Submitted:
13 December 2023
Posted:
14 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bioinformatics, databases, and structural analysis
2.2. Constructs, peptides, and proteins
2.3. Pull-down and fluorescence dot blot analysis
2.4. Fluorescence polarization
2.5. Trandfection and immunoprecipitation analysis
3. Results
3.1. Sequence-structure-function classification of human SH3 domains
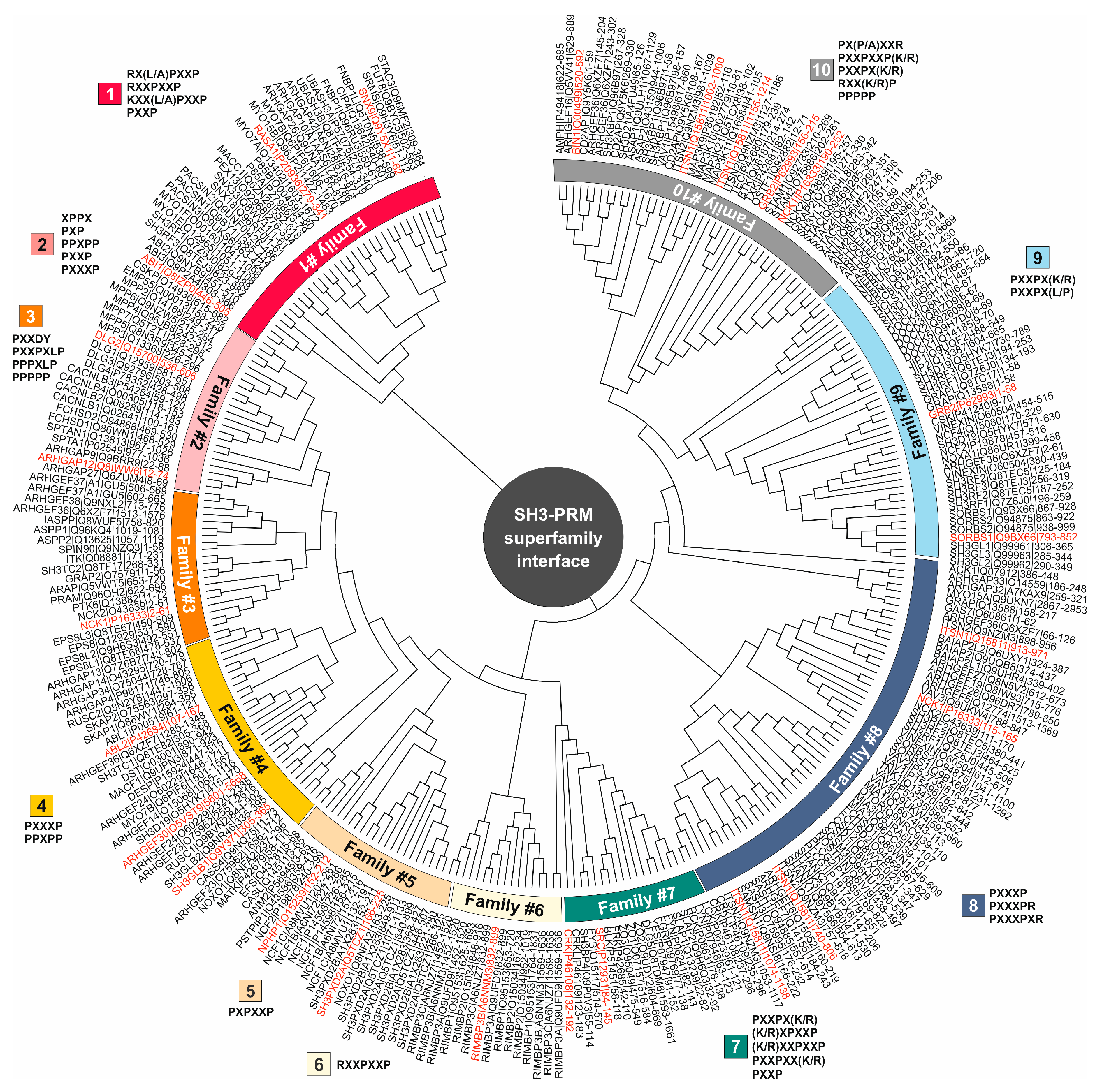
3.2. PRM selectivities of different SH3 families
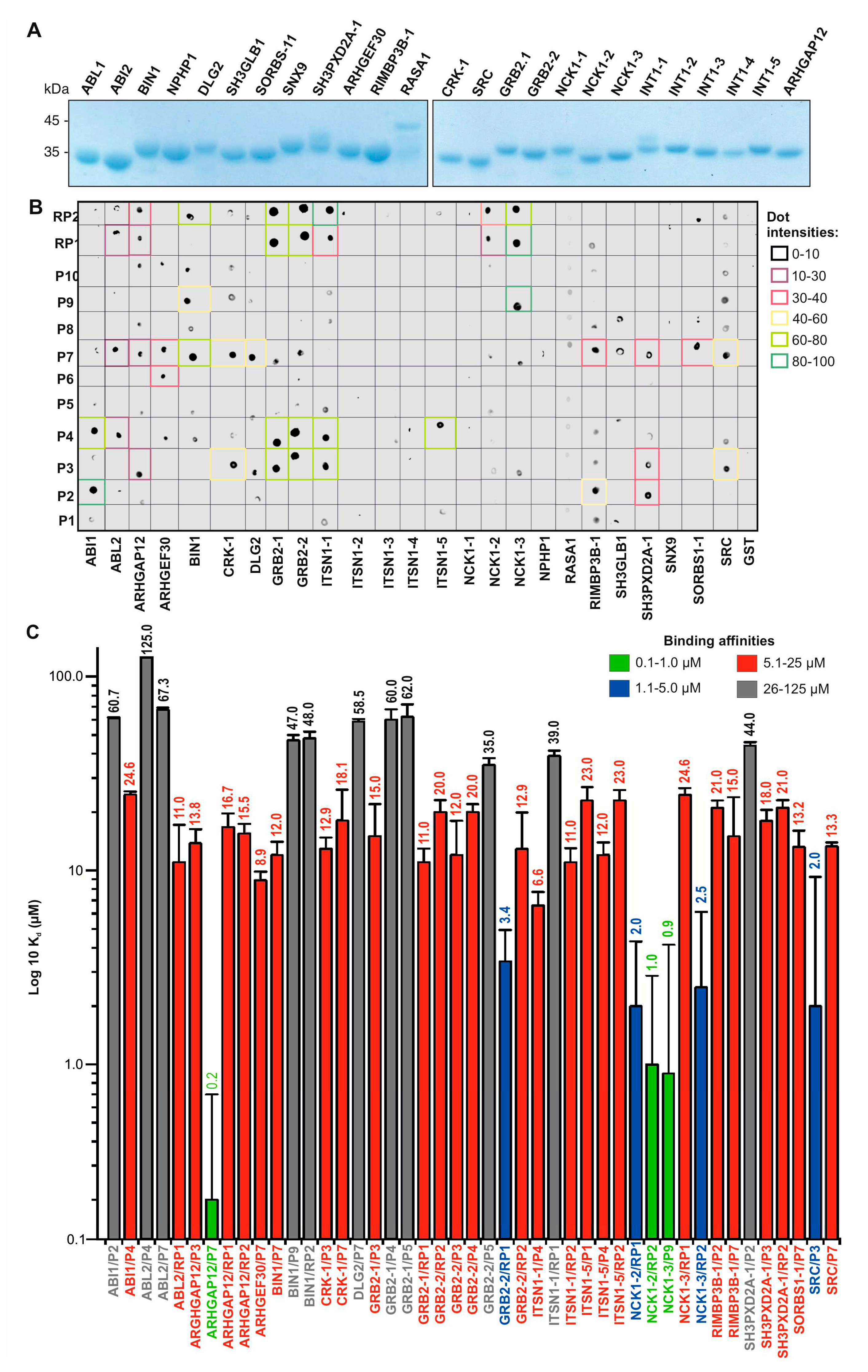
3.3. Affinity and selectivity of the SH3 family proteins for SOS1 PRP
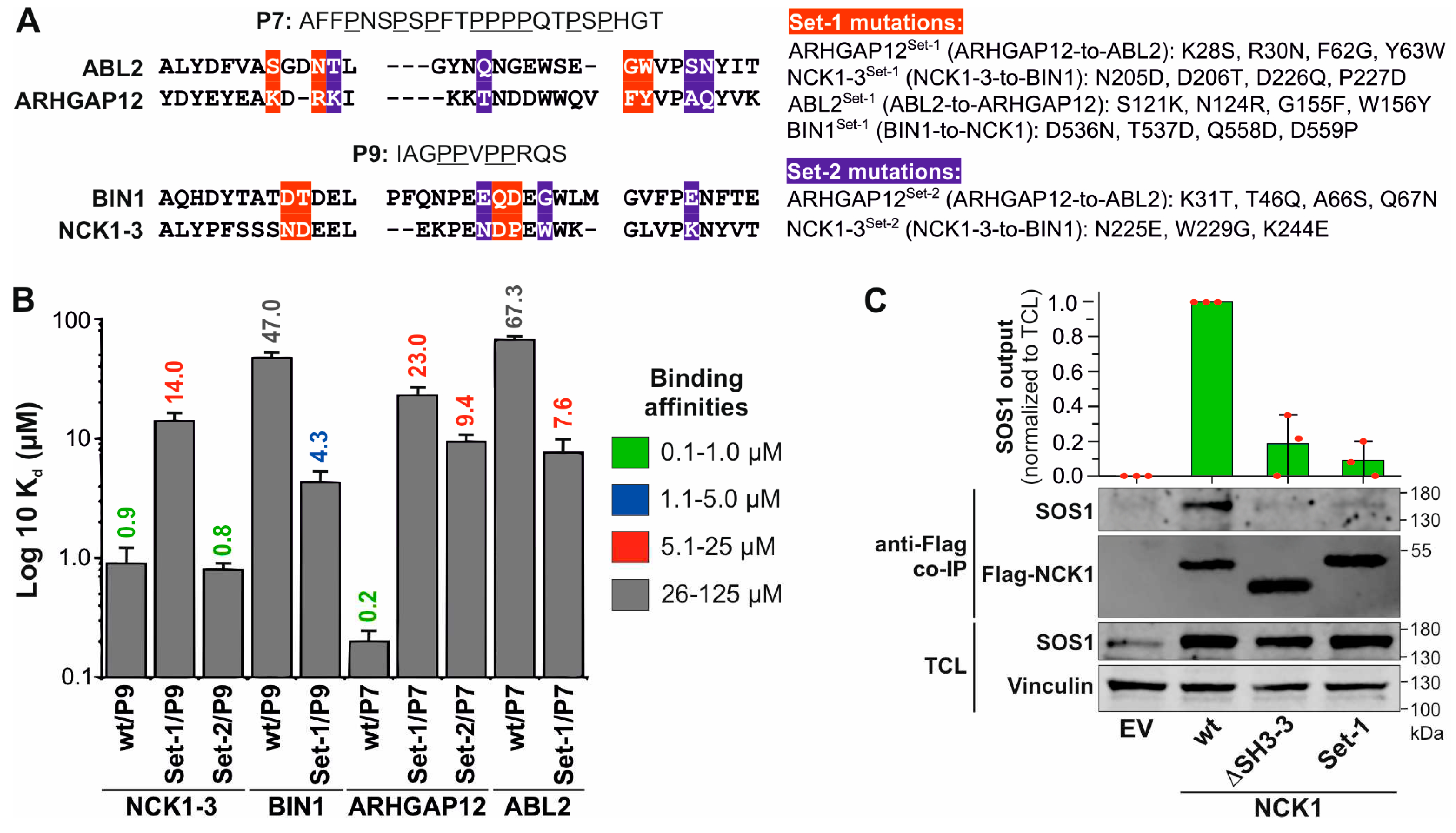
