1. Introduction
The Myzostomida, as originally classified by Graff in 1877, represent a distinctive group of diminutive and highly specialized symbiotic organisms primarily associated with crinoids. They exhibit a unique body plan, markedly diverging from other Annelida taxa, and their specialized adaptations have been highlighted by Eeckhaut and Lanterbecq (2005). These organisms are globally distributed, predominantly found in subtidal tropical zones, which are known to host diverse crinoid populations, as observed by Fishelson (1974). The morphological diversity within Myzostomida is intricately linked to their symbiotic relationships with crinoid hosts and displays convergence across phylogenetic lineages, as noted by Summers and Rouse (2014). Despite their apparent diversity, the taxonomic representation of Myzostomida remains notably underrepresented in scientific literature, with a substantial number of undescribed species, a situation emphasized by Summers, Al-Hakim, and Rouse (2014) through their ongoing species identification efforts that continually unveil new taxa. Presently, approximately 180 species of Myzostomida, classified into seven families, are known, with 159 species attributed to the genus Myzostoma (Kolbasova, Mekhova, 2019; Read, Fauchald, 2023). The majority of myzostomids are ectocommensal on crinoids, although some inhabit the host’s body, forming cysts, galls, or residing within the digestive system (Kolbasova, Mekhova, 2021). Myzostomids are also found associated with Asteroidea, Ophiuroidea (Lanterbeqc et al., 2006), and Antipataria (Terrana, Eeckhaut, 2017), and they are reported from diverse oceanic regions, from subtidal zones to depths exceeding 3000 meters.
Biogeographical boundaries have been a topic of discussion, with some doubts raised (Briggs, 1995; Ekman, 1953). However, recent empirical evidence (Kreft et al., 2010; Procheş et al., 2012; Holt et al., 2013; Rueda et al., 2013; Ficetola et al., 2017) supports the existence of clearly definable biogeographical boundaries in the ocean. These boundaries are evident even among sets of up to 65 thousand species (Costello et al., 2017) and suggest barriers to gene flow, particularly for benthic organisms lacking long-lived planktonic larvae. Given the relatively rapid development of the Myzostomida trochophore (Eeckhaut et al., 2003), it is reasonable to expect high realm specificity and speciation within myzostomid fauna.
In the coastal waters of China, only a fraction of the global Myzostomida species (7 out of 180 identified species) has been documented. These records are primarily confined to the coastal area near Hong Kong and Shenzhen City (Sun et al. 2008). Noteworthy species, such as Myzostoma attenuatum (Grygier, 1989), M. antennatum (Graff, 1884), M. bocki (Jägersten, 1937), M. dodecaphalcis (Grygier, 1992), M. nasonovi (Fedotov, 1938), M. cf. pallidum (Graff, 1877), and M. lobatum (Graff, 1877), have been previously recorded from this area. However, these areas represent only a small fraction of the rich marine biodiversity along China’s coastal waters, where at least 44 species of crinoids have been reported (Yulin & Ning, 2011), suggesting the presence of a broader spectrum of Myzostomida species yet to be recorded.
Hainan Island experiences significant temporal and spatial fluctuations in its shoreline and shallow-water community structures, leading to notable shifts in species composition and densities. It is recognized as a critical biodiversity hotspot within the South China Sea (Xiao et al., 2001). Positioned at the ecological confluence of the China-Japan subtropical subregion and the Indo-Malaysian tropical subregion within the Indo-West Pacific Warm-water Biotic Region, Hainan Island marks the southernmost point of the cold current’s surface flow along the Guangdong province’s shoreline (Liu, 2013). Despite its ecological significance, there is a conspicuous lack of documented Myzostomida diversity records for this region.
The primary objective of this investigation was to conduct a comprehensive integrative survey of the symbiotic fauna of macroinvertebrates residing within the shallow-water communities along the coast of the South China Sea. This study focused on the integrative taxonomy of poorly studied and enigmatic crinoid-associated Myzostomida worms that are common in tropical coral reef ecosystems.
2. Materials and Methods
2.1. Sample Collection and Processing
