Submitted:
30 November 2023
Posted:
01 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
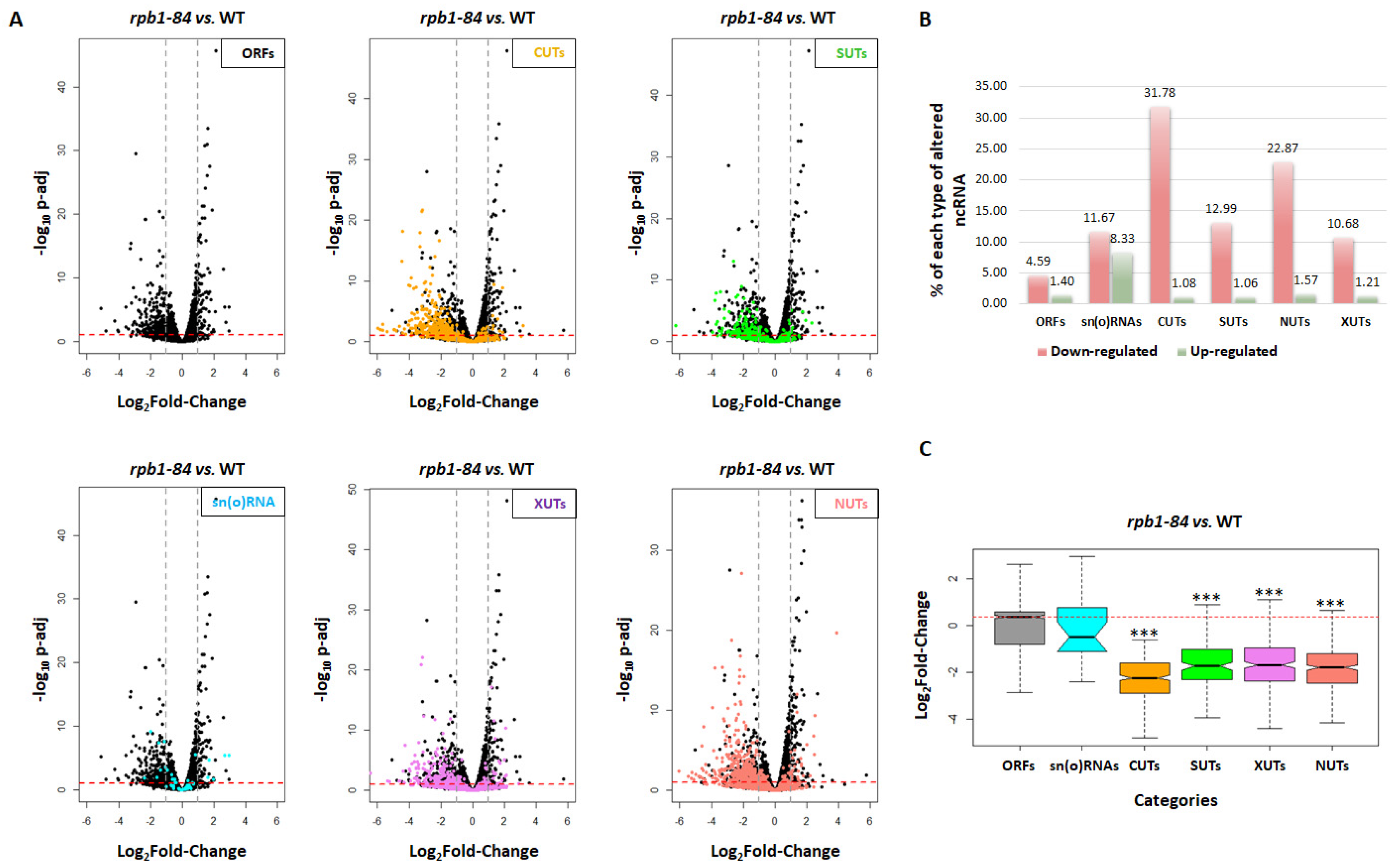
2.1. Altering assembly of RNA pol II affects ncRNA accumulation
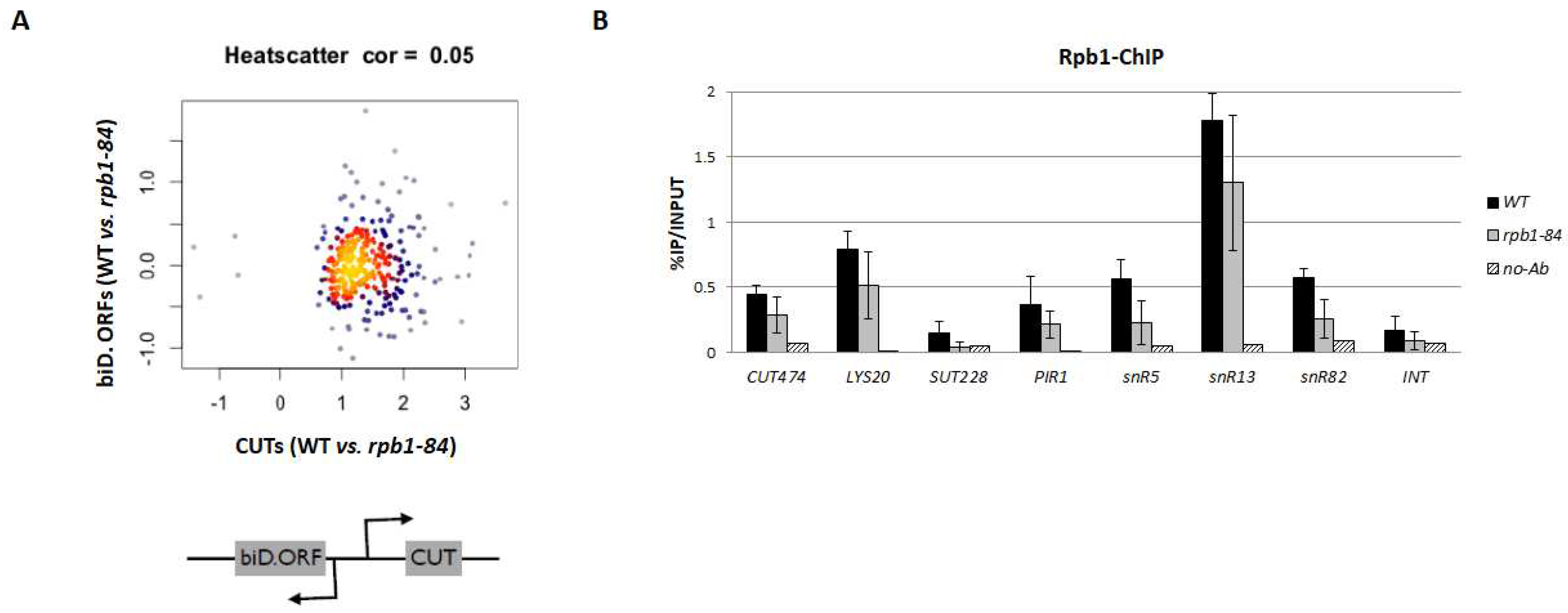
2.2. The decrease in CUTs seems to be independent of bidirectional transcription
