Submitted:
27 July 2023
Posted:
28 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Materials
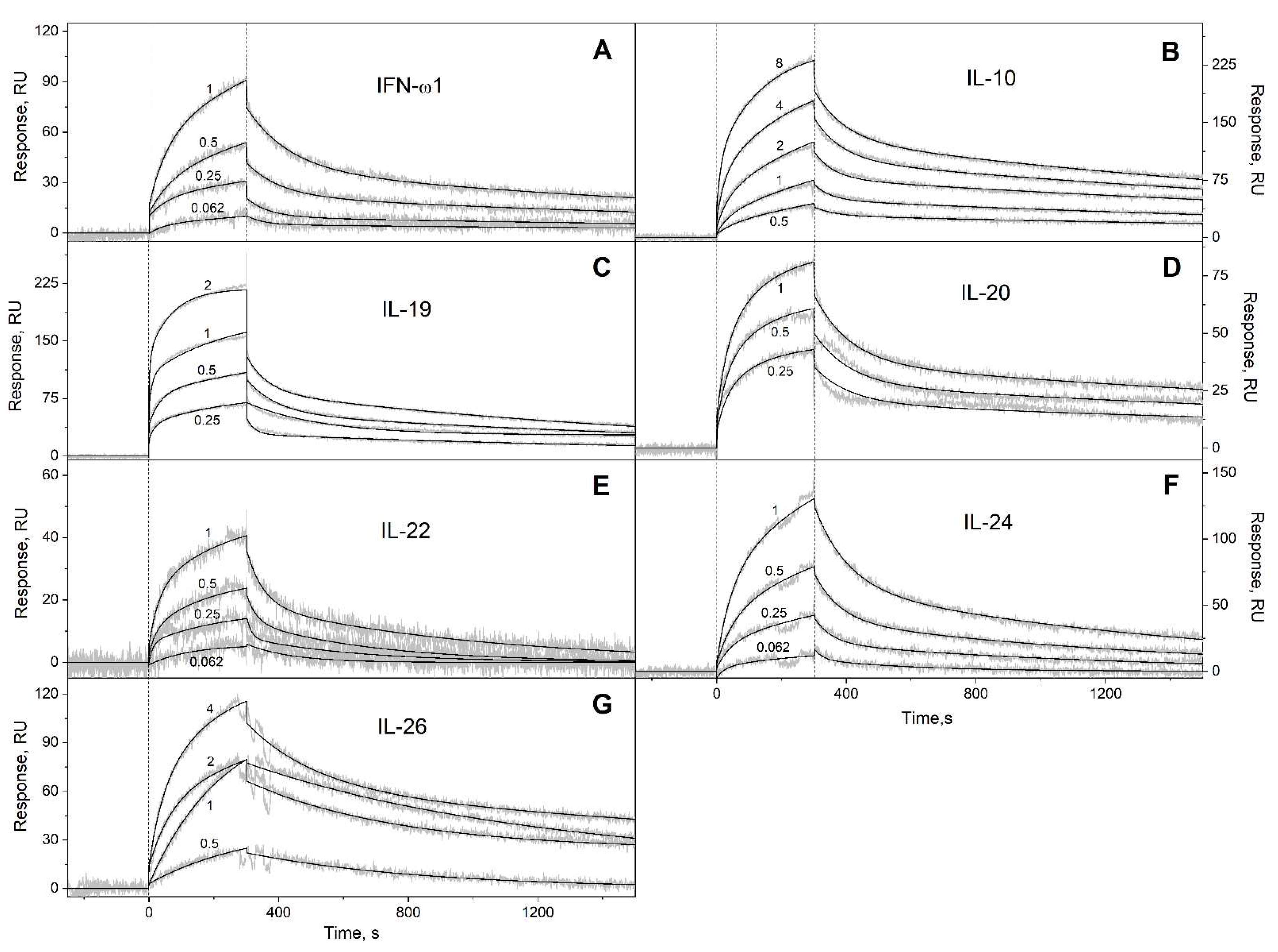
2.2. Surface plasmon resonance studies
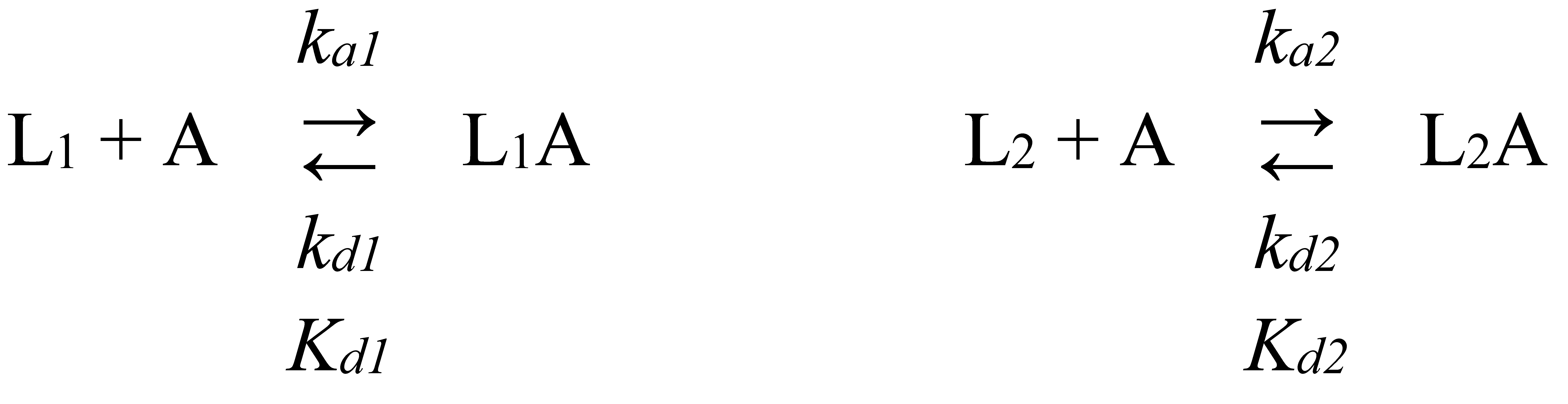
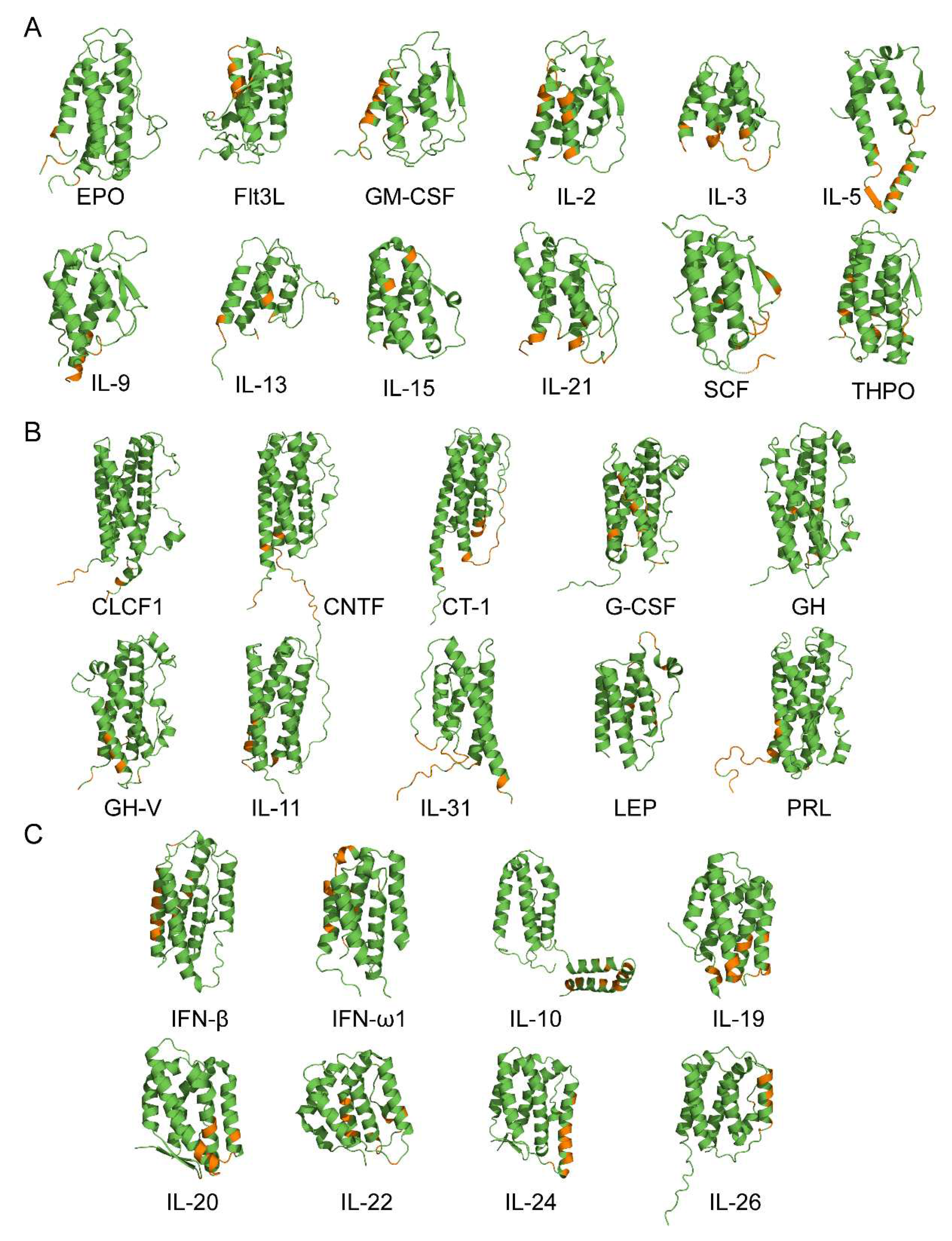
2.3. Structural classification of cytokines
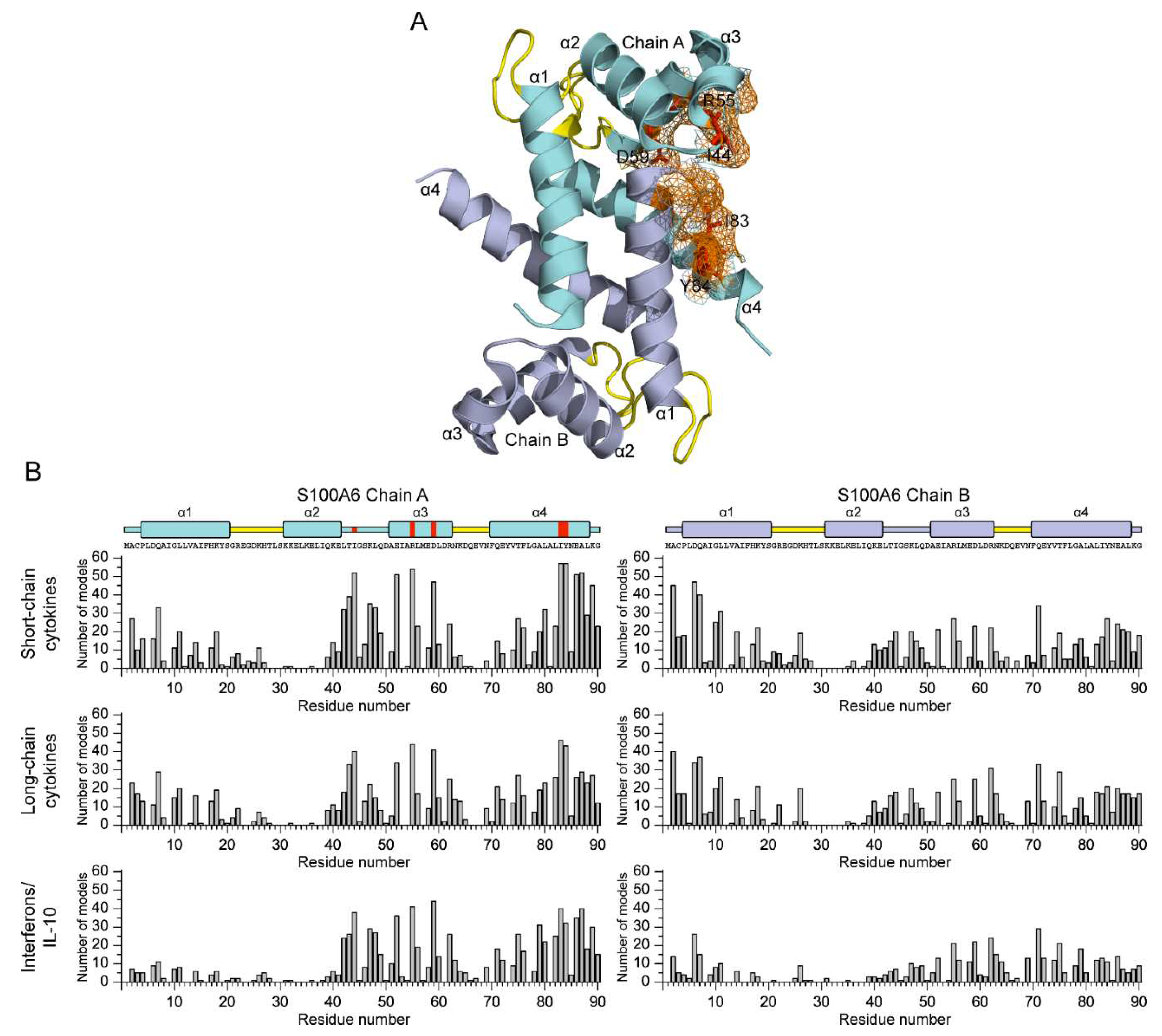
2.4. Structural modeling of the S100A6-cytokine complexes
3. Results and discussion
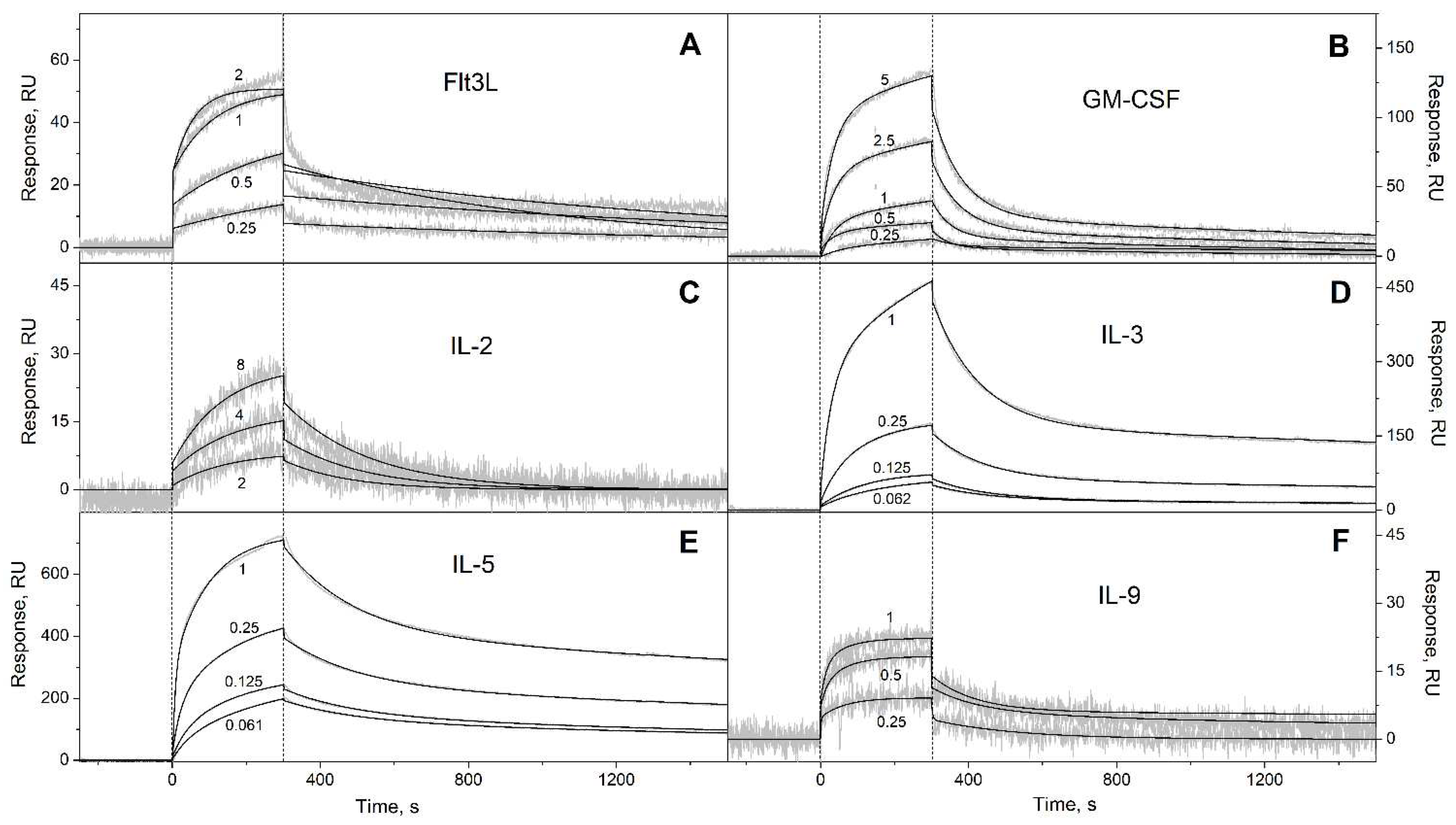
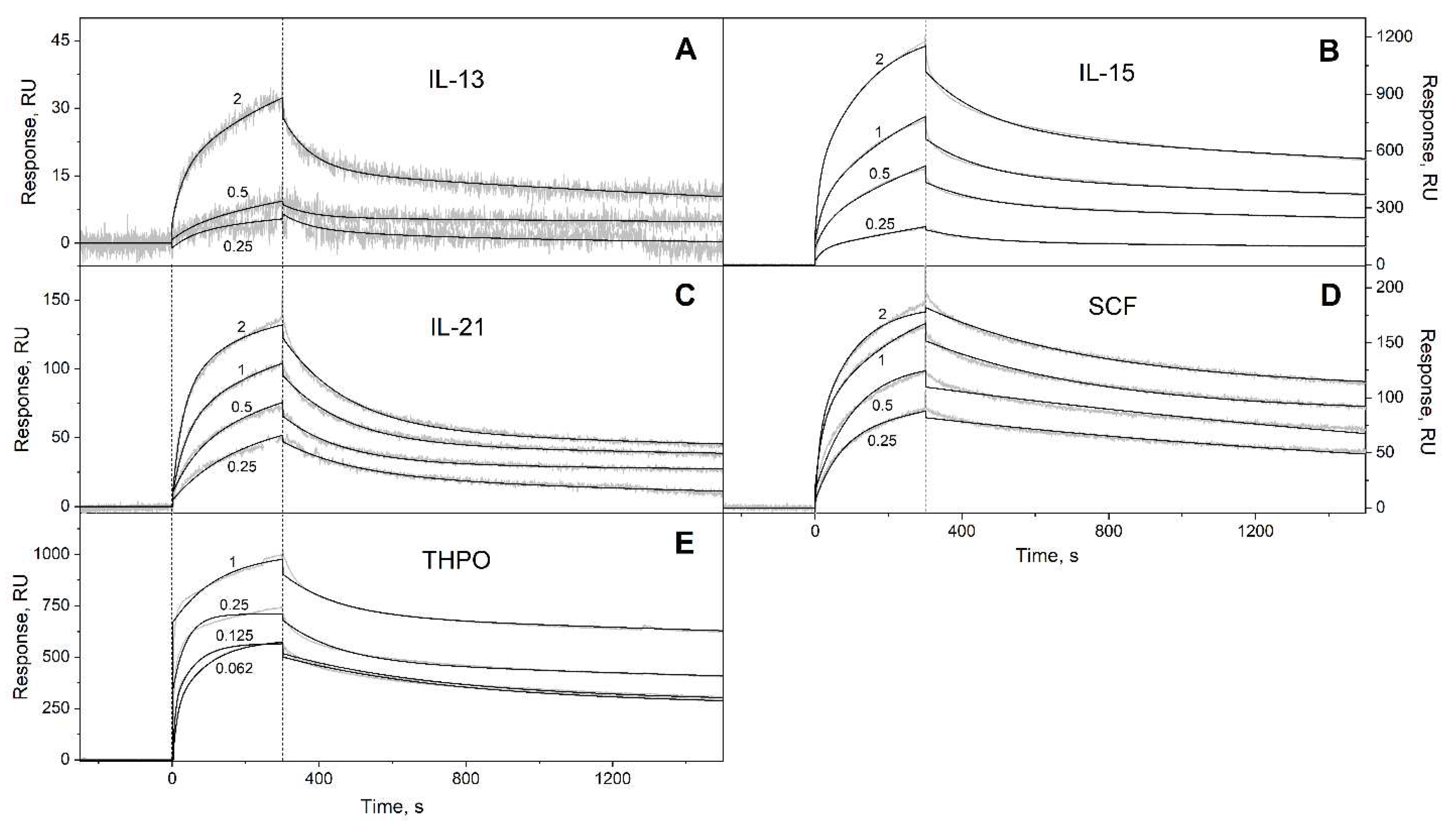
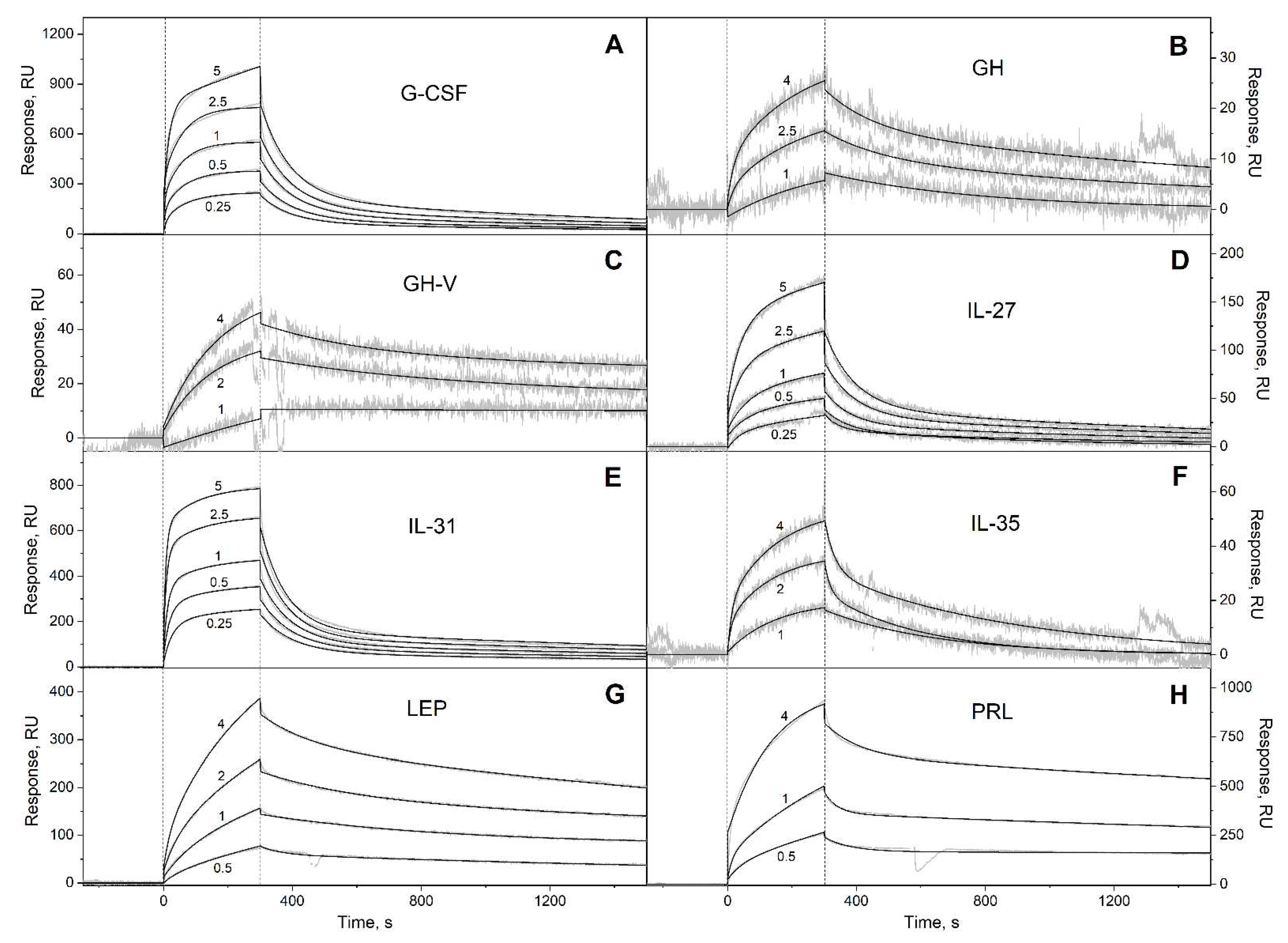
3.1. Selectivity of S100A6 binding to the four-helical cytokines
3.2. Structural modeling of the S100A6-cytokine complexes
3.3. Potential physiological significance of the S100A6 interactions with the four-helical cytokines
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr Mol Med 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. S100 family proteins in inflammation and beyond. Adv Clin Chem 2020, 98, 173–231. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef]
- Zimmer, D.B.; Eubanks, J.O.; Ramakrishnan, D.; Criscitiello, M.F. Evolution of the S100 family of calcium sensor proteins. Cell Calcium 2013, 53, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc.Natl.Acad.Sci.U.S.A 1972, 69, 581–584. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Fritz, G.; Heizmann, C.W. 3D Structures of the Calcium and Zinc Binding S100 Proteins. In Handbook of Metalloproteins, John Wiley & Sons, L., Ed. 2004. 2004. [Google Scholar] [CrossRef]
- Gilston, B.A.; Skaar, E.P.; Chazin, W.J. Binding of transition metals to S100 proteins. Sci China Life Sci 2016, 59, 792–801. [Google Scholar] [CrossRef]
- Streicher, W.W.; Lopez, M.M.; Makhatadze, G.I. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys Chem 2010, 151, 181–186. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D. , et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic acids research 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat Rev Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Cristóvão, J.S.; Gomes, C.M. S100 Proteins in Alzheimer's Disease. Front Neurosci-Switz 2019, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, D.; Tenbrock, K.; Roth, J. Alarmins of the S100-Family in Juvenile Autoimmune and Auto-Inflammatory Diseases. Front Immunol 2019, 10, 182–182. [Google Scholar] [CrossRef] [PubMed]