In the scope of our research, we directed our attention towards myzostomids associated with two specific crinoid species:
Comanthus parvicirrus (Müller, 1841) and
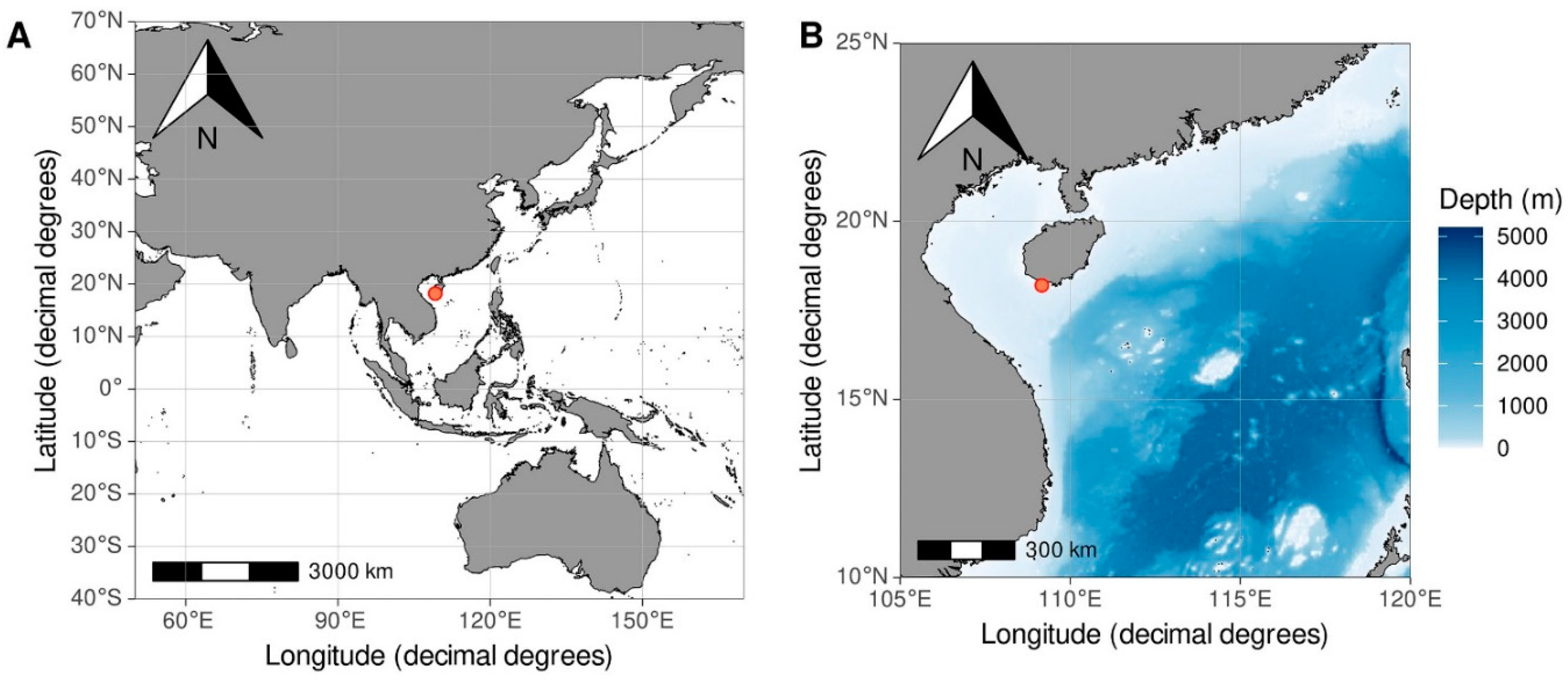
Comaster nobilis (Carpenter, 1884). These crinoids fall under the family Comatulidae, which is categorized within the order Comatulida. Our investigation focused exclusively on free-living external symbionts. The specimens that were found on these host crinoids were collected by V.N. Ivanenko and T.A. Britayev employed local boat and conducted SCUBA diving in the coastal waters near Sanya City, located on Hainan Island. Precise coordinates for the collection site were N 18°12.279’ E 109°30.197’ (
Figure 1). The crinoids were collected at depths ranging from 5 to 8 meters and isolated in resealable plastic bags underwater. Upon retrieval to the surface, a 10% ethanol solution was added to the bags. After 30 minutes of shaking the bags to ensure proper mixing, the liquid was passed through a sieve with a 100-micrometer mesh diameter. Following this procedure, the samples were preserved in 95% ethanol. The preserved specimens were then stored at a controlled temperature of approximately -20°C, ensuring their preservation for subsequent analyses. The maps utilized in this study were generated employing the ‘ggplot2’ package within R, version 2.1.1, with the assistance of the Plot Data on Oceanographic Maps tool developed by Vihtakari in 2023.
Our study encompassed a total of six distinct samples, which collectively included the two crinoid species mentioned earlier. This comprehensive examination involved the scrutiny of 80 individual myzostomid specimens. Detailed information pertaining to these specimens can be found in
Table 1. The selected preservation method was instrumental in maintaining the specimens’ structural integrity, facilitating their utilization in future morphological and molecular investigations.
All examined materials are currently housed and preserved within the collections of the Zoological Museum of Moscow State University, Russia (ZMMU), and the Faculty of Biology at MSU-BIT Shenzhen University, China. Museum specimen voucher numbers and NCBI GenBank Accession Numbers for our samples have been cataloged in
Table 2 for reference.
2.2. Morphological Analysis
For the purpose of morphological observation, a dissecting stereomicroscope Cnoptec SZ650 (China). Each collected specimen underwent a comprehensive external morphological examination, encompassing a thorough assessment of various attributes such as coloration, body morphology, surface texture, and appendage characteristics. This detailed examination was carried out under the scrutiny of a compound microscope Olympus BX43FC (Japan).
Species categorization and identification were executed with a reliance on the most up-to-date published descriptions of myzostomid species. From each of the identified species, a judicious selection process was undertaken to choose specimens with the most pristine and intact external morphological features. These selected specimens were subjected to a detailed photographic documentation process utilizing a high-resolution imaging stereomicroscope. Additionally, images were captured using a scanning electron microscope (SEM) KYKY EM6200 (China).
The SEM specimen preparation procedure involved a meticulous protocol, including the drying of specimens within a freeze dryer (Labconco FreeZone) maintained at a temperature of -50°C and a pressure of 0.03 mbar for a duration of four hours. Subsequently, a one-minute gold coating was applied to the specimens using a vacuum sputter.
2.3. Molecular analysis
For brief molecular species identity analysis, we used a shortened most informative fragment of mitochondrial cytochrome c oxidase subunit I (COI) gene (Rennstam et al. 2018). DNA was extracted on magnetic beads using OMEGA Bio-Tek E.Z.N.A. Mag-Bind Blood & Tissue DNA HDQ 96 Kit, according to manufacturer’s protocol. Target COI fragment was amplified by Sauron-S878 (Rennstam et al. 2018) and jgHCO2198 (Geller et al. 2013) with 5′v M13 (Messing, 1983) sequencing adapters (M13F_Sauron-S878 tgtaaaacgacggccagtGGDRCWGGWTGAACWGTWTAYCCNCC; M13R_jgHCO2198 caggaaacagctatgacTAIACYTCIGGRTGICCRAARAAYCA) primer pair. Reactions were performed using Clontech TaKaRa cellartis Premium Taq PCR kit. Amplicones were sequenced in both directions with M13F(-21) (TGTAAAACGACGGCCAGT) and M13R(-27) (CAGGAAACAGCTATGAC) sequencing primers (Messing, 1983) at Sangon Biotech (Shanhai, China).