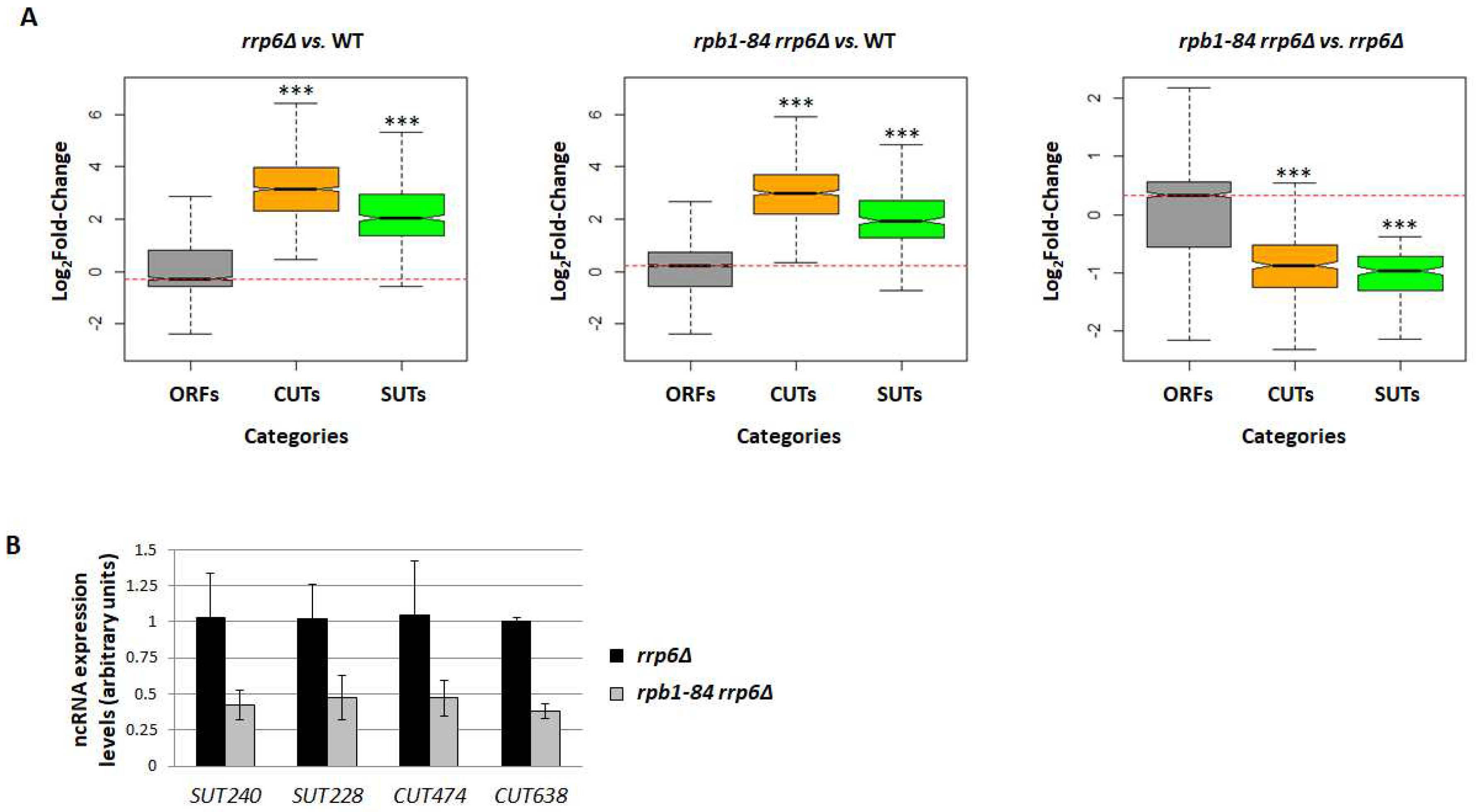
2.3. The decrease in ncRNA transcription observed in the rpb1-84 mutant does not appear to result from a malfunction of nuclear exosome
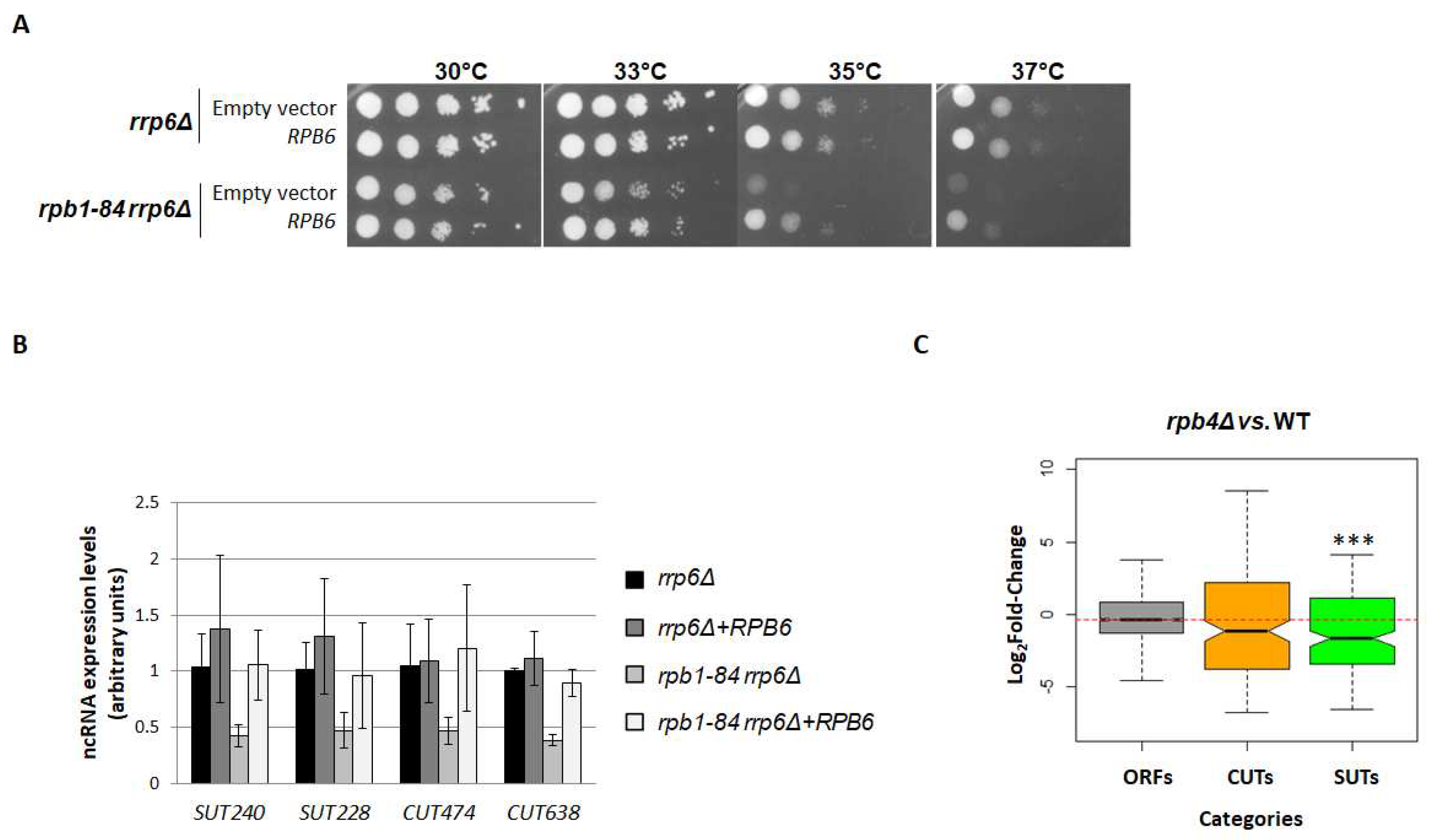
2.4. Correcting RNA pol II assembly overcomes the decrease in pervasive transcription
2.5. The drop in the ncRNA levels in the rpb1-84 mutant may be linked with the alteration in ncRNA transcription termination
3. Discussion
4. Materials and Methods
4.1. Yeast Strains, Plasmid, Genetic Manipulations and Media
4.2. RNA extraction, sequencing and bioinformatic analysis
4.3. Reverse transcription and qRT-PCR
4.4. Chromatin immunoprecipitation
4.5. Chromatin-enriched fractions and western blot analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Werner, F.; Grohmann, D., Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011, 9, (2), 85-98. [CrossRef]
- Barba-Aliaga, M.; Alepuz, P.; Pérez-Ortín, J. E., Eukaryotic RNA Polymerases. 2021.