3.4. Non-conserved residues define the selectivity and affinity of SH3-PRM interactions
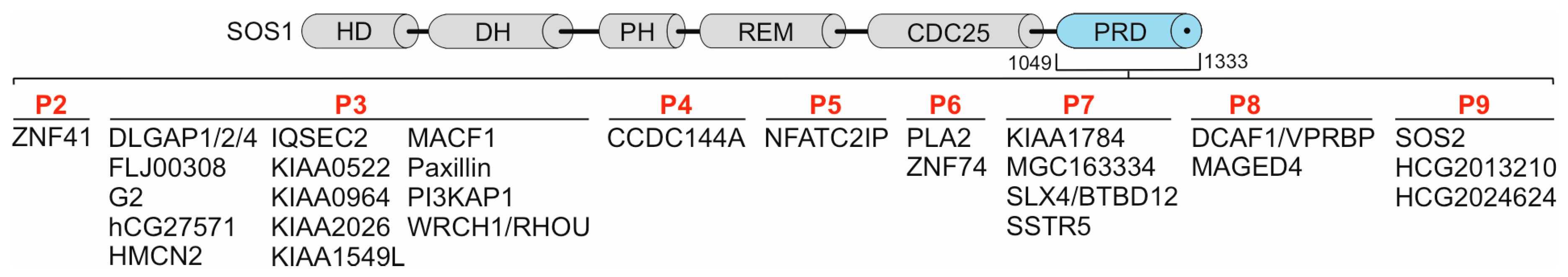
3.5. SH3-PRP relationships beyond SOS1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mabonga, L. and A.P. Kappo, Protein-protein interaction modulators: advances, successes and remaining challenges. Biophysical Reviews, 2019. 11(4): p. 559-581.
- Musacchio, A., et al., Crystal structure of a Src-homology 3 (SH3) domain. Nature, 1992. 359(6398): p. 851-855.
- Stahl, M.L., et al., Sequence similarity of phospholipase C with the non-catalytic region of src. Nature, 1988. 332(6161): p. 269-272.
- Mehrabipour, M., et al., A Systematic Compilation of Human SH3 Domains: A Versatile Superfamily in Cellular Signaling. Cells, 2023. 12(16): p. 2054.