- Sattar, Z.; Lora, A.; Jundi, B.; Railwah, C.; Geraghty, P. The S100 Protein Family as Players and Therapeutic Targets in Pulmonary Diseases. Pulm Med 2021, 2021, 5488591. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim Biophys Acta Mol Cell Res 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Allgower, C.; Kretz, A.L.; von Karstedt, S.; Wittau, M.; Henne-Bruns, D.; Lemke, J. Friend or Foe: S100 Proteins in Cancer. Cancers (Basel) 2020, 12, 2037. [Google Scholar] [CrossRef]
- Bresnick, A.R. S100 proteins as therapeutic targets. Biophys Rev 2018, 10, 1617–1629. [Google Scholar] [CrossRef]
- Engelkamp, D.; Schafer, B.W.; Mattei, M.G.; Erne, P.; Heizmann, C.W. Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci U S A 1993, 90, 6547–6551. [Google Scholar] [CrossRef]
- Schafer, B.W.; Wicki, R.; Engelkamp, D.; Mattei, M.G.; Heizmann, C.W. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 1995, 25, 638–643. [Google Scholar] [CrossRef]
- Marenholz, I.; Lovering, R.C.; Heizmann, C.W. An update of the S100 nomenclature. Biochim Biophys Acta 2006, 1763, 1282–1283. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Mayorov, S.A.; Deryusheva, E.I.; Avkhacheva, N.V.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Permyakov, E.A.; Permyakov, S.E. Highly specific interaction of monomeric S100P protein with interferon beta. Int J Biol Macromol 2020, 143, 633–639. [Google Scholar] [CrossRef]
- Hsieh, H.L.; Schafer, B.W.; Cox, J.A.; Heizmann, C.W. S100A13 and S100A6 exhibit distinct translocation pathways in endothelial cells. J Cell Sci 2002, 115, 3149–3158. [Google Scholar] [CrossRef]
- Jurewicz, E.; Wyroba, E.; Filipek, A. Tubulin-dependent secretion of S100A6 and cellular signaling pathways activated by S100A6-integrin beta1 interaction. Cell Signal 2018, 42, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Jurewicz, E.; Góral, A.; Filipek, A. S100A6 is secreted from Wharton's jelly mesenchymal stem cells and interacts with integrin β1. The International Journal of Biochemistry & Cell Biology 2014, 55, 298–303. [Google Scholar] [CrossRef]
- Rumpret, M.; von Richthofen, H.J.; van der Linden, M.; Westerlaken, G.H.A.; Talavera Ormeno, C.; Low, T.Y.; Ovaa, H.; Meyaard, L. Recognition of S100 proteins by Signal Inhibitory Receptor on Leukocytes-1 negatively regulates human neutrophils. Eur J Immunol 2021, 51, 2210–2217. [Google Scholar] [CrossRef]
- Moller, A.; Jauch-Speer, S.L.; Gandhi, S.; Vogl, T.; Roth, J.; Fehler, O. The roles of toll-like receptor 4, CD33, CD68, CD69, or CD147/EMMPRIN for monocyte activation by the DAMP S100A8/S100A9. Front Immunol 2023, 14, 1110185. [Google Scholar] [CrossRef] [PubMed]
