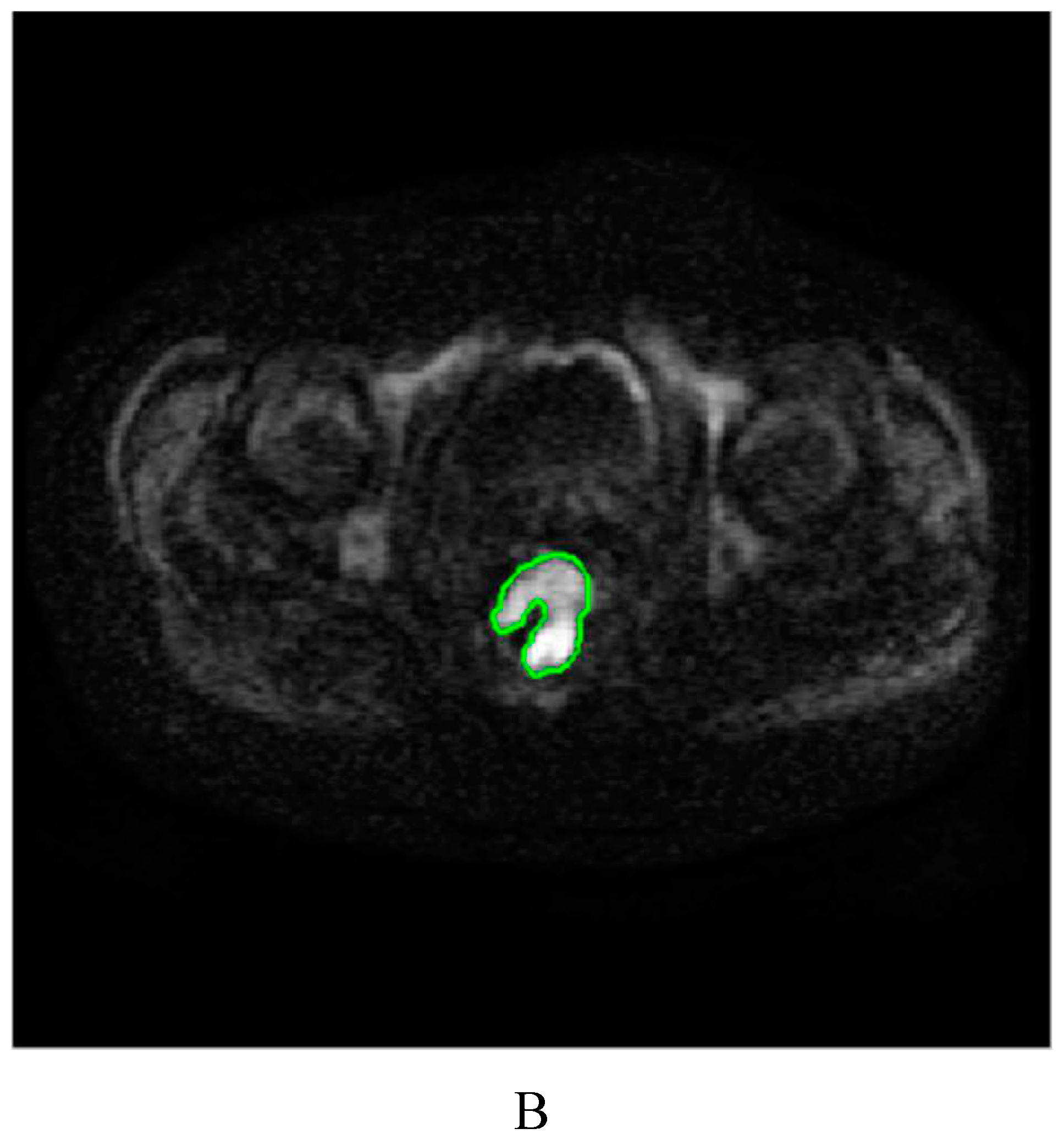
4.3. Image segmentation and feature extraction
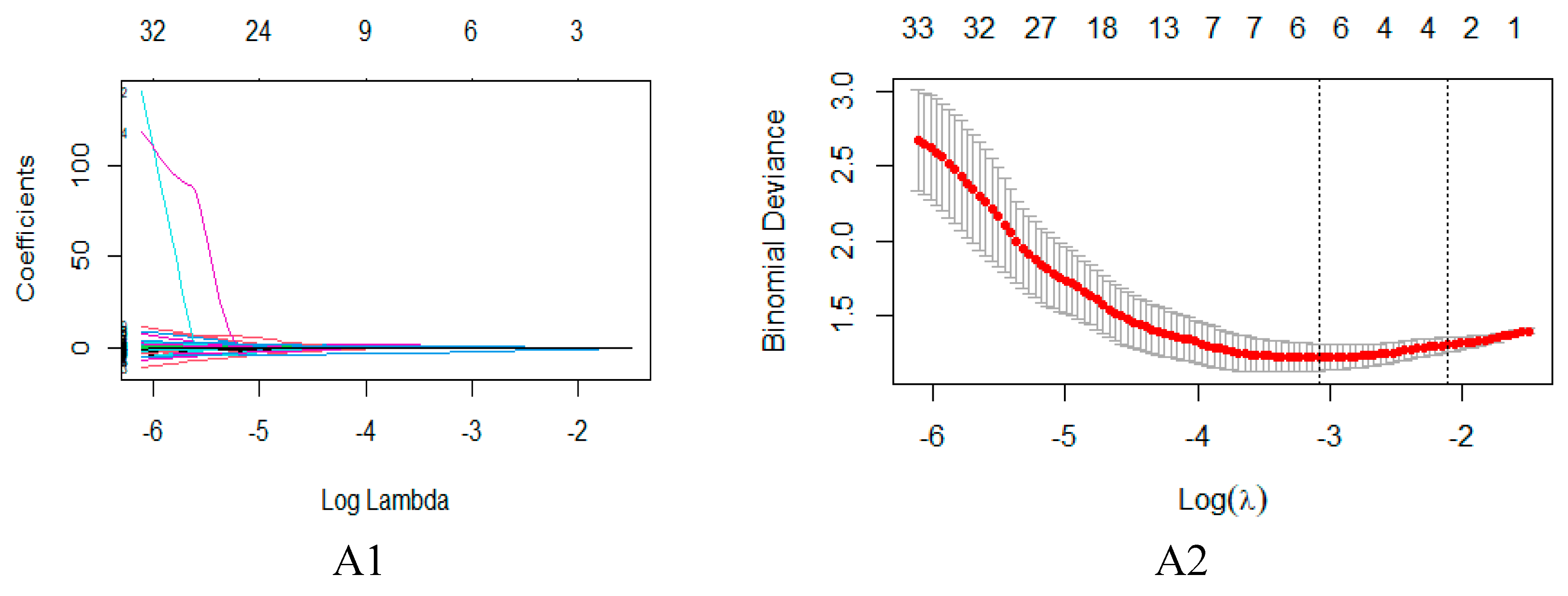
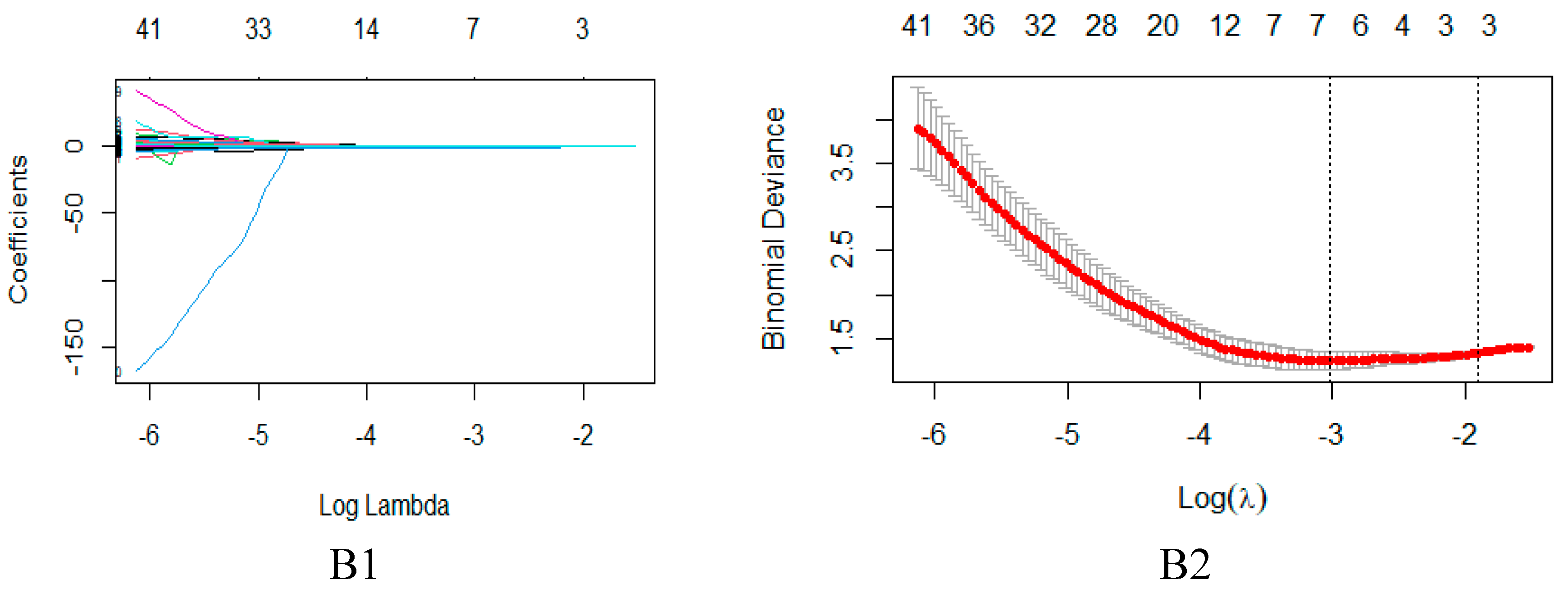
4.4. Feature dimensionality reduction and selection
4.5. Model construction and evaluation
4.6. Statistical methods
5. Conclusions
Supplementary Materials
References
- Martinou, E.; Moller-Levet, C.; Karamanis, D.; Bagwan, I.; Angelidi, A.M. HOXB9 Overexpression Promotes Colorectal Cancer Progression and Is Associated with Worse Survival in Liver Resection Patients for Colorectal Liver Metastases. Int. J. Mol. Sci. 2022, 23, 2281. [Google Scholar] [CrossRef]
- Delattre, J.-F.; Erdogan, A.S.O.; Cohen, R.; Shi, Q.; Emile, J.-F.; Taieb, J.; Tabernero, J.; André, T.; Meyerhardt, J.A.; Nagtegaal, I.D.; et al. A comprehensive overview of tumour deposits in colorectal cancer: Towards a next TNM classification. Cancer Treat. Rev. 2021, 103, 102325. [Google Scholar] [CrossRef]
- Akgül Ö., Çetinkaya E., Ersöz Ş., Tez M. Role of surgery in colorectal cancer liver metastases. World J Gastroenterol 2014;20(20): 6113-22. [CrossRef]