Consensus sequences for each sample were assembled from raw reads in both directions and checked for ambiguities and low-quality base identifications in Geneious R10 (Kearse et al., 2012). Combined sequences were checked for putative contamination by using BLAST-n algorithm at GenBank nr/nt database (Altschul et al., 1990). Original data and publicly available sequences for genus Myzostoma were aligned with the MUSCLE (Edgar, 2004) algorithm implemented in MEGA7 (Kumar et al., 2016). Other Myzostomids were used as outgroups. Additionally, all sequences were translated into amino acids to verify reading frames and check for stop-codons and avoid pseudo-gene sequence contamination. A total of 152 sequences were used, including outgroups, resulting alignment length was 622bp.
The Bayesian phylogenetic reconstruction of phylogeny (BI) was performed in MrBayes 3.2 (Ronquist & Huelsenbeck, 2003). Markov chains were sampled at intervals of 500 generations. The analysis was initiated with a random starting tree and ran for 5x107 generations. Maximum likelihood phylogeny inference (ML) was performed in the HPC-PTHREADS version of RaxML 8.2.12 (Stamatakis, 2014) with 400 pseudoreplicates of fast bootstrap under the GTRCAT model of nucleotide evolution. Number of sufficient bootstrap pseudoreplicates was determined by AutoMRE approach (Pattengale et al. 2009). Final phylogenetic tree images were rendered in FigTree 1.4.0 and further modified in Inkscape 1.1. Attribution to marine biogeographic realms followed borders from Costello et al. (2017) and used ranges described in Summers et al. 2014 and data from original species descriptions.
3. Results
The Myzostomida community of three specimens of Comanthus parvicirrus consisted of only one species – Myzostoma polycyclus Atkins, 1927. Among the myzostomid specimens collected from three specimens of Comaster nobilis four species have been found: M. sp. 1; M. sp. 2; Myzostoma sp. 3. and Myzostoma sp. nov.
3.1. Taxonomy
Myzostoma scopus sp. nov.
Holotype: (ZMMU Pl-4886) 1 specimen in 96% ethanol. Sampling site is in the vicinity of Sanya city, Hainan Island (N 18°12,279′ E 109°30′197), depth 5-8 m. Collected using SCUBA on 25 November 2009 by T.A. Britayev and V.N. Ivanenko.
Host: Comaster nobilis (Carpenter, 1884).
Etymology: The name “scopus” is derived from the Latin word “scopus,” which means “target,” owing to its color pattern that resembles an archery target
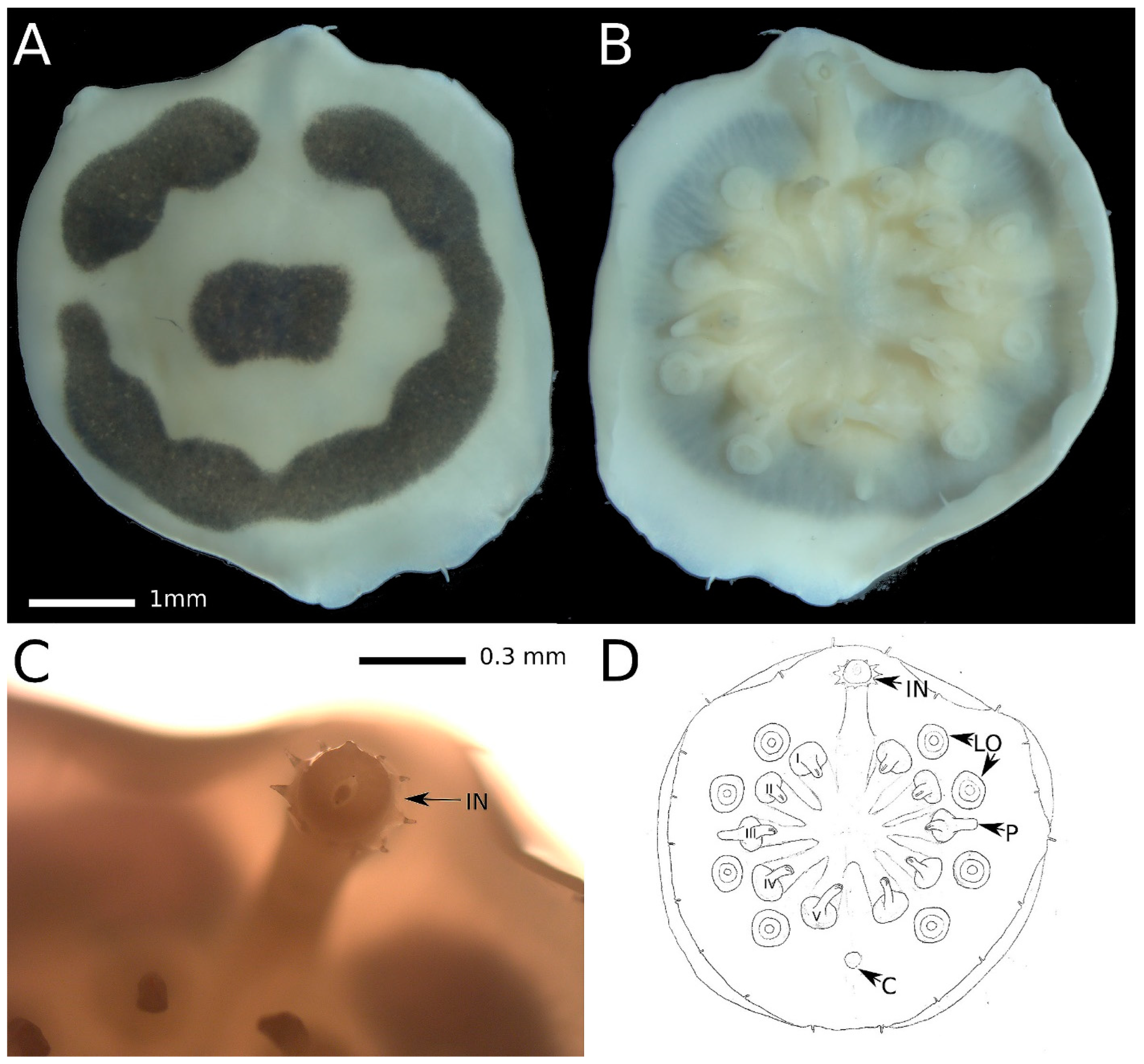
Description and diagnosis: The body is circular, approximately 5 mm in diameter. The dorsal surface exhibits a smooth texture and is of ivory color, featuring a black laterally elongated rectangular spot at its center, surrounded by an interrupted ring of the same black color as the central spot (
Figure 2A). The ventral surface is uniformly ivory in color (
Figure 2B,D). The centers of parapodial bases are positioned approximately 40% along the path from the center of the ventral side to the body margin. The parapodia are around 0.6-0.7 mm long and consist of two segments of roughly equal length: a stout conical base with a diameter of about 0.35 mm in its proximal part and a less massive cylindrical distal part with a rounded tip. Each parapodium is adorned with a hook. Parapodia are evenly distributed around the circular body. In our specimen, the parapodia bend toward the center, revealing the absence of median cirri. The outer side of the third parapodia carries a penis, measuring approximately 0.4 mm in length and exhibiting a similar shape to the distal part of the parapodium. Prominently visible large (approximately 0.3 mm) lateral organs, situated at about 30% along the path from the outer side of the parapodia to the body margin, alternate between the parapodia. The everted introvert (
Figure 2C) is 3.5 times as long as it is wide and bears ten acute papillae. The cloaca is positioned at the same distance from the body center and margin as the lateral organs. The body margin is smooth, thin, and semitransparent, hosting 20 very short (0.15-0.18 mm) tiny cirri of the same digitiform shape. The extreme edge of the body margin slightly curves on the ventral side, possibly due to ethanol fixation. The cirri are evenly distributed around the body margin.
