Submitted:
30 November 2023
Posted:
01 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Transcriptome sequencing and De novo assembly
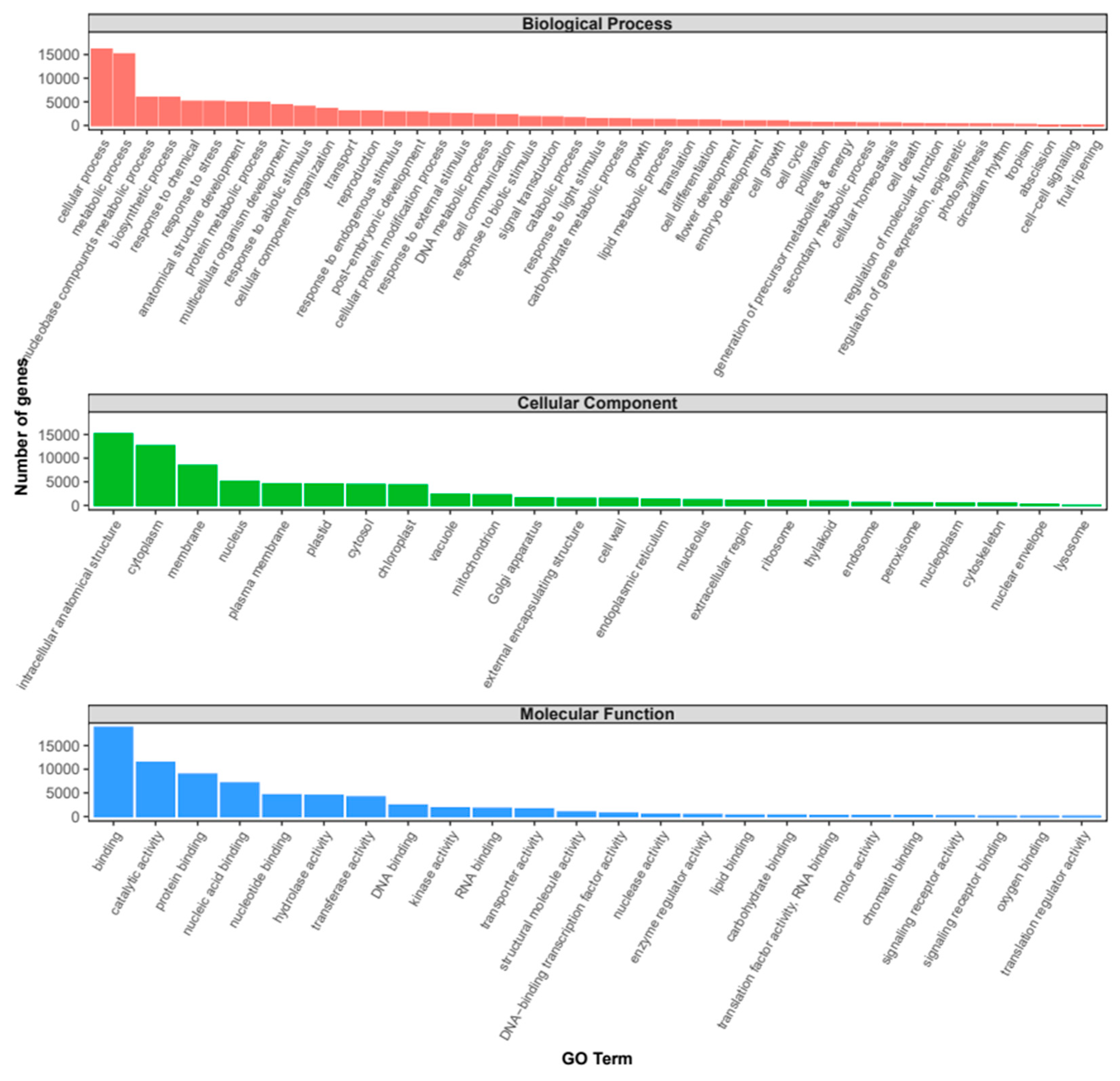
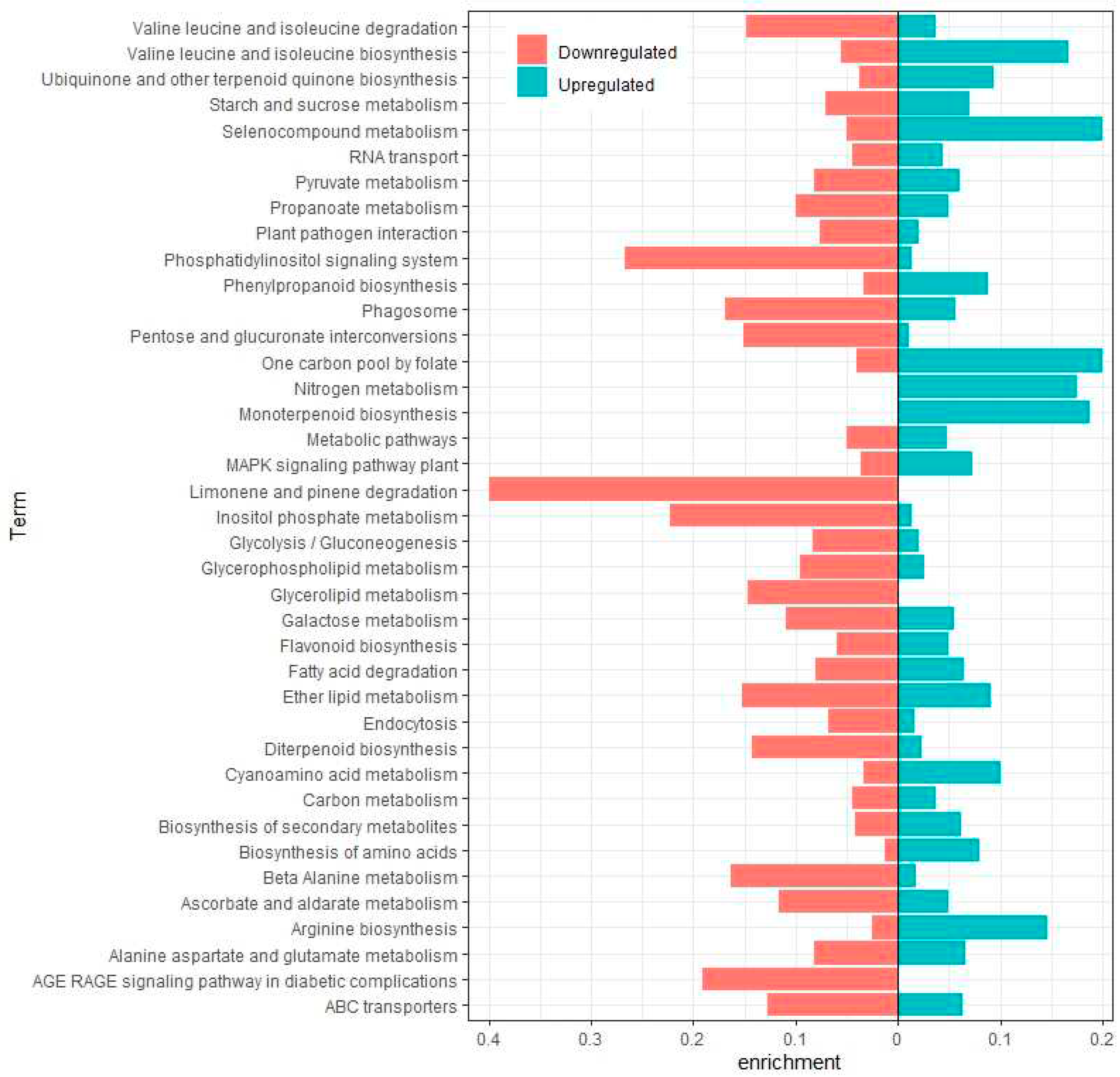
2.2. Annotation and differential expression analysis
2.3. Search of DEGs within QTLs intervals for autofertility
3. Discussion
3.1. Sequencing, assembly and annotation.
3.2. DEGs within QTL intervals previously described for autofertility
4. Materials and Methods
4.1. Plant materials and sample collection
4.2. RNA extraction, sequencing and De novo assembly
4.3. Annotation and differential expression analysis
4.4. Search of DEGs within QTL intervals previously described for autofertility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heslop-Harrison, Y.; Shivanna, K.R. The receptive surface of the angiosperm stigma. Ann Bot. 1977, 41, 1233–1258. [Google Scholar] [CrossRef]
- Heslop-Harrison, Y. Stigma characteristics and angiosperm taxonomy. Nord J Bot. 1981, 1, 401–420. [Google Scholar] [CrossRef]
- McInnis, S.M.; Costa, L.M.; Gutiérrez-Marcos, J.F.; Henderson, C.A.; Hiscock, S.J. Isolation and characterization of a polymorphic stigma-specific class III peroxidase gene from Senecio squalidus L. (Asteraceae). Plant Mol Biol. 2005, 57, 659–677. [Google Scholar] [CrossRef] [PubMed]
- Hiscock, S.J.; Allen, A.M. Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol. 2008, 179, 286–317. [Google Scholar] [CrossRef] [PubMed]
- Swanson, R.; Edlund, A.F.; Preuss, D. Species specificity in pollen-pistil interactions. Annu Rev Genet. 2004, 38, 793–818. [Google Scholar] [CrossRef] [PubMed]
- Zinkl, G.M.; Zwiebel, B.I.; Grier, D.G.; Preuss, D. Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999, 126, 5431–5440. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.; Sollapura, V.; Couroux, P.; Sprott, D.; Ravensdale, M.; Routly, E.; et al. The Brassica mature pollen and stigma proteomes: preparing to meet. Plant J. 2021, 107, 1546–1568. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.F.; Swanson, R.; Preuss, D. Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell. 2004, 16 Suppl: S84-97. [CrossRef]
- Allen, A.M.; Lexer, C.; Hiscock, S.J. Comparative analysis of pistil transcriptomes reveals conserved and novel genes expressed in dry, wet, and semidry stigmas. Plant Physiol. 2010, 154, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.L.; Xu, M.; Ma, F.F.; Chen, H.; Xu, X.H.; Gao, X.-Q.; et al. Comparative proteomic analysis reveals similar and distinct features of proteins in dry and wet stigmas. Proteomics. 2012, 12, 1983–1998. [Google Scholar] [CrossRef]
- Kenicer, G. Legumes of the World. Edited by G. Lewis, B. Schrire, B. MacKinder & M. Lock. Royal Botanic Gardens, Kew. 2005. xiv + 577pp., colour photographs & line drawings. ISBN 1 900 34780 6. £55.00 (hardback). Edin Jnl of Bot. 2006, 62, 195–196. [Google Scholar] [CrossRef]