- Rada, M.; Kapelanski-Lamoureux, A.; Petrillo, S.; Tabariès, S.; Siegel, P.; Reynolds, A.R.; Lazaris, A.; Metrakos, P. Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases. Commun. Biol. 2021, 4, 1–15. [Google Scholar] [CrossRef]
- De’angelis, N.; Baldini, C.; Brustia, R.; Pessaux, P.; Sommacale, D.; Laurent, A.; Le Roy, B.; Tacher, V.; Kobeiter, H.; Luciani, A.; et al. Surgical and regional treatments for colorectal cancer metastases in older patients: A systematic review and meta-analysis. PLOS ONE 2020, 15, e0230914. [Google Scholar] [CrossRef]
- Engstrand J., Nilsson H., Strömberg C., Jonas E., Freedman J. Colorectal cancer liver metastases-a population-based study on incidence, management and survival. BMC Cancer 2018;18(1): 78. [CrossRef]
- Poturnajova, M.; Furielova, T.; Balintova, S.; Schmidtova, S.; Kucerova, L.; Matuskova, M. Molecular features and gene expression signature of metastatic colorectal cancer (Review). Oncol. Rep. 2021, 45, 1–1. [Google Scholar] [CrossRef]
- Bhullar, D.S.; Barriuso, J.; Mullamitha, S.; Saunders, M.P.; O'Dwyer, S.T.; Aziz, O. Biomarker concordance between primary colorectal cancer and its metastases. EBioMedicine 2019, 40, 363–374. [Google Scholar] [CrossRef]
- Xu J., Fan J., Qin X., Cai J., Gu J., Wang S., Wang X., Zhang S., Zhang Z., China CRLM Guideline Group. Chinese guidelines for the diagnosis and comprehensive treatment of colorectal liver metastases (version 2018). J Cancer Res Clin Oncol 2019;145(3): 725-736. [CrossRef]
- Ren L., Zhu D., Benson AB 3rd., Nordlinger B., Koehne CH., Delaney CP., Kerr D., Lenz HJ., Fan J., Wang J., et al. Shanghai international consensus on diagnosis and comprehensive treatment of colorectal liver metastases (version 2019). Eur J Surg Oncol 2020;46(6): 955-966. [CrossRef]
- Loo JM., Scherl A., Nguyen A., Man FY., Weinberg E., Zeng Z., Saltz L., Paty PB., Tavazoie SF. Extracellular metabolic energetics can promote cancer progression. Cell 2015;160(3): 393-406. [CrossRef]
- Li Y., Eresen A., Shangguan J., Yang J., Lu Y., Chen D., Wang J., Velichko Y., Yaghmai V., Zhang Z. Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am J Cancer Res 2019;9(11): 2482-2492.