Remarks: Myzostoma scopus sp. nov. displays several distinguishing characteristics, although there are a few myzostomid species that bear some resemblance to it, particularly in terms of having a circular body adorned with 20 small finger-like cirri around its margin. A detailed morphological comparison is provided below.
Myzostoma scopus sp. nov. is closely related to M. susanae, originally described from Comaster schlegelii found at a depth of 6 meters at Lizard Island Reef, Australia. However, unlike M. susanae, it exhibits a distinctive pigmentation pattern on its dorsal side and possesses more prominent cirri.
Another species that shares similarities is M. coriacium Graff, 1884, collected from Colobometra perspinosa at depths of 7.5-10 meters in Port Denison, Australia. According to Graff’s description, M. coriacium is not transparent and appears darkish brown in alcohol. It also bears lateral organs located halfway between the center and margin of the body. Graff’s illustration of M. coriacium’s cross-section depicts a body that is deeply ventrally concave and dorsally convex. In contrast, M. scopus sp. nov. can be distinguished by its color, a semitransparent body margin that is slightly bent ventrally, and lateral organs positioned closer to the body margin.
M. scopus sp. nov. shares a color pattern resembling that of M. horologium Graff, 1884, but with a different shape. It can be differentiated from M. horologium by its notably shorter parapodia, which do not extend beyond the body margin.
In comparison to M. brevipes, another species with a circular body, M. scopus sp. nov. can be distinguished by its smooth dorsal side, the absence of a dorsomedial ridge, and the absence of radial folds on the dorsal side.
When compared to M. seymourcollegiorum Rouse & Grygier, 2005, found on Cenolia trichoptera (Müller) and Cenolia glebosus Rowe et al. in southern Australia, M. scopus sp. nov. can be distinguished by the absence of parapodial cirri.
Differentiating from both M. pallidum Graff, 1877 (recorded from Comatula solaris and Comanthus parvicirrus) and M. triste Graff, 1877 (recorded from Comanthus parvicirrus), both from Bohol strait, M. scopus sp. nov. is characterized by its notably shorter parapodia, finger-like cirri with thin bases, and distinct coloration.
Myzostoma polycyclus Atkins, 1927
Studied material: 33 specimens from C. parvicirrus.
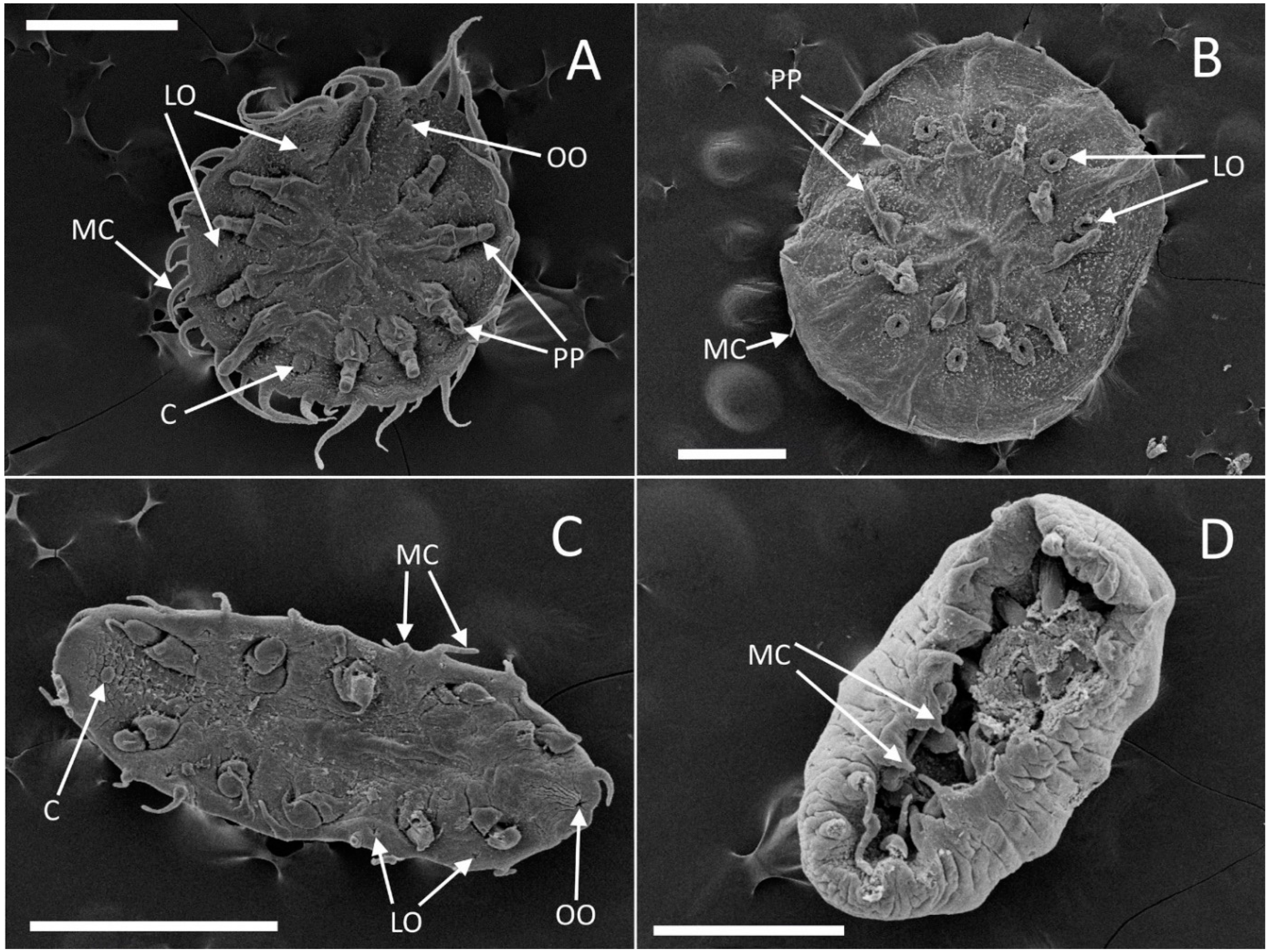
External morphology (
Figure 3A): Examined material conforms to original (Atkins, 1927) and consequent descriptions (Eeckhaut et al., 1998; Lanterbecq, Eeckhaut, 2003). Specimens from various samples exhibited varying colors, which correlated with the coloration of their respective hosts (beige or dark blue). However, no morphological differences were observed among them, consistent with previous findings.
Type locality: Pacific: Torres strait; Host: Comanthus parvicirrus; depth: missing in the original description (Atkins, 1927).
Other records, hosts, and depth: Comanthus suavius Rowe, Hoggett, Birtles, and Vail, 1986, Capillaster multiradiatus (Linnaeus, 1758), Clarkcomanthus littoralis (Carpenter, 1888). North Sulawesi, Papua New Guinea, Fiji, Japan. Depth 2-9 m.
Remarks: The specimens have undergone pigmentation from the host while being stored in ethanol.
Myzostoma sp. 1
Studied material: 8 specimens from C. nobilis.
External morphology: The body is circular (