- Chen, W.; Stoddard, F.L.; Baldwin, T.C. Developmental Regulation of Mannan, Arabinogalactan-Protein, and Pectic Epitopes in Pistils of Vicia faba (Faba Bean). Int J Plant Sci. 2006, 167, 919–932. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, L.; Nan, Z.; Wang, Y. Comparative transcriptional profiling provides insights into the evolution and development of the zygomorphic flower of Vicia sativa (Papilionoideae). PLoS ONE. 2013, 8, e57338. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Datta, S.; Shaw, A.K.; Thimmaiah, M.R.; Satya, P.; Mitra, J.; et al. De novo transcriptome of a bast fibre crop Crotalaria juncea reveals T2 ribonuclease genes to investigate late-acting self-rejection of pollens. Industrial Crops and Products 2023, 202, 117064. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.; Heslop-Harrison, Y. Pollen-Stigma Interaction in the Leguminosae: the Organization of the Stigma in Trifolium pratense L. Ann Bot. 1983, 51, 571–583. [Google Scholar] [CrossRef]
- Lord, E.M.; Heslop-Harrison, Y. Pollen-Stigma Interaction in the Leguminosae: Stigma Organization and the Breeding System in Vicia faba L. Ann Bot. 1984, 54, 827–836. [Google Scholar] [CrossRef]
- Costa, M.F.B.; Paulino, J.V.; Marinho, C.R.; Leite, V.G.; Pedersoli, G.D.; Teixeira, S.P. Stigma Diversity in Tropical Legumes with Considerations on Stigma Classification. Bot Rev. 2014, 80, 1–29. [Google Scholar] [CrossRef]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa Ariane, R.; Pinto, R.B.; et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny – The Legume Phylogeny Working Group (LPWG). Taxon. 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Hedhly, A.; Hormaza, J.I.; Herrero, M. Global warming and sexual plant reproduction. Trends Plant Sci. 2009, 14, 30–36. [Google Scholar] [CrossRef]
- Stoddard, F.L. Climate change can affect crop pollination in unexpected ways. J Exp Bot. 2017, 68, 1819–1821. [Google Scholar] [CrossRef]
- Rasmont, P.; Franzen, M.; Lecocq, T.; Harpke, A.; Roberts, S.; Biesmeijer, K.; et al. Climatic risk and distribution atlas of european bumblebees. BR. 2015, 10, 1–236. [Google Scholar] [CrossRef]
- Memmott, J.; Craze, P.G.; Waser, N.M.; Price, M.V. Global warming and the disruption of plant-pollinator interactions. Ecol Lett. 2007, 10, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Settele, J.; Bishop, J.; Potts, S.G. Climate change impacts on pollination. Nat Plants. 2016, 2, 16092. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, F.L.; Bond, D.A. The pollination requirements of the faba bean. Bee World. 1987, 68, 144–152. [Google Scholar] [CrossRef]
- Adcock, M.E.; Lawes, D.A. Self-fertility and the distribution of seed yield in Vicia faba L. Euphytica. 1976, 25, 89–96. [Google Scholar] [CrossRef]