- Kijima, S.; Sasaki, T.; Nagata, K.; Utano, K.; Lefor, A.T.; Sugimoto, H. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J. Gastroenterol. 2014, 20, 16964–75. [Google Scholar] [CrossRef]
- Gaitanidis, A.; Alevizakos, M.; Tsaroucha, A.; Tsalikidis, C.; Pitiakoudis, M. Predictive Nomograms for Synchronous Distant Metastasis in Rectal Cancer. J. Gastrointest. Surg. 2018, 22, 1268–1276. [Google Scholar] [CrossRef]
- Wu, J.-B.; Sarmiento, A.L.; Fiset, P.-O.; Lazaris, A.; Petrillo, S.; Metrakos, P.; Gao, Z.-H. Histologic features and genomic alterations of primary colorectal adenocarcinoma predict growth patterns of liver metastasis. World J. Gastroenterol. 2019, 25, 3408–3425. [Google Scholar] [CrossRef]
- Liu M., Ma X., Shen F., Xia Y., Jia Y., Lu J. MRI-based radiomics nomogram to predict synchronous liver metastasis in primary rectal cancer patients. Cancer Med 2020;9(14): 5155-5163. [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Zhang X., Zhang Y., Zhang G., Qiu X., Tan W., Yin X., Liao L. Deep Learning With Radiomics for Disease Diagnosis and Treatment: Challenges and Potential. Front Oncol 2022;12: 773840. [CrossRef]
- Mayerhoefer ME., Materka A., Langs G., Häggström I., Szczypiński P., Gibbs P., Cook G. Introduction to Radiomics. J Nucl Med 2020;61(4): 488-495. [CrossRef]
- Yip SS., Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016;61(13): R150-66. [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Liu N., Wu Y., Tao Y., Zheng J., Huang X., Yang L., Zhang X. Differentiation of Hepatocellular Carcinoma from Intrahepatic Cholangiocarcinoma through MRI Radiomics. Cancers (Basel) 2023;15(22):5373. [CrossRef]
- Gong XQ., Liu N., Tao YY., Li L., Li ZM., Yang L., Zhang XM. Radiomics models based on multisequence MRI for predicting PD-1/PD-L1 expression in hepatocellular carcinoma. Sci Rep 2023;13(1):7710. [CrossRef]
- Mao Q., Zhou MT., Zhao ZP., Liu N., Yang L., Zhang XM. Role of radiomics in the diagnosis and treatment of gastrointestinal cancer. World J Gastroenterol 2022;28(42):6002-6016. [CrossRef]
- Alshohoumi, F.; Al-Hamdani, A.; Hedjam, R.; AlAbdulsalam, A.; Al Zaabi, A. A Review of Radiomics in Predicting Therapeutic Response in Colorectal Liver Metastases: From Traditional to Artificial Intelligence Techniques. Healthcare 2022, 10, 2075. [Google Scholar] [CrossRef]
- Staal, F.; Taghavi, M.; van der Reijd, D.; Gomez, F.; Imani, F.; Klompenhouwer, E.; Meek, D.; Roberti, S.; de Boer, M.; Lambregts, D.; et al. Predicting local tumour progression after ablation for colorectal liver metastases: CT-based radiomics of the ablation zone. Eur. J. Radiol. 2021, 141, 109773. [Google Scholar] [CrossRef]
- Tharmaseelan, H.; Hertel, A.; Tollens, F.; Rink, J.; Woźnicki, P.; Haselmann, V.; Ayx, I.; Nörenberg, D.; Schoenberg, S.O.; Froelich, M.F. Identification of CT Imaging Phenotypes of Colorectal Liver Metastases from Radiomics Signatures—Towards Assessment of Interlesional Tumor Heterogeneity. Cancers 2022, 14, 1646. [Google Scholar] [CrossRef]
- de la Pinta, C.; Castillo, M.E.; Collado, M.; Galindo-Pumariño, C.; Peña, C. Radiogenomics: Hunting Down Liver Metastasis in Colorectal Cancer Patients. Cancers 2021, 13, 5547. [Google Scholar] [CrossRef]
- Han Y., Chai F., Wei J., Yue Y., Cheng J., Gu D., Zhang Y., Tong T., Sheng W., Hong N., et al. Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis. Front Oncol 2020;10: 1363. [CrossRef]
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Setola, S.V.; Aversana, F.D.; Ottaiano, A.; Avallone, A.; Nasti, G.; Grassi, F.; et al. Contrast MR-Based Radiomics and Machine Learning Analysis to Assess Clinical Outcomes following Liver Resection in Colorectal Liver Metastases: A Preliminary Study. Cancers 2022, 14, 1110. [Google Scholar] [CrossRef]
- Shi R., Chen W., Yang B., Qu J., Cheng Y., Zhu Z., Gao Y., Wang Q., Liu Y., Li Z., et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res 2020;10(12): 4513-4526.