Figure 3B) and flat, measuring 1-1.2 mm in diameter. It exhibits a greyish-beige coloration with a darker region in the center of the dorsal side. The body is rounded evenly and is adorned with twenty prominent finger-like cirri, each measuring 0.08-0.1 mm in length. The dorsal surface displays a median ridge and radial folds, while the ventral surface features scattered ciliary bundles. Parapodia are positioned at approximately 40% of the distance from the center of the body to the body margin, measuring 0.2 mm in length. Lateral organs are
located roughly a quarter of the distance from the parapodial bases to the body margin and have a diameter of about 0.1 mm.
Remarks: The specimens bear a resemblance to Myzostoma brevipes Graff, 1884, which was originally described from Crinometra brevipinna (Pourtalès, 1868) (as Antedon pourtalèsi Carpenter, 1888) in the Caribbean Sea at a depth of 300 meters. While no significant contradictions with Graff’s description have been identified, the studied material does not align with the type locality, depth, and host associated with the species mentioned above. The synonymy of this species warrants further clarification. Additionally, our specimens share some similarities with M. pallidum Graff, 1877, described from Comatula solaris Lamarck, 1816, and Comanthus parvicirrus from shallow waters in the Philippines. However, they differ from the aforementioned species by featuring thin finger-like marginal cirri, in contrast to the triangular cirri found in Myzostoma pallidum.
Myzostoma sp. 2
Studied material: 28 specimens from three specimens of C. nobilis.
External morphology: The body of examined 20 specimens is elongated (
Figure 3C), measuring between 0.8-1.2 mm, and oval, with a length approximately twice its width and a very slight convexity. The dorsal surface appears smooth and is colored yellowish grey, occasionally exhibiting a lighter dorsomedian stripe in certain specimens. On the ventral side, parapodia emerge from cavities and can extend to the body margin, sometimes even surpassing the margin, making them visible from a dorsal view. The length of the parapodia is approximately one-third of the maximum body width. There are twenty finger-like cirri, each measuring about one-fifth the length of the maximum body width. Four pairs of lateral organs are situated on low mounds, positioned midway between the base of the parapodia and the body margin. These lateral organs and parapodia alternate in arrangement. The everted introvert is roughly half the body width in length and is adorned with four distal papillae. The oral opening is ventral and oriented toward the anterior body margin.
Remarks: The observed specimens share some similarities with Myzostoma longitergum Eeckhaut, Grygier, Deheyn, 1998. However, they lack cirri 2-9 grouped in doublets flanking lateral organs, as mentioned in the original description. Further studies are needed for a conclusive identification of these specimens.
Myzostoma sp. 3
Studied material: seven specimens from C. nobilis.
External morphology: (
Figure 3D). The body is elongated, measuring approximately 1 mm in length, and exhibits a pronounced lateral compression, with a height equal to or sometimes even greater than its width, approximately 0.5 mm. The body surface appears opaque and smooth, displaying a beige coloration. There are twenty short cirri, each measuring between 0.08 to 0.1 mm in length, with a broad base that tapers towards the distal end. Additionally, the body margin is folded over the ventral side.