- Kambal, A.E.; Bond, D.A.; Toynbee-Clarke, G. A study on the pollination mechanism in field beans ( Vicia faba L.). J Agric Sci. 1976, 87, 519. [Google Scholar] [CrossRef]
- Torres, A.M.; Moreno, M.T.; Cubero, J.I. Genetics of Six Components of Autofertility in Vicia faba. Plant Breeding. 1993, 110, 220–228. [Google Scholar] [CrossRef]
- Drayner, J.M. Self- and cross-fertility in field beans (Vicia faba Linn.). J Agric Sci. 1959, 53, 387–403. [Google Scholar] [CrossRef]
- Chen, W. Pollination, Fertilization and Floral Traits Co-Segregating with Autofertility in Faba Bean. Journal of New Seeds 2009, 10, 14–30. [Google Scholar] [CrossRef]
- Aguilar-Benitez, D.; Casimiro-Soriguer, I.; Ferrándiz, C.; Torres, A.M. Study and QTL mapping of reproductive and morphological traits implicated in the autofertility of faba bean. BMC Plant Biol. 2022, 22, 175. [Google Scholar] [CrossRef]
- Aguilar-Benitez, D.; Casimiro-Soriguer, I.; Gutiérrez, N.; Torres, A.M. High-density linkage map and fine QTL mapping of architecture, phenology and yield related traits in faba bean (Vicia faba L.).
- Khan, M.A.; Alghamdi, S.S.; Ammar, M.H.; Sun, Q.; Teng, F.; Migdadi, H.M.; et al. Transcriptome profiling of faba bean (Vicia faba L.) drought-tolerant variety hassawi-2 under drought stress using RNA sequencing. Electron J Biotechnol. 2019, 39, 15–29. [Google Scholar] [CrossRef]
- Lyu, J.I.; Ramekar, R.; Kim, J.M.; Hung, N.N.; Seo, J.S.; Kim, J.-B.; et al. Unraveling the complexity of faba bean (Vicia faba L.) transcriptome to reveal cold-stress-responsive genes using long-read isoform sequencing technology. Sci Rep. 2021, 11, 21094. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, S.; Seoane, P.; Bautista, R.; Palomino, C.; Claros, G.M.; Torres, A.M.; et al. Large-Scale Transcriptome Analysis in Faba Bean (Vicia faba L.) under Ascochyta fabae Infection. PLoS ONE. 2015, 10, e0135143. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Perdomo, E.; Vidal, A.; Kreplak, J.; Duborjal, H.; Leveugle, M.; Duarte, J.; et al. Development of new genetic resources for faba bean (Vicia faba L.) breeding through the discovery of gene-based SNP markers and the construction of a high-density consensus map. Sci Rep. 2020, 10, 6790. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Chen, H.; Liu, C.; Li, L.; Liu, L.; Han, X.; et al. Transcriptome profile analysis of two Vicia faba cultivars with contrasting salinity tolerance during seed germination. Sci Rep. 2020, 10, 7250. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Bian, X.-C.; Yang, F.; Chen, M.-X.; Das, D.; Zhu, X.-R.; et al. Comprehensive transcriptome analysis of faba bean in response to vernalization. Planta 2019, 251, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, R.; Liu, Y.; Hou, W.; Wang, X.; Miao, Y.; et al. Development and application of the Faba_bean_130K targeted next-generation sequencing SNP genotyping platform based on transcriptome sequencing. Theor Appl Genet. 2021, 134, 3195–3207. [Google Scholar] [CrossRef] [PubMed]