- Teyra, J., et al., Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure, 2017. 25(10): p. 1598-1610. e3.
- Berndt, S., V.V. Gurevich, and T. Iverson, Crystal structure of the SH3 domain of human Lyn non-receptor tyrosine kinase. Plos one, 2019. 14(4): p. e0215140.
- Bircher, J.E. and A.J. Koleske, Trio family proteins as regulators of cell migration and morphogenesis in development and disease – mechanisms and cellular contexts. Journal of Cell Science, 2021. 134(3): p. jcs248393.
- Hildebrandt, F., et al., A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet, 1997. 17(2): p. 149-53.
- Saunier, S., et al., Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet, 2000. 66(3): p. 778-89.
- Wu, T., Z. Shi, and T. Baumgart, Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization. PloS one, 2014. 9(4): p. e93060.
- Hohendahl, A., A. Roux, and V. Galli, Structural insights into the centronuclear myopathy-associated functions of BIN1 and dynamin 2. J Struct Biol, 2016. 196(1): p. 37-47.
- Manso, J.A., et al., PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders. Cell Mol Life Sci, 2022. 79(2): p. 131.
- Starnes, T.W., et al., The F-BAR protein PSTPIP1 controls extracellular matrix degradation and filopodia formation in macrophages. Blood, 2014. 123(17): p. 2703-14.