- Cramer, P., Organization and regulation of gene transcription. Nature 2019, 573, (7772), 45-54. [CrossRef]
- Werner, M.; Thuriaux, P.; Soutourina, J., Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol 2009, 19, (6), 740-5. [CrossRef]
- Pawlicki, J. M.; Steitz, J. A., Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol 2010, 20, (1), 52-61. [CrossRef]
- Kornienko, A. E.; Guenzl, P. M.; Barlow, D. P.; Pauler, F. M., Gene regulation by the act of long non-coding RNA transcription. BMC Biol 2013, 11, 59. [CrossRef]
- Tuck, A. C.; Tollervey, D., RNA in pieces. Trends Genet 2011, 27, (10), 422-32. [CrossRef]
- Jacquier, A., The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nature Reviews Genetics 2009, 10, (12), 833-844. [CrossRef]
- Jensen, T. H.; Jacquier, A.; Libri, D., Dealing with pervasive transcription. Molecular cell 2013, 52, (4), 473-484. [CrossRef]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Münster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L. M., Bidirectional promoters generate pervasive transcription in yeast. Nature 2009, 457, (7232), 1033-1037. [CrossRef]
- Bertone, P.; Stolc, V.; Royce, T. E.; Rozowsky, J. S.; Urban, A. E.; Zhu, X.; Rinn, J. L.; Tongprasit, W.; Samanta, M.; Weissman, S., Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, (5705), 2242-2246. [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C., The transcriptional landscape of the mammalian genome. science 2005, 309, (5740), 1559-1563. [CrossRef]
- David, L.; Huber, W.; Granovskaia, M.; Toedling, J.; Palm, C. J.; Bofkin, L.; Jones, T.; Davis, R. W.; Steinmetz, L. M., A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A 2006, 103, (14), 5320-5. [CrossRef]
- Li, L.; Wang, X.; Stolc, V.; Li, X.; Zhang, D.; Su, N.; Tongprasit, W.; Li, S.; Cheng, Z.; Wang, J., Genome-wide transcription analyses in rice using tiling microarrays. Nature genetics 2006, 38, (1), 124-129. [CrossRef]
- Stolc, V.; Gauhar, Z.; Mason, C.; Halasz, G.; Van Batenburg, M. F.; Rifkin, S. A.; Hua, S.; Herreman, T.; Tongprasit, W.; Barbano, P. E., A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 2004, 306, (5696), 655-660. [CrossRef]
- Novačić, A.; Vučenović, I.; Primig, M.; Stuparević, I., Non-coding RNAs as cell wall regulators in Saccharomyces cerevisiae. Critical reviews in microbiology 2020, 46, (1), 15-25. [CrossRef]
- Neil, H.; Malabat, C.; d’Aubenton-Carafa, Y.; Xu, Z.; Steinmetz, L. M.; Jacquier, A., Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 2009, 457, (7232), 1038-1042. [CrossRef]
- Van Dijk, E. L.; Chen, C. L.; d’Aubenton-Carafa, Y.; Gourvennec, S.; Kwapisz, M.; Roche, V.; Bertrand, C.; Silvain, M.; Legoix-Ne, P.; Loeillet, S., XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 2011, 475, (7354), 114-117. [CrossRef]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.-C.; Dufour, M.-E.; Boulay, J.; Régnault, B.; Devaux, F.; Namane, A.; Séraphin, B., Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly (A) polymerase. Cell 2005, 121, (5), 725-737. [CrossRef]
- Schulz, D.; Schwalb, B.; Kiesel, A.; Baejen, C.; Torkler, P.; Gagneur, J.; Soeding, J.; Cramer, P., Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 2013, 155, (5), 1075-1087. [CrossRef]
- Venkatesh, S.; Li, H.; Gogol, M. M.; Workman, J. L., Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nature communications 2016, 7, (1), 13610. [CrossRef]
- Lardenois, A.; Liu, Y.; Walther, T.; Chalmel, F.; Evrard, B.; Granovskaia, M.; Chu, A.; Davis, R. W.; Steinmetz, L. M.; Primig, M., Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proceedings of the National Academy of Sciences 2011, 108, (3), 1058-1063. [CrossRef]
- Wery, M.; Szachnowski, U.; Andjus, S.; de Andres-Pablo, A.; Morillon, A., The RNA helicases Dbp2 and Mtr4 regulate the expression of Xrn1-sensitive long non-coding RNAs in yeast. Frontiers in RNA research 2023, 1. [CrossRef]
- Wery, M.; Descrimes, M.; Vogt, N.; Dallongeville, A.-S.; Gautheret, D.; Morillon, A., Nonsense-mediated decay restricts LncRNA levels in yeast unless blocked by double-stranded RNA structure. Molecular cell 2016, 61, (3), 379-392. [CrossRef]
- Porrua, O.; Libri, D. Porrua, O.; Libri, D., Transcription termination and the control of the transcriptome: why, where and how to stop. Nature reviews Molecular cell biology 2015, 16, (3), 190-202. [CrossRef]
- Tudek, A.; Porrua, O.; Kabzinski, T.; Lidschreiber, M.; Kubicek, K.; Fortova, A.; Lacroute, F.; Vanacova, S.; Cramer, P.; Stefl, R., Molecular basis for coordinating transcription termination with noncoding RNA degradation. Molecular cell 2014, 55, (3), 467-481. [CrossRef]