- Hou W, Zhang X, Liu Y, Liu Y, Feng BL. RNA-Seq and genetic diversity analysis of faba bean (Vicia faba L.) varieties in China. PeerJ. 2023, 11, e14259. [CrossRef] [PubMed]
- Wang, H.; Qi, X.; Chen, S.; Feng, J.; Chen, H.; Qin, Z.; et al. An integrated transcriptomic and proteomic approach to dynamically study the mechanism of pollen-pistil interactions during jasmine crossing. J Proteomics. 2021, 249, 104380. [Google Scholar] [CrossRef]
- He, Y.; Song, Q.; Chen, S.; Wu, Y.; Zheng, G.; Feng, J.; et al. Transcriptome analysis of self- and cross-pollinated pistils revealing candidate unigenes of self-incompatibility in Camellia oleifera. The Journal of Horticultural Science and Biotechnology 2020, 95, 19–31. [Google Scholar] [CrossRef]
- Quiapim, A.C.; Brito, M.S.; Bernardes, L.A.S.; Dasilva, I.; Malavazi, I.; DePaoli, H.C.; et al. Analysis of the Nicotiana tabacum stigma/style transcriptome reveals gene expression differences between wet and dry stigma species. Plant Physiol. 2009, 149, 1211–1230. [Google Scholar] [CrossRef]
- Gorelova, V.; Ambach, L.; Rébeillé, F.; Stove, C.; Van Der Straeten, D. Folates in plants: research advances and progress in crop biofortification. Front Chem. 2017, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Dötterl, S.; Gershenzon, J. Chemistry, biosynthesis and biology of floral volatiles: roles in pollination and other functions. Nat Prod Rep. 2023. [CrossRef] [PubMed]
- Chen, C.; Song, Q. Responses of the pollinating wasp Ceratosolen solmsi marchali to odor variation between two floral stages of Ficus hispida. J Chem Ecol. 2008, 34, 1536–1544. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; et al. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Bruce, T.J.A.; Martin, J.L.; Smart, L.E.; Pickett, J.A. Development of semiochemical attractants for monitoring bean seed beetle, Bruchus rufimanus. Pest Manag Sci. 2011, 67, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol Plant. 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Jamshed, M.; Sankaranarayanan, S.; Abhinandan, K.; Samuel, M.A. Stigma receptivity is controlled by functionally redundant MAPK pathway components in arabidopsis. Mol Plant. 2020, 13, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- McInnis, S.M.; Desikan, R.; Hancock, J.T.; Hiscock, S.J. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? New Phytol. 2006, 172, 221–228. [Google Scholar] [CrossRef]