- Qin, Y., et al., A recurrent SHANK1 mutation implicated in autism spectrum disorder causes autistic-like core behaviors in mice via downregulation of mGluR1-IP3R1-calcium signaling. Mol Psychiatry, 2022. 27(7): p. 2985-2998.
- Leblond, C.S., et al., Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS genetics, 2012. 8(2): p. e1002521.
- Ball, L.J., et al., Recognition of Proline-Rich Motifs by Protein–Protein-Interaction Domains. Angewandte Chemie International Edition, 2005. 44(19): p. 2852-2869.
- Li, S.S.-C., Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochemical Journal, 2005. 390(3): p. 641-653.
- Cesareni, G. and M. Gimona, Modular Protein Domains. 2005.
- Cámara-Artigas, A., et al., 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops. Journal of structural biology, 2014. 186(1): p. 195-203.
- Carducci, M., et al., The protein interaction network mediated by human SH3 domains. Biotechnology advances, 2012. 30(1): p. 4-15.
- Panni, S., L. Dente, and G. Cesareni, In vitro evolution of recognition specificity mediated by SH3 domains reveals target recognition rules. Journal of Biological Chemistry, 2002. 277(24): p. 21666-21674.
- Kazemein Jasemi, N.S., et al., The intramolecular allostery of GRB2 governing its interaction with SOS1 is modulated by phosphotyrosine ligands. Biochemical Journal, 2021. 478(14): p. 2793-2809.
- Mayer, B.J., SH3 domains: complexity in moderation. Journal of cell science, 2001. 114(7): p. 1253-1263.
- Kay, B.K., SH3 domains come of age. FEBS letters, 2012. 586(17): p. 2606-2608.
- Landgraf, C., et al., Protein interaction networks by proteome peptide scanning. PLoS Biol, 2004. 2(1): p. e14.
- Bae, D.J., et al., ArhGAP12 plays dual roles in Stabilin-2 mediated efferocytosis: Regulates Rac1 basal activity and spatiotemporally turns off the Rac1 to orchestrate phagosome maturation. Biochim Biophys Acta Mol Cell Res, 2019. 1866(10): p. 1595-1607.
- Hibino, H., et al., RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca(2+) channels. Neuron, 2002. 34(3): p. 411-23.
- Fan, P.-D. and S.P. Goff, Abl interactor 1 binds to sos and inhibits epidermal growth factor-and v-Abl-induced activation of extracellular signal-regulated kinases. Molecular and cellular biology, 2000. 20(20): p. 7591-7601.
- Tong, X.K., et al., The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. Embo j, 2000. 19(6): p. 1263-71.
- Qian, X., et al., The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. Embo j, 2000. 19(4): p. 642-54.
- Hu, Q., D. Milfay, and L.T. Williams, Binding of NCK to SOS and activation of ras-dependent gene expression. Mol Cell Biol, 1995. 15(3): p. 1169-74.
- Chardin, P., et al., Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science, 1993. 260(5112): p. 1338-1343.
- Birge, R.B., et al., Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Communication and Signaling, 2009. 7(1): p. 13.
- Rufer, A.C., et al., Isoform-selective interaction of the adaptor protein Tks5/FISH with Sos1 and dynamins. J Mol Biol, 2009. 390(5): p. 939-50.
- Sini, P., et al., Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nature Cell Biology, 2004. 6(3): p. 268-274.
- Wilkinson, B., J. Li, and M.P. Coba, Synaptic GAP and GEF complexes cluster proteins essential for GTP signaling. Scientific Reports, 2017. 7(1): p. 5272.
- Nakamoto, T., et al., CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol Cell Biol, 2000. 20(5): p. 1649-58.
- Salem, D. and R.J. Fecek, Role of microtubule actin crosslinking factor 1 (MACF1) in bipolar disorder pathophysiology and potential in lithium therapeutic mechanism. Translational Psychiatry, 2023. 13(1): p. 221.
- Rasmussen, A.H., H.B. Rasmussen, and A. Silahtaroglu, The DLGAP family: neuronal expression, function and role in brain disorders. Molecular Brain, 2017. 10(1): p. 43.