- Vasiljeva, L.; Kim, M.; Mutschler, H.; Buratowski, S.; Meinhart, A., The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 2008, 15, (8), 795-804. [CrossRef]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L., The regulation and functions of the nuclear RNA exosome complex. Nature Reviews Molecular Cell Biology 2016, 17, (4), 227-239. [CrossRef]
- Malabat, C.; Feuerbach, F.; Ma, L.; Saveanu, C.; Jacquier, A., Quality control of transcription start site selection by nonsense-mediated-mRNA decay. Elife 2015, 4, e06722. [CrossRef]
- Marquardt, S.; Hazelbaker, D. Z.; Buratowski, S., Distinct RNA degradation pathways and 3′extensions of yeast non-coding RNA species. Transcription 2011, 2, (3), 145-154. [CrossRef]
- Sohrabi-Jahromi, S.; Hofmann, K. B.; Boltendahl, A.; Roth, C.; Gressel, S.; Baejen, C.; Soeding, J.; Cramer, P., Transcriptome maps of general eukaryotic RNA degradation factors. Elife 2019, 8, e47040. [CrossRef]
- Smith, J. E.; Alvarez-Dominguez, J. R.; Kline, N.; Huynh, N. J.; Geisler, S.; Hu, W.; Coller, J.; Baker, K. E., Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell reports 2014, 7, (6), 1858-1866. [CrossRef]
- Collin, P.; Jeronimo, C.; Poitras, C.; Robert, F., RNA Polymerase II CTD Tyrosine 1 Is Required for Efficient Termination by the Nrd1-Nab3-Sen1 Pathway. Molecular cell 2019, 73, (4), 655-669. e7. [CrossRef]
- Mosley, A. L.; Pattenden, S. G.; Carey, M.; Venkatesh, S.; Gilmore, J. M.; Florens, L.; Workman, J. L.; Washburn, M. P., Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 2009, 34, (2), 168-78. [CrossRef]
- Hsu, P. L.; Yang, F.; Smith-Kinnaman, W.; Yang, W.; Song, J.-E.; Mosley, A. L.; Varani, G., Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. Journal of molecular biology 2014, 426, (16), 2970-2981. [CrossRef]
- Hunter, G. O.; Fox, M. J.; Smith-Kinnaman, W. R.; Gogol, M.; Fleharty, B.; Mosley, A. L., Phosphatase Rtr1 regulates global levels of serine 5 RNA polymerase II C-terminal domain phosphorylation and cotranscriptional histone methylation. Molecular and cellular biology 2016, 36, (17), 2236-2245. [CrossRef]
- Victorino, J. F.; Fox, M. J.; Smith-Kinnaman, W. R.; Justice, S. A. P.; Burriss, K. H.; Boyd, A. K.; Zimmerly, M. A.; Chan, R. R.; Hunter, G. O.; Liu, Y., RNA Polymerase II CTD phosphatase Rtr1 fine-tunes transcription termination. PLoS genetics 2020, 16, (3), e1008317. [CrossRef]
- Kim, H.; Erickson, B.; Luo, W.; Seward, D.; Graber, J. H.; Pollock, D. D.; Megee, P. C.; Bentley, D. L., Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 2010. [CrossRef]
- Egloff, S.; O’Reilly, D.; Chapman, R. D.; Taylor, A.; Tanzhaus, K.; Pitts, L.; Eick, D.; Murphy, S., Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007, 318, (5857), 1777-9. [CrossRef]
- Egloff, S.; Zaborowska, J.; Laitem, C.; Kiss, T.; Murphy, S., Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 2012, 45, (1), 111-22. [CrossRef]
- Egloff, S., Role of Ser7 phosphorylation of the CTD during transcription of snRNA genes. RNA biology 2012, 9, (8), 1033-1038. [CrossRef]
- Garrido-Godino, A. I.; Cuevas-Bermúdez, A.; Gutiérrez-Santiago, F.; Mota-Trujillo, M. d. C.; Navarro, F., The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae. International journal of molecular sciences 2022, 23, (4), 2002. [CrossRef]
- Garrido-Godino, A. I.; Gutiérrez-Santiago, F.; Navarro, F., Biogenesis of RNA Polymerases in Yeast. Frontiers in Molecular Biosciences 2021, 8. [CrossRef]
- Gómez-Navarro, N.; Estruch, F., Different pathways for the nuclear import of yeast RNA polymerase II. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2015, 1849, (11), 1354-1362. [CrossRef]
- Forget, D.; Lacombe, A. A.; Cloutier, P.; Lavallee-Adam, M.; Blanchette, M.; Coulombe, B., Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res 2013, 41, (14), 6881-91. [CrossRef]
- Garcia-Lopez, M. C.; Miron-Garcia, M. C.; Garrido-Godino, A. I.; Mingorance, C.; Navarro, F., Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr Genet 2010, 56, (3), 251-63. [CrossRef]
- Garrido-Godino, A. I.; Garcia-Lopez, M. C.; Navarro, F., Correct Assembly of RNA Polymerase II Depends on the Foot Domain and Is Required for Multiple Steps of Transcription in Saccharomyces cerevisiae. Mol Cell Biol 2013, 33, (18), 3611-26. [CrossRef]
- Cross, F. R., ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 1997, 13, (7), 647-53.