- Ruma, I.M.; Putranto, E.W.; Kondo, E.; Murata, H.; Watanabe, M.; Huang, P.; Kinoshita, R.; Futami, J.; Inoue, Y.; Yamauchi, A. , et al. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-κB and ROS formation upon ligand binding. Clin Exp Metastasis 2016, 33, 609–627. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamamoto, M.; Miyai, M.; Maeda, T.; Hiruma, J.; Murata, H.; Kinoshita, R.; Winarsa Ruma, I.M.; Putranto, E.W.; Inoue, Y. , et al. Identification of an S100A8 Receptor Neuroplastin-beta and its Heterodimer Formation with EMMPRIN. J Invest Dermatol 2016, 136, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Tomonobu, N.; Kinoshita, R.; Sakaguchi, M. S100 Soil Sensor Receptors and Molecular Targeting Therapy Against Them in Cancer Metastasis. Transl Oncol 2020, 13, 100753. [Google Scholar] [CrossRef]
- Pankratova, S.; Klingelhofer, J.; Dmytriyeva, O.; Owczarek, S.; Renziehausen, A.; Syed, N.; Porter, A.E.; Dexter, D.T.; Kiryushko, D. The S100A4 Protein Signals through the ErbB4 Receptor to Promote Neuronal Survival. Theranostics 2018, 8, 3977–3990. [Google Scholar] [CrossRef]
- Warner-Schmidt, J.L.; Flajolet, M.; Maller, A.; Chen, E.Y.; Qi, H.; Svenningsson, P.; Greengard, P. Role of p11 in cellular and behavioral effects of 5-HT4 receptor stimulation. J Neurosci 2009, 29, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Klingelhofer, J.; Moller, H.D.; Sumer, E.U.; Berg, C.H.; Poulsen, M.; Kiryushko, D.; Soroka, V.; Ambartsumian, N.; Grigorian, M.; Lukanidin, E.M. Epidermal growth factor receptor ligands as new extracellular targets for the metastasis-promoting S100A4 protein. Febs J 2009, 276, 5936–5948. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.K.; Yu, C. The IL1alpha-S100A13 heterotetrameric complex structure: a component in the non-classical pathway for interleukin 1alpha secretion. J Biol Chem 2011, 286, 14608–14617. [Google Scholar] [CrossRef] [PubMed]
- Carreira, C.M.; LaVallee, T.M.; Tarantini, F.; Jackson, A.; Lathrop, J.T.; Hampton, B.; Burgess, W.H.; Maciag, T. S100A13 is involved in the regulation of fibroblast growth factor-1 and p40 synaptotagmin-1 release in vitro. Journal of Biological Chemistry 1998, 273, 22224–22231. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.A.; Chou, R.H.; Li, H.C.; Yang, L.W.; Yu, C. Structural insights into the interaction of human S100B and basic fibroblast growth factor (FGF2): Effects on FGFR1 receptor signaling. Bba-Proteins Proteom 2013, 1834, 2606–2619. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Zemskova, M.Y.; Rystsov, G.K.; Vologzhannikova, A.A.