- Maruoka, M., et al., Identification of B cell adaptor for PI3-kinase (BCAP) as an Abl interactor 1-regulated substrate of Abl kinases. FEBS Lett, 2005. 579(14): p. 2986-90.
- Kim, E., et al., GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol, 1997. 136(3): p. 669-78.
- Bustelo, X.R., V. Sauzeau, and I.M. Berenjeno, GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays, 2007. 29(4): p. 356-70.
- Risse, S.L., et al., SH3-mediated targeting of Wrch1/RhoU by multiple adaptor proteins. Biol Chem, 2013. 394(3): p. 421-32.
- López-Colomé, A.M., et al., Paxillin: a crossroad in pathological cell migration. Journal of Hematology & Oncology, 2017. 10(1): p. 50.
- Webb, D.J., et al., Paxillin phosphorylation sites mapped by mass spectrometry. Journal of Cell Science, 2005. 118(21): p. 4925-4929.
- Kreienkamp, H.-J., et al., Physiology of somatostatin receptors: from genetics to molecular analysis. Somatostatin, 2004: p. 185-202.
- Verschueren, E., et al., Evolution of the SH3 domain specificity Landscape in Yeasts. PloS one, 2015. 10(6): p. e0129229.
- Cesareni, G., et al., Can we infer peptide recognition specificity mediated by SH3 domains? FEBS letters, 2002. 513(1): p. 38-44.
- Chook, Y.M., et al., The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. Journal of Biological Chemistry, 1996. 271(48): p. 30472-30478.
- Herrero-Garcia, E. and J.P. O'Bryan, Intersectin scaffold proteins and their role in cell signaling and endocytosis. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2017. 1864(1): p. 23-30.
- Alfaidi, M., M.L. Scott, and A.W. Orr, Sinner or Saint?: Nck adaptor proteins in vascular biology. Frontiers in Cell and Developmental Biology, 2021. 9: p. 688388.
- Yu, X., et al., Inhibitory short peptides targeting EPS8/ABI1/SOS1 tri-complex suppress invasion and metastasis of ovarian cancer cells. BMC Cancer, 2019. 19(1): p. 878.
- Hahn, S. and D. Kim, Transient protein-protein interaction of the SH3-peptide complex via closely located multiple binding sites. PLoS One, 2012. 7(3): p. e32804.
- Mayer, B.J. and K. Saksela, SH3 domains. Modular protein domains, 2005: p. 37-58.
- Dionne, U., et al., SRC homology 3 domains: Multifaceted binding modules. Trends in Biochemical Sciences, 2022.
- Lettau, M., et al., The adapter protein Nck: role of individual SH3 and SH2 binding modules for protein interactions in T lymphocytes. Protein Sci, 2010. 19(4): p. 658-69.
- Zarrinpar, A., S.-H. Park, and W.A. Lim, Optimization of specificity in a cellular protein interaction network by negative selection. Nature, 2003. 426(6967): p. 676-680.
- Ahmad, M., W. Gu, and V. Helms, Mechanism of fast peptide recognition by SH3 domains. Angewandte Chemie International Edition, 2008. 47(40): p. 7626-7630.
- Yu, H., et al., Structural basis for the binding of proline-rich peptides to SH3 domains. Cell, 1994. 76(5): p. 933-945.
- McDonald, C.B., et al., SH3 domains of Grb2 adaptor bind to PXψPXR motifs within the Sos1 nucleotide exchange factor in a discriminate manner. Biochemistry, 2009. 48(19): p. 4074-4085.
- Jaiswal, M., et al., Functional cross-talk between ras and rho pathways: a Ras-specific GTPase-activating protein (p120RasGAP) competitively inhibits the RhoGAP activity of deleted in liver cancer (DLC) tumor suppressor by masking the catalytic arginine finger. Journal of Biological Chemistry, 2014. 289(10): p. 6839-6849.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