- Xie, J.; Méndez, J.D.; Méndez-Valenzuela, V.; Aguilar-Hernández, M.M. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Janse van Rensburg, H.C.; Medvedev, S.; Demidchik, V.; et al. Glycation of Plant Proteins: Regulatory Roles and Interplay with Sugar Signalling? Int J Mol Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.-W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem J. 2009, 421, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Thole, J.M.; Nielsen, E. Phosphoinositides in plants: novel functions in membrane trafficking. Curr Opin Plant Biol. 2008, 11, 620–631. [Google Scholar] [CrossRef]
- Preuss, M.L.; Schmitz, A.J.; Thole, J.M.; Bonner, H.K.S.; Otegui, M.S.; Nielsen, E. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol. 2006, 172, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yan, A.; Feijó, J.A.; Furutani, M.; Takenawa, T.; Hwang, I.; et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell. 2010, 22, 4031–4044. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C. LARP6 proteins in plants. Biochem Soc Trans. 2021, 49, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; et al. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jiang, B.; Xie, R.; Xu, M.; Li, H.; Liu, K. Genome-wide identification and analysis of Medicago truncatula Small auxin upregulated RNA (SAUR) gene family uncover their roles in nodule formation. J Plant Biochem Biotechnol. 2020. [CrossRef]
- Bultreys, A.; Trombik, T.; Drozak, A.; Boutry, M. Nicotiana plumbaginifolia plants silenced for the ATP-binding cassette transporter gene NpPDR1 show increased susceptibility to a group of fungal and oomycete pathogens. Mol Plant Pathol. 2009, 10, 651–663. [Google Scholar] [CrossRef]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell. 2010, 22, 1483–1497. [Google Scholar] [CrossRef]
- Zhu, B.; Li, H.; Xia, X.; Meng, Y.; Wang, N.; Li, L.; et al. ATP-Binding Cassette G Transporters SGE1 and MtABCG13 Control Stigma Exsertion. Plant Physiol. 2020, 184, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.T.; Choi, H.; Palmgren, M.; Martinoia, E.; Hwang, J.-U.; Lee, Y. Arabidopsis ABCG28 is required for the apical accumulation of reactive oxygen species in growing pollen tubes. Proc Natl Acad Sci USA. 2019, 116, 12540–12549. [Google Scholar] [CrossRef]
- Aguilar-Benitez, D.; Casimiro-Soriguer, I.; Torres, A.M. First approach to pod dehiscence in faba bean: genetic and histological analyses. Sci Rep. 2020, 10, 17678. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Benitez, D.; Casimiro-Soriguer, I.; Maalouf, F.; Torres, A.M. Linkage mapping and QTL analysis of flowering time in faba bean. Sci Rep. 2021, 11, 13716. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Bucchini F, Del Cortona A, Kreft Ł, Botzki A, Van Bel M, Vandepoele K. TRAPID 2.0: a web application for taxonomic and functional analysis of de novo transcriptomes. Nucleic Acids Res. 2021, 49, e101. [CrossRef] [PubMed]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010, 26, 139–140. [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. 2022.