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N. , et al. Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity. Int J Mol Sci 2022, 23, 15956. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Deryusheva, E.I.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Erythropoietin Interacts with Specific S100 Proteins. Biomolecules 2022, 12, 120. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sofin, A.D.; Avkhacheva, N.V.; Denesyuk, A.I.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N. , et al. Interferon Beta Activity Is Modulated via Binding of Specific S100 Proteins. International journal of molecular sciences 2020, 21, 9473. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sofin, A.D.; Avkhacheva, N.V.; Deryusheva, E.I.; Rastrygina, V.A.; Permyakova, M.E.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Interferon-β Activity Is Affected by S100B Protein. International journal of molecular sciences 2022, 23, 1997. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Specific cytokines of interleukin-6 family interact with S100 proteins. Cell Calcium 2022, 101, 102520. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Deryusheva, E.I.; Permyakova, M.E.; Sokolov, A.S.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Calcium-Bound S100P Protein Is a Promiscuous Binding Partner of the Four-Helical Cytokines. Int J Mol Sci 2022, 23, 12000. [Google Scholar] [CrossRef]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic acids research 2020, 48, D376–D382. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.A.; Ecsedi, P.; Kovacs, G.M.; Poti, A.L.; Remenyi, A.; Kardos, J.; Gogl, G.; Nyitray, L. High-throughput competitive fluorescence polarization assay reveals functional redundancy in the S100 protein family. The FEBS journal 2020, 287, 2834–2846. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.A.; Bartus, É.; Mag, B.; Boros, E.; Roszjár, L.; Gógl, G.; Travé, G.; Martinek, T.A.; Nyitray, L. Promiscuity mapping of the S100 protein family using a high-throughput holdup assay. Sci Rep 2022, 12, 5904. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, W.; Filipek, A. S100A6 Protein-Expression and Function in Norm and Pathology. Int J Mol Sci 2023, 24, 1341. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Brocker, C.; Thompson, D.; Matsumoto, A.; Nebert, D.W.; Vasiliou, V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum Genomics 2010, 5, 30–55. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Otterbein, L.R.; Kordowska, J.; Witte-Hoffmann, C.; Wang, C.L.; Dominguez, R. Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure 2002, 10, 557–567. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A. , et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Sokolov, A.S.; Vologzhannikova, A.A.; Permyakova, M.E.; Khorn, P.A.; Ismailov, R.G.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Baksheeva, V.E. , et al. Interleukin-11 binds specific EF-hand proteins via their conserved structural motifs. J Biomol Struct Dyn 2017, 35, 78–91. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Rastrygina, V.A.; Solovyev, V.V.; Ismailov, R.G.; Mikhailov, R.V.; Ulitin, A.B.; Yakovenko, A.R.; Mirzabekov, T.A.; Permyakov, E.A. , et al. High-affinity interaction between interleukin-11 and S100P protein. Biochem Biophys Res Commun 2015, 468, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E. Measuring binding of S100 proteins to RAGE by surface plasmon resonance. Methods Mol Biol 2013, 963, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Fritz, G.; Weibel, M.; Heizmann, C.W.; Galichet, A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem 2007, 282, 31317–31331. [Google Scholar] [CrossRef]
- Mohan, S.K.; Gupta, A.A.; Yu, C. Interaction of the S100A6 mutant (C3S) with the V domain of the receptor for advanced glycation end products (RAGE). Biochem Biophys Res Commun 2013, 434, 328–333. [Google Scholar] [CrossRef]
- Loosen, S.H.; Benz, F.; Niedeggen, J.; Schmeding, M.; Schuller, F.; Koch, A.; Vucur, M.; Tacke, F.; Trautwein, C.; Roderburg, C. , et al. Serum levels of S100A6 are unaltered in patients with resectable cholangiocarcinoma. Clin Transl Med 2016, 5, 39. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Denesyuk, A.I.; Denessiouk, K.A.; Permyakova, M.E.; Kazakov, A.S.; Ismailov, R.G.; Rastrygina, V.A.; Sokolov, A.S.; Permyakov, E.A. Monomeric state of S100P protein: Experimental and molecular dynamics study. Cell Calcium 2019, 80, 152–159. [Google Scholar] [CrossRef]
- Guzel, C.; van den Berg, C.B.; Duvekot, J.J.; Stingl, C.; van den Bosch, T.P.P.; van der Weiden, M.; Steegers, E.A.P.; Steegers-Theunissen, R.P.M.; Luider, T.M. Quantification of Calcyclin and Heat Shock Protein 90 in Sera from Women with and without Preeclampsia by Mass Spectrometry. Proteomics Clin Appl 2019, 13, e1800181. [Google Scholar] [CrossRef]
- Wang, T.; Liang, Y.; Thakur, A.; Zhang, S.; Yang, T.; Chen, T.; Gao, L.; Chen, M.; Ren, H. Diagnostic significance of S100A2 and S100A6 levels in sera of patients with non-small cell lung cancer. Tumour Biol 2016, 37, 2299–2304. [Google Scholar] [CrossRef]