- Nonet, M.; Scafe, C.; Sexton, J.; Young, R., Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 1987, 7, (5), 1602-11. [CrossRef]
- Martínez-Fernández, V.; Cuevas-Bermúdez, A.; Gutiérrez-Santiago, F.; Garrido-Godino, A. I.; Rodríguez-Galán, O.; Jordán-Pla, A.; Lois, S.; Triviño, J. C.; de la Cruz, J.; Navarro, F., Prefoldin-like Bud27 influences the transcription of ribosomal components and ribosome biogenesis in Saccharomyces cerevisiae. RNA 2020, rna. 075507.120. [CrossRef]
- Liao, Y.; Smyth, G. K.; Shi, W., featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, (7), 923-930. [CrossRef]
- Love, M. I.; Huber, W.; Anders, S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014, 15, (12), 1-21. [CrossRef]
- Garrido-Godino, A. I.; Gupta, I.; Gutiérrez-Santiago, F.; Martínez-Padilla, A. B.; Alekseenko, A.; Steinmetz, L. M.; Pérez-Ortín, J. E.; Pelechano, V.; Navarro, F., Rpb4 and Puf3 imprint and post-transcriptionally control the stability of a common set of mRNAs in yeast. RNA biology 2021, 18, (8), 1206-1220. [CrossRef]
- Cuevas-Bermúdez, A.; Garrido-Godino, A. I.; Navarro, F., A novel yeast chromatin-enriched fractions purification approach, yChEFs, for the chromatin-associated protein analysis used for chromatin-associated and RNA-dependent chromatin-associated proteome studies from Saccharomyces cerevisiae. Gene Reports 2019, 16, 100450. [CrossRef]
- Cuevas-Bermúdez, A.; Garrido-Godino, A. I.; Gutiérrez-Santiago, F.; Navarro, F., A Yeast Chromatin-enriched Fractions Purification Approach, yChEFs, from Saccharomyces cerevisiae. Bio-Protocol 2020, 10, (1). [CrossRef]
- Garrido-Godino, A.; García-López, M.; García-Martínez, J.; Pelechano, V.; Medina, D.; Pérez-Ortín, J.; Navarro, F., Rpb1 foot mutations demonstrate a major role of Rpb4 in mRNA stability during stress situations in yeast. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2016, 1859, (5), 731-743. [CrossRef]
- Pelechano, V.; Wilkening, S.; Jarvelin, A. I.; Tekkedil, M. M.; Steinmetz, L. M., Genome-wide polyadenylation site mapping. Methods Enzymol 2012, 513, 271-96. [CrossRef]
- Wilkening, S.; Pelechano, V.; Jarvelin, A. I.; Tekkedil, M. M.; Anders, S.; Benes, V.; Steinmetz, L. M., An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res 2013, 41, (5), e65. [CrossRef]
- Robinson, M. D.; Grigull, J.; Mohammad, N.; Hughes, T. R., FunSpec: a web-based cluster interpreter for yeast. BMC bioinformatics 2002, 3, (1), 35. [CrossRef]
- Lykke-Andersen, S.; Jensen, T. H., Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nature reviews Molecular cell biology 2015, 16, (11), 665-677. [CrossRef]
- Mayer, A.; Heidemann, M.; Lidschreiber, M.; Schreieck, A.; Sun, M.; Hintermair, C.; Kremmer, E.; Eick, D.; Cramer, P., CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012, 336, (6089), 1723-5. [CrossRef]
- García-López, M. C.; Navarro, F., RNA polymerase II conserved protein domains as platforms for protein-protein interactions. Transcription 2011, 2, (4), 193-7. [CrossRef]