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- O’Sullivan, D.M.; Angra, D.; Harvie, T.; Tagkouli, V.; Warsame, A. A genetic toolbox for Vicia faba improvement. 2019.
- Khazaei, H.; O’Sullivan, D.M.; Stoddard, F.L.; Adhikari, K.N.; Paull, J.G.; Schulman, A.H.; et al. Recent advances in faba bean genetic and genomic tools for crop improvement. Legume Science 2021, 3, e75. [Google Scholar] [CrossRef]
- Jayakodi, M.; Golicz, A.A.; Kreplak, J.; Fechete, L.I.; Angra, D.; Bednář, P.; et al. The giant diploid faba genome unlocks variation in a global protein crop. Nature 2023, 615, 652–659. [Google Scholar] [CrossRef] [PubMed]
| Sample | Raw reads | Number of bases (Gb) | Clean reads |
|---|---|---|---|
| AF27.18 | 133,319,602 | 19,997 | 121,359,412 |
| AF27.28 | 65,839,438 | 9,875 | 60,315,740 |
| AF27.5 | 61,476,838 | 9,221 | 56,683,402 |
| AF44.1 | 55,001,874 | 8,250 | 49,763,678 |
| AF44.4 | 65,871,434 | 9,880 | 59,782,290 |
| AF44.14 | 64,747,566 | 9,712 | 59,231,922 |
| AF14.2 | 56,963,200 | 8,544 | 43,002,584 |
| AF14.5 | 70,837,140 | 10,625 | 65,081,586 |
| AF14.16 | 63,834,224 | 9,575 | 58,881,026 |
| AS6.3 | 73,167,636 | 10,975 | 67,520,844 |
| AS6.5 | 57,074,180 | 8,561 | 52,288,334 |
| AS6.14 | 47,358,170 | 7,103 | 42,875,694 |
| AS96.1 | 68,186,700 | 10,228 | 62,004,170 |
| AS96.9 | 63,583,396 | 9,537 | 58,218,214 |
| AS96.12 | 57,687,624 | 8,653 | 51,643,386 |
| AS19.2 | 57,687,624 | 9,500 | 58,475,436 |
| AS19.3 | 63,147,560 | 9,472 | 57,224,622 |
| AS19.9 | 57,643,612 | 8,646 | 52,847,570 |
| Total | 1,189,079,630 | 1,077,199,910 |
| Trait | Chr | QTL marker | Gene | Transcript ID | Regulation | Organism | Protein ID | Protein description | Arabidopsis ID |
|---|---|---|---|---|---|---|---|---|---|
| PSF_2009/10 + PSC_2014/15 | CHR1 | AX_416823680 | 1g073840 | TRINITY_DN9113_c0_g1 | UR | P. sativum | XP_050883094.1 | protochlorophyllide reductase, chloroplastic | AT5G54190 |
| SSF_2012/13 | CHR1 | AX_181472988 | 1g097120 | TRINITY_DN15728_c0_g1 | DR | P. sativum | XP_050915769.1 | uncharacterized protein LOC127130849 isoform X1 | AT5G61750 |
| SL | CHR1 | AX_416737058 | 1g289680 | TRINITY_DN36521_c0_g1 | DR | P. sativum | XP_050886087.1 | PUTATIVE CLATHRIN ASSEMBLY PROTEIN | AT5G35200 |
| SL/FL | CHR1 | AX_181488729 | 1g290400 | TRINITY_DN21322_c0_g1 | DR | P. sativum | XP_050890085.1 | deSI-like protein At4g17486 | AT4G17486 |
| SL | CHR1 | AX_416788157 | 1g291400 | TRINITY_DN59629_c0_g1 | DR | P. sativum | XP_050920567.1 | UNCHARACTERIZED PROTEIN LOC127138220 | AT4G21215 |