Remarks: The morphology of the observed specimens closely resembles that of Myzostoma compressum Graff, 1884, originally collected from Bathycrinus aldrichianus Thomson, 1876 in the subantarctic Indian Ocean (Prince Edward Island) at a depth of 2.5 km. However, the discovery of this species on a different host in the shallow waters of the South China Sea raises questions about this similarity. It also bears some resemblance to M. nasonovi Fedotov, 1938, which was reported from Hong Kong, but the latter species lacks any cirri, whereas our specimens clearly possess them.
3.2. Molecular analysis
We obtained sequences of COI gene fragment for four myzostomid species (including new species described in this manuscript). GenBank accession numbers for all new sequences are listed in
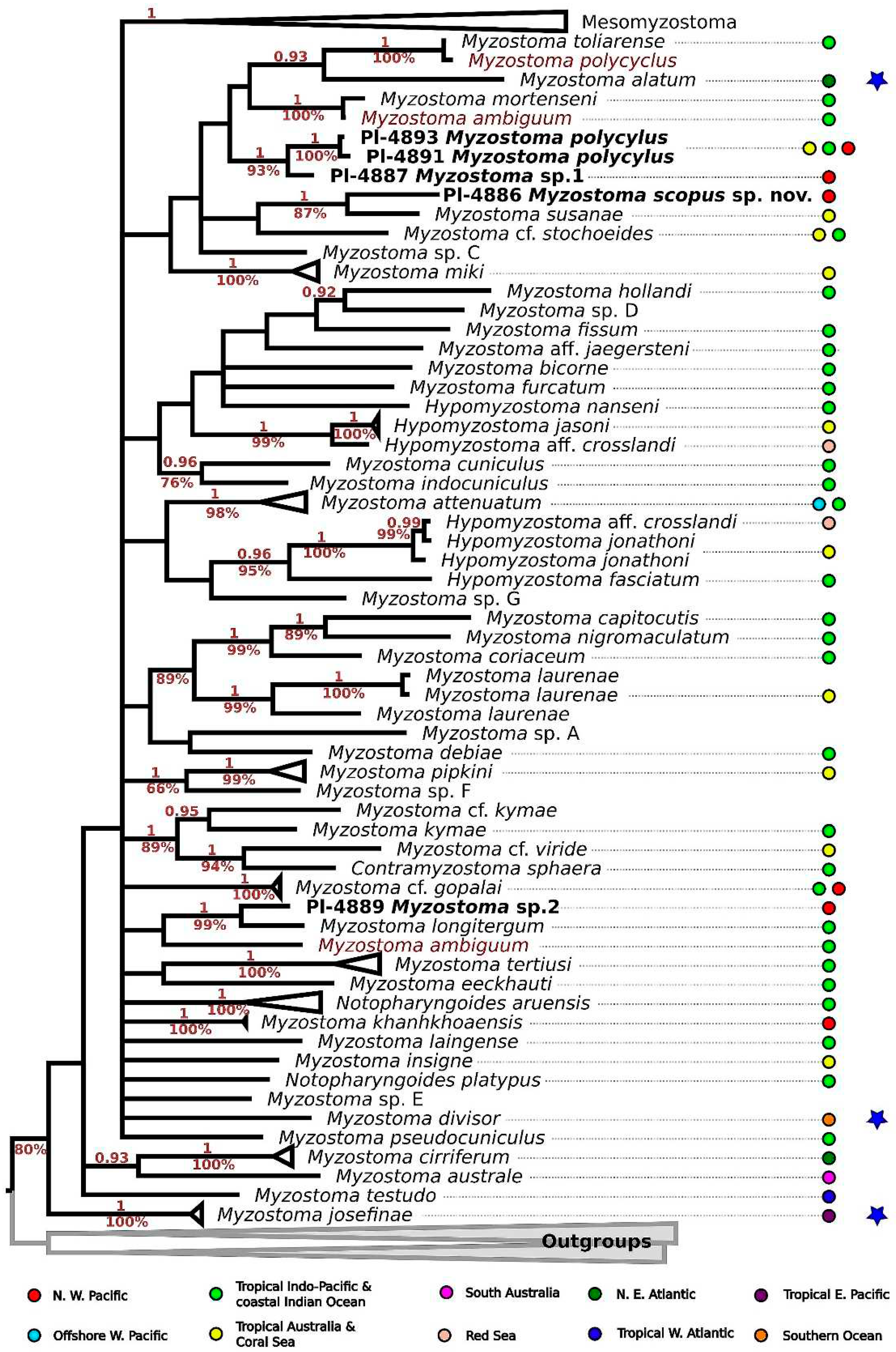
Table 2. Phylogenetic reconstruction based on all publicly available and original data for myzostomid COI (
Figure 4) is not completely resolved in both reconstruction methods. Uncollapsed phylogenies with accession numbers are available as supplementary material (
Figure S1,
Figure S2). The genus
Myzostoma is not reconstructed as a monophyletic clade and forms a clade with samples of
Hypomysostoma and
Mesomyzostoma with moderate support in ML reconstruction.
Myzostoma sp. 1 is reconstructed as sister to clade formed by our
M. polycyclus samples (1/100%) with high support in both approaches (1/93%).
M. polycyclus (DQ238202) is recovered in another clade (1/100%), together with
M. toliarense (DQ238201) and their COI sequences are almost identical.
M. scopus sp. nov. is recovered as sister to
M. susanae, with high BI support (1/87%). Our specimen of
M. sp. 2 is recovered in a well-supported clade (1/99%) with publicly available sequence of
M. longitergum (KM014193), but sequence overlapping fragments only have 94.7% identical bases.
4. Discussion
This study represents a significant contribution to the understanding of myzostomid fauna within Chinese coastal ecosystems. The discovery of five myzostomid species in this area, including the description of a novel species, extends their known geographical distribution and highlights the potential for further research. While our data broadens knowledge on symbiotic associations with C. parvicirrus, it is essential to acknowledge the limited sample size.