- Cai, X.Y.; Lu, L.; Wang, Y.N.; Jin, C.; Zhang, R.Y.; Zhang, Q.; Chen, Q.J.; Shen, W.F. Association of increased S100B, S100A6 and S100P in serum levels with acute coronary syndrome and also with the severity of myocardial infarction in cardiac tissue of rat models with ischemia-reperfusion injury. Atherosclerosis 2011, 217, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N. , et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 2014, 42, D358–363. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F. , et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Ismailov, R.G.; Xue, B.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Intrinsic disorder in S100 proteins. Mol Biosyst 2011, 7, 2164–2180. [Google Scholar] [CrossRef] [PubMed]
- Yatime, L.; Betzer, C.; Jensen, R.K.; Mortensen, S.; Jensen, P.H.; Andersen, G.R. The Structure of the RAGE:S100A6 Complex Reveals a Unique Mode of Homodimerization for S100 Proteins. Structure 2016, 24, 2043–2052. [Google Scholar] [CrossRef]
- Rousseau, F.; Gauchat, J.F.; McLeod, J.G.; Chevalier, S.; Guillet, C.; Guilhot, F.; Cognet, I.; Froger, J.; Hahn, A.F.; Knappskog, P.M. , et al. Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proceedings of the National Academy of Sciences of the United States of America 2006, 103, 10068–10073. [Google Scholar] [CrossRef]
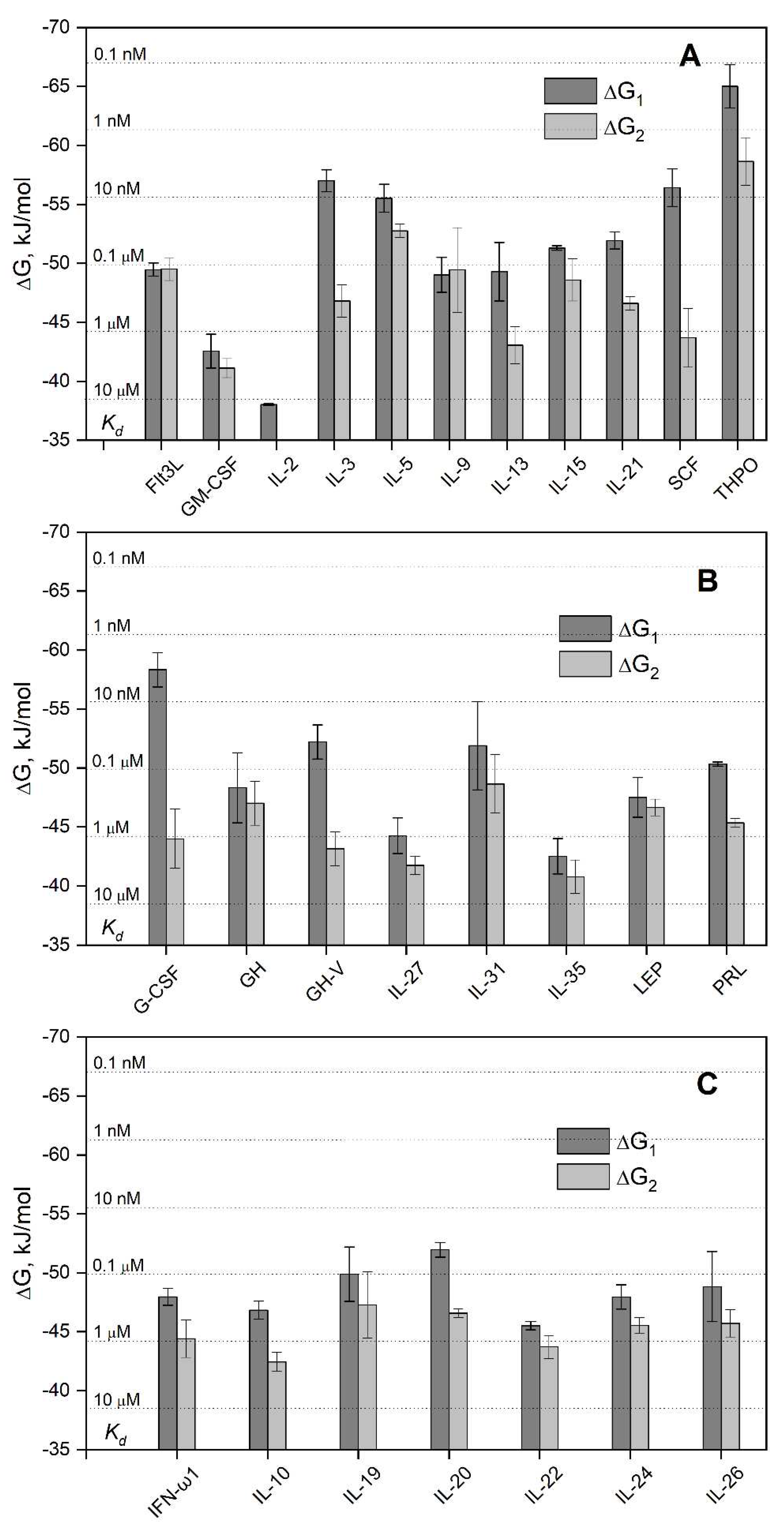
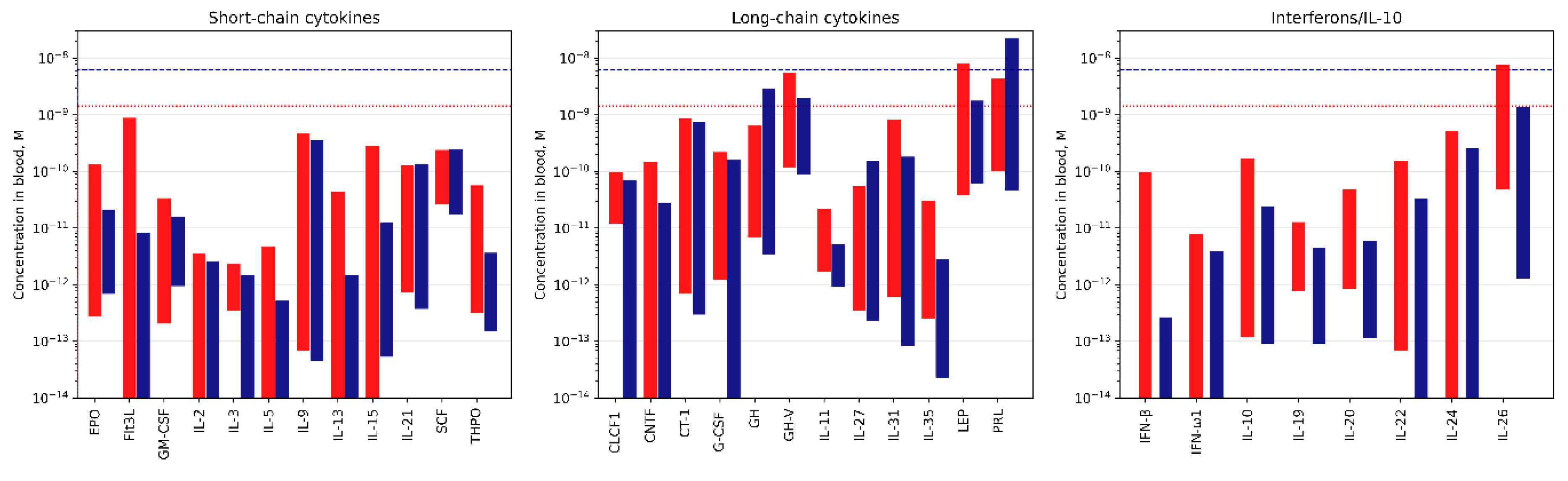
| Cytokine | Kd1, M | Kd2, M | Reference |
| Short-chain cytokines | |||
| EPO | (1.5 ± 0.3) × 10−7 | (6.5 ± 2.6) × 10−7 | [38] |
| Long-chain cytokines | |||
| CLCF1 | (1.1 ± 0.8) × 10-6 | (3.7 ± 0.8) × 10-6 | [41] |
| CNTF | (1.1 ± 1.0) × 10-7 | (2.07 ± 0.08) × 10-6 | [41] |
| CT-1 | (1.6 ± 0.5) × 10-6 | (1.2 ± 0.4) × 10-5 | [41] |
| IL-11 | (8.3 ± 1.9) × 10-6 | (7.9 ± 2.1) × 10-6 | [41] |
| Interferons/IL-10 | |||
| IFN-β | (8.2 ± 2.4) × 10-8 (0.70 ± 0.03) × 10-9 * |
(2.67 ± 0.57) × 10-7 (2.81 ± 0.49) × 10-7 * |
[39] |
| Full name | Abbreviation | UniProt ID | Manufacturer | Cat. number | Source |
| Short-chain cytokines | |||||
| Fms-related tyrosine kinase 3 ligand | Flt3L | P49771 | PeproTech | 300-19 | E.coli |
| Granulocyte-macrophage colony-stimulating factor | GM-CSF | P04141 | PeproTech | 300-03 | E.coli |
| Interleukin-2 | IL-2 | P60568 | PeproTech | AF-200-02 | E.coli |
| Interleukin-3 | IL-3 | P08700 | SCI-Store (Russia) | PSG160-10 | CHO |
| Interleukin-4 | IL-4 | P05112 | PeproTech | AF-200-04 | E.coli |