- Schulz, D.; Pirkl, N.; Lehmann, E.; Cramer, P., Rpb4 subunit functions mainly in mRNA synthesis by RNA polymerase II. Journal of Biological Chemistry 2014, 289, (25), 17446-17452. [CrossRef]
- Allepuz-Fuster, P.; Martínez-Fernández, V.; Garrido-Godino, A. I.; Alonso-Aguado, S.; Hanes, S. D.; Navarro, F.; Calvo, O., Rpb4/7 facilitates RNA polymerase II CTD dephosphorylation. Nucleic acids research 2014, 42, (22), 13674-13688. [CrossRef]
- Shalem, O.; Groisman, B.; Choder, M.; Dahan, O.; Pilpel, Y., Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet 2011, 7, (9), e1002273. [CrossRef]
- Fasken, M. B.; Leung, S. W.; Banerjee, A.; Kodani, M. O.; Chavez, R.; Bowman, E. A.; Purohit, M. K.; Rubinson, M. E.; Rubinson, E. H.; Corbett, A. H., Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. Journal of Biological Chemistry 2011, 286, (43), 37429-37445. [CrossRef]
- Ellison, M. A.; Lederer, A. R.; Warner, M. H.; Mavrich, T. N.; Raupach, E. A.; Heisler, L. E.; Nislow, C.; Lee, M. T.; Arndt, K. M., The Paf1 complex broadly impacts the transcriptome of Saccharomyces cerevisiae. Genetics 2019, 212, (3), 711-728. [CrossRef]
- Schneider, C.; Kudla, G.; Wlotzka, W.; Tuck, A.; Tollervey, D., Transcriptome-wide analysis of exosome targets. Molecular cell 2012, 48, (3), 422-433. [CrossRef]
- Goler-Baron, V.; Selitrennik, M.; Barkai, O.; Haimovich, G.; Lotan, R.; Choder, M., Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 2008, 22, (15), 2022-7. [CrossRef]
- Mitsuzawa, H.; Kanda, E.; Ishihama, A., Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts. Nucleic Acids Res 2003, 31, (16), 4696-701. [CrossRef]
- Kubicek, K.; Cerna, H.; Holub, P.; Pasulka, J.; Hrossova, D.; Loehr, F.; Hofr, C.; Vanacova, S.; Stefl, R., Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes & development 2012, 26, (17), 1891-1896. [CrossRef]
- Yurko, N.; Liu, X.; Yamazaki, T.; Hoque, M.; Tian, B.; Manley, J. L., MPK1/SLT2 links multiple stress responses with gene expression in budding yeast by phosphorylating Tyr1 of the RNAP II CTD. Molecular cell 2017, 68, (5), 913-925. e3. [CrossRef]
| Functional Category | p-value | |
|---|---|---|
| Up-regulated | purine nucleotide biosynthetic process [GO:0006164] | 7.81559e-05 |
| glycolysis [GO:0006096] | 0.000472358 | |
| response to stress [GO:0006950] | 0.000887445 | |
| metabolic process [GO:0008152] | 0.00134185 | |
| gluconeogenesis [GO:0006094] | 0.001615 | |
| cell adhesion [GO:0007155] | 0.00353101 | |
| manganese ion transport [GO:0006828] | 0.00466732 | |
| ‘de novo’ IMP biosynthetic process [GO:0006189] | 0.00594903 | |
| Down-regulated | transcription, DNA-dependent [GO:0006351] | 0.0009675 |
| sexual reproduction [GO:0019953] | 0.00111532 | |
| mating [GO:0007618] | 0.00111532 | |
| trehalose biosynthetic process [GO:0005992] | 0.00117173 | |
| regulation of transcription, DNA-dependent [GO:0006355] | 0.0014433 | |
| ATP-dependent chromatin remodeling [GO:0043044] | 0.004667 | |
| termination of RNA polymerase II transcription, exosome-dependent [GO:0030847] | 0.00500098 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