Our observations have identified Myzostoma polycyclus as the symbiont of C. parvicirrus, with notable prevalence. Importantly, this marks the first documentation of M. polycyclus in coastal waters of China, providing valuable insights into its distribution. In contrast, C. nobilis is known to host numerous myzostomid species, emphasizing the need to address gaps in faunal knowledge concerning echinoderm symbionts. Future research should focus on determining the phylogenetic placement of the newly described M. scopus sp. nov. and expanding the survey of myzostomid biodiversity in proximate regions. Comprehensive follow-up studies are crucial for a deeper understanding of myzostomid of other microscopic invertebrates diversity in Hainan and the broader South China Sea region.
Our study contributes to the available COI DNA barcodes for myzostomids. The obtained data revealed significant distinctions between the new species
M. scopus sp. nov. and its closest species
M. susanae (
Figure 4 and
Figure S1). The genetic analysis revealed intriguing patterns, such as the high genetic divergence within specimens morphologically like
M. longitergum, suggesting a potential species complex. Additionally, the COI sequence of
M. polycyclus, previously misattributed in a published study, has been correctly assigned for the first time in our research. This highlights the importance of accurate genetic data for species identification.
Despite these advancements, the overall genetic diversity of myzostomids remains underrepresented in public databases, and more extensive research is needed. The existing data are skewed towards Pacific shallow-water species, with limited coverage in other marine realms. This emphasizes the necessity of dedicated studies to obtain sequences from a broader taxonomic and geographical range. Considering the host specificity of myzostomids, it is plausible that a substantial number of undescribed species exist, particularly on crinoids, which host hundreds of species.
5. Conclusions
The findings of this study bring to the fore the concealed biodiversity residing within marine macroinvertebrates, emphasizing the pivotal importance of inventorying these and numerous other symbiotic relationships, especially when confronted with environmental challenges (Zeppilli et al. 2015; Korzhavina et al. 2023). It is noteworthy that a multitude of these microscopic invertebrate symbionts may assume parasitic or vector roles, particularly within ecosystem-influential taxonomic groups such as corals, echinoderms, and marine sponges.
This investigation underscores the pressing necessity for comprehensive integrative research endeavors that transcend mere species enumeration. The complexity of marine ecosystems demands multidisciplinary research to disentangle intricate relationships, offering essential insights for informed decision-making in an ever-changing global landscape.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Displays the uncollapsed Bayesian Inference (BI) phylogeny. Figure S2: Features the uncollapsed Maximum Likelihood (ML) phylogeny. Figure S3: Illustrates the distribution of myzostomids, as depicted in Figure 4, across global marine biogeographical realms, following the framework established by Costello et al. (2017) with modifications.
Author Contributions
Conceptualization, V.I., D.S. and T.B.; methodology, A.I., D.S. and V.I.; software, D.S.; validation, A.I., D.S., Y.Z., A.K., T.B. and V.I.; formal analysis, A.I., D.S. and V.I.; investigation, A.I. and Y.Z; resources, A.K.; writing—original draft preparation, A.I., D.S., and V.I.; writing—review and editing, A.I., D.S., Y.Z., A.K., T.B. and V.I.; visualization, D.S. and A.I.; supervision, T.B. and V.I.; project administration, A.K. and V.I.; funding acquisition, A.K.. All authors have read and agreed to the published version of the manuscript.
Funding
The project was supported by Shenzhen University Stability Support Program (Grant 210101006).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genetic data obtained in this study is available by Accession Numbers provided in
Table 2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. Journal of molecular biology 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Atkins, D. Report on the Myzostomida collected by Mr. F.A. Potts in Torres Strait, with a description of a species obtained by Professor J. Stanley Gardiner from the Maldives. Proceedings of the Zoological Society of London 1927, 97, 339–357. [Google Scholar] [CrossRef]
- Briggs, J.C. Global biogeography; Elsevier, Amsterdam, The Netherlands, 1995. 452 pp. ISBN 0-444-88297-9.
- Britayev, T.A.; Mekhova, E.S. Assessment of hidden diversity of crinoids and their symbionts in the Bay of Nhatrang, Vietnam. Org Divers Evol 2011, 11, 275–285. [Google Scholar] [CrossRef]
- Costello, M.J.; Tsai, P.; Wong, P.S.; Kwok Lun Cheung, A.; Basher, Z.; Chaudhary, C. Marine biogeographic realms and species endemicity. Nat Commun 2017, 8, 1057. [Google Scholar] [CrossRef] [PubMed]
- Eckman, S. Zoogeography of the Sea; Sidgwick and Jackson, London, UK, 1953. 417 p.
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Eeckhaut, I.; Fievez, L.; Mueller, C.M.M. () Larval development of Myzostoma cirriferum (Myzostomida). Journal of Morphology 2003, 258, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Eeckhaut, I.; Grygier, M. J.; Deheyn, D. Myzostomes from Papua New Guinea, with related Indo-West Pacific distribution records and description of five new species. Bulletin of Marine Science 1998, 62, 841–886. [Google Scholar]
- Eeckhaut, I.; Lanterbecq, D. Myzostomida: a review of the phylogeny and ultrastructure. Hydrobiologia 2005, 535/536, 253–275. [Google Scholar] [CrossRef]
- Fedotov, D.M. Spezialisation und Degradation im Körperbau der Myzostomiden in Abhängigkeit von der Lebensweise. Acta Zoologica 1938, 19, 353–385. [Google Scholar] [CrossRef]
- Ficetola, G. F.; Mazel, F.; Thuiller, W. Global determinants of zoogeographical boundaries. Nat. Ecol. Evol. 2017, 1, 0089. [Google Scholar] [CrossRef]
- Geller, J.; Meyer, C.; Parker, M.; Hawk, H. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Molecular ecology resources 2013, 13, 851–861. [Google Scholar] [CrossRef]
- von Graff, L. Das Genus Myzostoma (F.S. Leuckhart). Wilhelm Engelmann, Leipzig, Germany, 1877; 82 p.