| Interleukin-5 | IL-5 | P05113 | PeproTech | 200-05 | E.coli |
| Interleukin-7 | IL-7 | P13232 | SCI-Store (Russia) | PSG240-10 | CHO |
| Interleukin-9 | IL-9 | P15248 | PeproTech | 200-09 | E.coli |
| Interleukin-13 | IL-13 | P35225 | PeproTech | 200-13 | E.coli |
| Interleukin-15 | IL-15 | P40933 | PeproTech | 200-15 | E.coli |
| Interleukin-21 | IL-21 | Q9HBE4 | PeproTech | 200-21 | E.coli |
| Macrophage colony-stimulating factor 1 | M-CSF | P09603 | PeproTech | 300-25 | E.coli |
| Stem cell factor, soluble form | SCF | P21583 | PeproTech | 300-07 | E.coli |
| Thrombopoietin | THPO | P40225 | SCI-Store (Russia) | PSG090-10 | CHO |
| Thymic stromal lymphopoietin | TSLP | Q969D9 | PeproTech | 300-62 | E.coli |
| Long-chain cytokines | |||||
| Chorionic somatomammotropin hormone 1 | PL | P0DML2 | R&D Systems | 5757-PL/CF | CHO |
| Granulocyte colony-stimulating factor | G-CSF | P09919 | Pharmstandard (Russia) | n/a | E.coli |
| Growth hormone | GH | P01241 | PeproTech | AF-100-40 | E.coli |
| Growth hormone variant | GH-V | P01242 | R&D Systems | 7668-GH/CF | E.coli |
| Interleukin-12 | IL-12 | P29459* & P29460 | PeproTech | 200-12H | HEK293 |
| Interleukin-23 | IL-23 | Q9NPF7* & P29460 | PeproTech | 200-23 | Hi-5 |
| Interleukin-27 | IL-27 | Q8NEV9* & Q14213 | PeproTech | 200-38 | HEK293 |
| Interleukin-31 | IL-31 | Q6EBC2 | PeproTech | 200-31 | E.coli |
| Interleukin-35 | IL-35 | P29459* & Q14213 | PeproTech | 200-37 | HEK293 |
| Leptin | LEP | P41159 | PeproTech | AF-300-27 | E.coli |
| Prolactin | PRL | P01236 | PeproTech | 100-07 | E.coli |
| Interferons/IL-10 | |||||
| Interferon α-2 | IFN-α2 | P01563 | Vector-Medica (Russia) | n/a | E.coli |
| Interferon γ | IFN-γ | P01579 | Pharmaclon (Russia) | n/a | E.coli |
| Interferon ω-1 | IFN-ω1 | P05000 | PeproTech | 300-02J | E.coli |
| Interleukin-10 | IL-10 | P22301 | PeproTech | AF-200-10 | E.coli |
| Interleukin-19 | IL-19 | Q9UHD0 | PeproTech | 200-19 | E.coli |
| Interleukin-20 | IL-20 | Q9NYY1 | PeproTech | 200-20 | E.coli |
| Interleukin-22 | IL-22 | Q9GZX6 | PeproTech | 200-22 | E.coli |
| Interleukin-24 | IL-24 | Q13007 | PeproTech | 200-35 | CHO |
| Interleukin-26 | IL-26 | Q9NPH9 | R&D Systems | 1375-IL/CF | E.coli |
| Cytokine | kd1, s-1 | Kd1, M | kd2, s-1 | Kd2, M |
| Short-chain cytokines | ||||
| Flt3L | (8.83 ± 2.54) × 10-4 | (1.22 ± 0.27) × 10-7 | (8.32 ± 4.05) × 10-4 | (1.25 ± 0.46) × 10-7 |
| GM-CSF | (7.92 ± 4.02) × 10-4 | (2.26 ± 1.18) × 10-6 | (1.76 ± 0.49) × 10-2 | (3.63 ± 1.14) × 10-6 |