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Setola, S.V.; Dell’aversana, F.; Grassi, F.; Belli, A.; Silvestro, L.; Ottaiano, A.; et al. Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. La Radiol. medica 2022, 127, 763–772. [Google Scholar] [CrossRef]
- Taghavi M., Trebeschi S., Simões R., Meek DB., Beckers RCJ., Lambregts DMJ., Verhoef C., Houwers JB., van der Heide UA., Beets-Tan RGH., et al. Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. AbdomRadiol (NY) 2021;46(1): 249-256. [CrossRef]
- Becker, A.S.; Schneider, M.A.; Wurnig, M.C.; Wagner, M.; Clavien, P.A.; Boss, A. Radiomics of liver MRI predict metastases in mice. Eur. Radiol. Exp. 2018, 2, 1–10. [Google Scholar] [CrossRef]
- Rocca, A.; Brunese, M.C.; Santone, A.; Avella, P.; Bianco, P.; Scacchi, A.; Scaglione, M.; Bellifemine, F.; Danzi, R.; Varriano, G.; et al. Early Diagnosis of Liver Metastases from Colorectal Cancer through CT Radiomics and Formal Methods: A Pilot Study. J. Clin. Med. 2021, 11, 31. [Google Scholar] [CrossRef]
- Liang M., Cai Z., Zhang H., Huang C., Meng Y., Zhao L., Li D., Ma X., Zhao X. Machine Learning-based Analysis of Rectal Cancer MRI Radiomics for Prediction of Metachronous Liver Metastasis. Acad Radiol 2019;26(11): 1495-1504. [CrossRef]
- Li ZF., Kang LQ., Liu FH., Zhao M., Guo SY., Lu S., Quan S. Radiomics based on preoperative rectal cancer MRI to predict the metachronous liver metastasis. Abdom Radiol (NY) 2023;48(3): 833-843. [CrossRef]
- Zhu HQ., Wang DY., Xu LS., Chen JL., Chu EW., Zhou CJ. Diagnostic value of an enhanced MRI combined with serum CEA, CA19-9, CA125 and CA72-4 in the liver metastasis of colorectal cancer. World J Surg Oncol 2022;20(1): 401. [CrossRef]
- Zhang D., Yu M., Xu T., Xiong B. Predictive value of serum CEA, CA19-9 and CA125 in diagnosis of colorectal liver metastasis in Chinese population. Hepatogastroenterology 2013;60(126): 1297-301. [CrossRef]
- Li Y., Gong J., Shen X., Li M., Zhang H., Feng F., Tong T. Assessment of Primary Colorectal Cancer CT Radiomics to Predict Metachronous Liver Metastasis. Front Oncol 2022;12: 861892. [CrossRef]
- Lee S., Choe EK., Kim SY., Kim HS., Park KJ., Kim D. Liver imaging features by convolutional neural network to predict the metachronous liver metastasis in stage I-III colorectal cancer patients based on preoperative abdominal CT scan. BMC Bioinformatics 2020;21(Suppl 13): 382. [CrossRef]
- Landreau, P.; Drouillard, A.; Launoy, G.; Ortega-Deballon, P.; Jooste, V.; Lepage, C.; Faivre, J.; Facy, O.; Bouvier, A. Incidence and survival in late liver metastases of colorectal cancer. J. Gastroenterol. Hepatol. 2014, 30, 82–85. [Google Scholar] [CrossRef]