- von Graff, L. Report on the Myzostomida collected by H.M.S. Challenger during the Years 1873-1876. In Reports on the Scientific Results of the Voyage of H.M.S. Challenger during the years 1873–76, Zoology. 1884; Volume 10, 82 pp.
- Grygier, M.J. Three new species of Myzostoma (Myzostomida). Proceedings of the Biological Society of Washington 1989, 102, 793–804. [Google Scholar]
- Holt, B.G.; Lessard, J.-P.; Borregaard, M.K.; Fritz, S.A.; Araujo, M.B.; Dimitrov, D.; Fabre, P.-H.; Graham, C.H.; Graves, G.R.; Jønsson, K.A.; et al. An update of Wallace’s zoogeographic regions of the world. Science 2013, 339, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kolbasova, G.D.; Mekhova, E.S. Myzostoma khanhkhoaensis (Myzostomida), a new myzostomid species from the Nhatrang Bay, Vietnam. Zootaxa 2019, 4691, 235–249. [Google Scholar] [CrossRef]
- Korzhavina, O.A.; Grishina, D.Y.; Chen, X.; Fontaneto, D.; Ivanenko, V.N. Diving into Diversity: Copepod Crustaceans in Octocoral Associations. Diversity 2023, 15, 1140. [Google Scholar] [CrossRef]
- Kreft, H.; Jetz, W. A framework for delineating biogeographical regions based on species distributions. J. Biogeog 2010, 37, 2029–2053. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution 2018, 35, 1547. [Google Scholar] [CrossRef]
- Lanterbecq, D.; Eeckhaut, I. Myzostomida from Madagascar, with the description of two new species. Hydrobiologia 2003, 496, 115–123. [Google Scholar] [CrossRef]
- Lanterbecq, D.; Rouse, G.W.; Milinkovitch, M.C.; Eeckhaut, I. Molecular phylogenetic analyses indicate multiple independent emergences of parasitism in Myzostomida (Protostomia). Systematic Biology 2006, 55, 208–227. [Google Scholar] [CrossRef]
- Liao, Y.; Xiao, N. Species composition and faunal characteristics of echinoderms in China Seas. Biodiversity Science 2011, 19, 729–736. [Google Scholar]
- Liu, J. Y. Liu, J. Y. Status of Marine Biodiversity of the China Seas. PLoS ONE 2013, 8(1): e50719. [CrossRef]
- McClendon, J.F. The myzostomes of the ‘Albatross’ Expedition to Japan. Bulletin of the American Museum of Natural History 1906, 21, 119–130. [Google Scholar]
- Messing, J. New M13 vectors for cloning. In Methods in enzymology (Vol. 101, pp. 20–78). Academic Press, 1983.
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R..; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? In Research in Computational Molecular Biology: 13th Annual International Conference, RECOMB 2009, Tucson, AZ, USA, May 18-21, 2009. Springer, Berlin, Heidelberg, 2009. pp. 184–200.
- Procheş, Ş.; Ramdhani, S. The world’s zoogeographical regions confirmed by cross-taxon analyses. Bioscience 2012, 62, 260–270. [Google Scholar] [CrossRef]
- Read, G.; Fauchald, K. (Ed.) World Polychaeta Database. Myzostomida. 2023. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=233983 (Accessed on 19 November 2023).
- Rennstam, R.O.; Sint, D.; Horngacher, N.; Traugott, M. A broadly applicable COI primer pair and an efficient single-tube amplicon library preparation protocol for metabarcoding. Ecol Evol. 2018, 8, 12335–12350. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rouse, G.W.; Grygier, M.J. Myzostoma seymourcollegiorum n. sp. (Myzostomida) from southern Australia, with a description of its larval development. Zootaxa 2005, 1010, 53–64. [Google Scholar] [CrossRef]
- Rueda, M.; Rodriguez, M. A.; Hawkins, B. A. Identifying global zoogeographical regions: lessons from Wallace. J. Biogeog. 2013, 40, 2215–2225. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Summers, M.M.; Al-Hakim, I.I.; Rouse, G.W. Turbo-taxonomy: 21 new species of Myzostomida (Annelida). Zootaxa, 2014, 3873, 301–344. [Google Scholar] [CrossRef]
- Summers, M.M.; Rouse, G.W. Phylogeny of Myzostomida (Annelida) and their relationships with echinoderm hosts. BMC Evolutionary Biology 2014, 14, 170. [Google Scholar] [CrossRef]
- Sun, R.; Lei, Y.; Zhou, J. Phylum Annelida. In Checklist of Marine Biota of China Seas; Liu, R., Ed. Science Press, Academia Sinica, Beijing, China, 2008; pp. 405–452.
- Terrana, L.; Eeckhaut, I. Taxonomic description and 3D modelling of a new species of myzostomid (Annelida, Myzostomida) associated with black corals from Madagascar. Zootaxa 2017, 4244, 277–295. [Google Scholar] [CrossRef]
- Vihtakari, M. ggOceanMaps: Plot Data on Oceanographic Maps using ‘ggplot2’. R package version 2.1.1. 2023. Available online: https://CRAN.R-project.org/package=ggOceanMaps (Accessed on 19 November 2023).
- Xiao, Y.; Chen, S.B.; Zhang, L.; Yue, P.; Ouyang, Z.Y.; Liu, X.C. Designing nature reserve systems in Hainan Island based on ecosystem services. Acta Ecologica Sinica 2011, 31, 7357–7369. [Google Scholar]
- Yulin, L.; Ning, X. Species composition and faunal characteristics of echinoderms in China seas. Biodiversity Science 2011, 19, 729–736. [Google Scholar] [CrossRef]
- Zeppilli, D.; Sarrazin, J.; Leduc, D.; et al. Is the meiofauna a good indicator for climate change and anthropogenic impacts? Mar Biodiv 2015, 45, 505–535. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).