| IL-2* | (4.97 ± 0.78) × 10-3 | (1.20 ± 0.04) × 10-5 | n/a | n/a |
| IL-3 | (2.71 ± 0.78) × 10-4 | (6.11 ± 2.19) × 10-9 | (7.28 ± 0.65) × 10-3 | (4.02 ± 2.03) × 10-7 |
| IL-5 | (2.47 ± 0.45) × 10-4 | (1.16 ± 0.52) × 10-8 | (5.07 ± 0.20) × 10-3 | (3.23 ± 0.73) × 10-8 |
| IL-9 | (5.97 ± 2.79) × 10-3 | (1.69 ± 0.90) × 10-7 | (5.85 ± 2.72) × 10-3 | (2.71 ± 2.42) × 10-7 |
| IL-13 | (2.64 ± 0.98) × 10-4 | (1.97 ± 1.50) × 10-7 | (1.36 ± 0.06) × 10-2 | (1.91 ± 1.07) × 10-6 |
| IL-15 | (1.62 ± 0.24) × 10-4 | (5.70 ± 0.46) × 10-8 | (6.29 ± 0.26) × 10-3 | (2.15 ± 1.32) × 10-7 |
| IL-21 | (1.88 ± 1.33) × 10-4 | (4.57 ± 1.30) × 10-8 | (5.68 ± 0.34) × 10-3 | (3.87 ± 0.86) × 10-7 |
| SCF | (2.57 ± 0.92) × 10-4 | (8.72 ± 4.95) × 10-9 | (4.51 ± 1.52) × 10-3 | (1.90 ± 1.45) × 10-6 |
| THPO | (6.00 ± 3.29) × 10-5 | (2.89 ± 1.83) × 10-10 | (4.47 ± 2.46) × 10-3 | (3.96 ± 2.65) × 10-9 |
| Long-chain cytokines | ||||
| G-CSF | (7.72 ± 0.31) × 10-4 | (3.91 ± 2.06) × 10-9 | (1.04 ± 0.04) × 10-2 | (1.68 ± 1.29) × 10-6 |
| GH | (3.73 ± 2.36) × 10-4 | (3.42 ± 2.85) × 10-7 | (5.87 ± 1.16) × 10-3 | (4.16 ± 2.65) × 10-7 |
| GH-V | (7.92 ± 3.35) × 10-5 | (4.62 ± 2.41) × 10-8 | (3.06 ± 1.27) × 10-3 | (1.78 ± 0.92) × 10-6 |
| IL-27# | (9.61 ± 4.18) × 10-4 | (1.16 ± 0.63) × 10-6 | (1.47 ± 0.62) × 10-2 | (2.80 ± 0.85) × 10-6 |
| IL-31 | (4.64 ± 0.13) × 10-4 | (1.06 ± 0.96) × 10-7 | (1.11 ± 0.12) × 10-2 | (2.56 ± 1.95) × 10-7 |
| IL-35# | (2.59 ± 0.81) × 10-3 | (2.34 ± 1.26) × 10-6 | (3.91 ± 1.55) × 10-2 | (4.65 ± 2.42) × 10-6 |
| LEP | (3.18 ± 2.17) × 10-3 | (3.25 ± 1.92) × 10-7 | (2.87 ± 1.00) × 10-4 | (3.85 ± 1.06) × 10-7 |
| PRL | (1.59 ± 0.23) × 10-4 | (8.40 ± 0.58) × 10-8 | (1.55 ± 0.82) × 10-2 | (6.35 ± 0.96) × 10-7 |
| Interferons/IL-10 | ||||
| IFN-ω1 | (4.71 ± 0.22) × 10-4 | (2.28 ± 0.64) × 10-7 | (1.29 ± 0.55) × 10-2 | (1.11 ± 0.63) × 10-6 |
| IL-10 | (3.98 ± 0.28) × 10-4 | (3.61 ± 1.09) × 10-7 | (1.50 ± 0.37) × 10-2 | (2.13 ± 0.67) × 10-6 |
| IL-19 | (6.28 ± 0.86) × 10-4 | (1.47 ± 1.08) × 10-7 | (2.77 ± 1.69) × 10-2 | (4.96 ± 4.04) × 10-7 |
| IL-20 | (3.08 ± 0.86) × 10-4 | (4.51 ± 1.12) × 10-8 | (9.14 ± 2.51) × 10-3 | (3.85 ± 0.59) × 10-7 |
| IL-22 | (3.57 ± 2.56) × 10-3 | (5.93 ± 0.84) × 10-7 | (3.26 ± 2.67) × 10-2 | (1.31 ± 0.49) × 10-6 |
| IL-24 | (5.73 ± 0.85) × 10-4 | (2.39 ± 0.94) × 10-7 | (7.79 ± 1.72) × 10-3 | (6.01 ± 1.57) × 10-7 |
| IL-26 | (5.98 ± 2.92) × 10-4 | (2.78 ± 2.31) × 10-7 | (2.56 ± 1.39) × 10-3 | (6.00 ± 2.63) × 10-7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