- Huang, E.P.; O’connor, J.P.B.; McShane, L.M.; Giger, M.L.; Lambin, P.; Kinahan, P.E.; Siegel, E.L.; Shankar, L.K. Criteria for the translation of radiomics into clinically useful tests. Nat. Rev. Clin. Oncol. 2022, 20, 69–82. [Google Scholar] [CrossRef]
- Amin MB, Greene FL, journal SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA: a cancer journal for clinicians. 2017;67(2):93-9. [CrossRef]
| Sequence | TR/TE (ms) | ST (mm) | Matrix (mm2) | FOV (mm2) | FA (°) |
| (oblique) axial T2WI | 1700-5050/110-120 | 4-6 | 320-384×256 | 200-360×200-360 | 90 |
| (oblique) axial DWI | 3000-7000/50-80 | 4-6 | 128-160×192 | 340-380×340–380 | 90 |
| Training | Validation | |||||
| CRLM(+) (n =39) |
CRLM(-) (n =44) |
P value | CRLM(+) (n =18) |
CRLM(-) (n =19) |
P value | |
| Age/yr (Mean±SD) | 64.97±11.76 | 59.50±12.89 | 0.052 | 60.06±13.95 | 57.58±12.73 | 0.565 |
| Sex (%) | 0.228 | 0.419 | ||||
| Male | 28(71.8) | 26(59.1) | 16(88.9) | 15(78.9) | ||
| Female | 11(28.2) | 18(40.9) | 2(11.1) | 4(21.1) | ||
| MRT stage (%) | 0.988 | 0.810 | ||||
| T1 | 0(0) | 1(2.3) | 0(0) | 0(0) | ||
| T2 | 0(0) | 9(20.5) | 1(5.6) | 2(10.5) | ||
| T3 | 26(66.7) | 24(54.5) | 11(61.1) | 10(52.6) | ||
| T4 | 13(33.3) | 10(22.7) | 6(33.3) | 7(36.8) | ||
| MRN stage (%) | 0.887 | 0.890 | ||||
| N0 | 8(20.5) | 11(25.0) | 5(27.8) | 6(31.6) | ||
| N1 | 11(28.2) | 12(27.3) | 5(27.8) | 4(21.0) | ||
| N2 | 20(51.3) | 21(47.7) | 8(44.4) | 9(47.4) | ||
| CEA level (%) | <0.001 | 0.001 | ||||
| Normal | 7(17.9) | 31(70.5) | 2(11.1) | 14(73.7) | ||
| Elevated | 32(82.1) | 13(29.5) | 16(88.9) | 5(26.3) | ||
| CA19-9 level (%) | <0.001 | 0.999 | ||||
| Normal | 19(48.7) | 42(95.5) | 9(50.0) | 19(100) | ||
| Elevated | 20(51.3) | 2(4.5) | 9(50.0) | 0(0) | ||
| Prediction model | Feature category | CRLM (+) vs. CRLM (-) | |
| T2WI model | Texture features | GLCM | X0.4 Inverse Variance |
| Shape features | Shape | Compactness 1 | |
| Max3D Diameter | |||
| DWI model | Texture features | GLCM | X45.7 Information MeasureCorr1 |
| X135.7 Information MeasureCorr1 | |||
| Shape features | Shape | Max3D Diameter |
| Cohort | Prediction model | AUC | Sen | Spe | PPV | NPV | ACC | F1-score |
| Training cohort | ||||||||
| T2WI model | 0.811 | 0.795 | 0.718 | 0.761 | 0.757 | 0.759 | 0.778 | |
| DWI model | 0.803 | 0.750 | 0.769 | 0.786 | 0.732 | 0.759 | 0.767 | |
| M model | 0.824 | 0.773 | 0.769 | 0.791 | 0.750 | 0.771 | 0.782 | |
| U model | 0.899 | 0.795 | 0.744 | 0.778 | 0.763 | 0.772 | 0.787 | |
| Validation cohort | ||||||||
| T2WI model | 0.795 | 0.842 | 0.778 | 0.800 | 0.824 | 0.811 | 0.821 | |
| DWI model | 0.798 | 0.737 | 0.778 | 0.778 | 0.737 | 0.757 | 0.757 | |
| M model | 0.813 | 0.789 | 0.778 | 0.789 | 0.778 | 0.784 | 0.789 | |
| U model | 0.889 | 0.895 | 0.833 | 0.850 | 0.882 | 0.865 | 0.872 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