| TOTALS + NORMALQ + NORMAL% +RATIO_PSIZE | CHR2 | AX_416731323 | 2g022600 | TRINITY_DN12936_c0_g1 | UR | Medicago truncatula | XP_013454609.1 | DCN1-like protein 4 | AT1G15860 |
| PSC_2008/09 | CHR4 | AX_416818743 | 4g037560 | TRINITY_DN1719_c0_g2 | UR | P. sativum | XP_050882017.1 | DNA damage-repair/toleration protein DRT100-like | AT3G12610 |
| PSC_2008/09 | CHR4 | AX_416752208 | 4g039440 | TRINITY_DN27765_c0_g4 | DR | P. sativum | XP_050873114.1 | phosphatidylinositol 4-phosphate 5-kinase 6-like | AT3G07960 |
| PSF_2006/07 | CHR4 | AX_181454844 | 4g050680 | TRINITY_DN4382_c0_g1 | DR | P. sativum | XP_050873622.1 | probable serine/threonine-protein kinase WNK10 isoform X1 | AT3G19910 |
| PSF_2006/07 | CHR4 | AX_416805445 | 4g051160 | TRINITY_DN59133_c0_g1 | DR | P. sativum | XP_050873638.1 | AAA-ATPase At2g46620 | AT2G46620 |
| PSF_2006/07 | CHR4 | AX_416752041 | 4g052320 | TRINITY_DN4671_c0_g1 | UR | P. sativum | XP_050873656.1 | NAC domain-containing protein 43 | AT2G46770 |
| PSF_2006/07 | CHR4 | AX_181491519 | 4g054640 | TRINITY_DN2527_c0_g1 | UR | P. sativum | XP_050873724.1 | protein STRUBBELIG-RECEPTOR FAMILY 6-like | AT1G53730 |
| PSF_2010/11 | CHR4 | AX_181454259 | 4g074360 | TRINITY_DN15070_c0_g1 | UR | P. sativum | XP_050874209.1 | putative disease resistance protein RGA3 | AT3G14470 |
| PSF_2010/11 | CHR4 | AX_416818256 | 4g079840 | TRINITY_DN41039_c0_g1 | UR | P. sativum | XP_050874330.1 | hydroquinone glucosyltransferase-like | AT4G01070 |
| PSF_2010/11 | CHR4 | AX_416818256 | 4g079840 | TRINITY_DN51281_c0_g1 | UR | P. sativum | XP_050874330.1 | hydroquinone glucosyltransferase-like | AT4G01070 |
| PSF_2010/11 | CHR4 | AX_181179181 | 4g082280 | TRINITY_DN15138_c0_g1 | DR | P. sativum | XP_050919587.1 | peroxidase 31-like | AT5G47000 |
| PSF_2010/11 | CHR4 | AX_181179181 | 4g082280 | TRINITY_DN57808_c0_g1 | DR | P. sativum | XP_050919587.1 | peroxidase 31-like | AT5G47000 |
| PSF_2010/11 | CHR4 | AX_181179181 | 4g082280 | TRINITY_DN15138_c0_g2 | DR | P. sativum | XP_050919587.1 | peroxidase 31-like | AT5G47000 |
| PSF_2010/11 | CHR4 | AX_181467359 | 4g085240 | TRINITY_DN23725_c0_g1 | UR | P. sativum | XP_050872529.1 | transcription factor SRM1-like | AT5G08520 |
| PSF_2010/11 | CHR4 | AX_181147515 | 4g094400 | TRINITY_DN60717_c0_g1 | DR | P. sativum | XP_050872421.1 | probable calcium-binding protein CML18 | AT1G32250 |
| PSF_2010/11 | CHR4 | AX_181183310 | 4g117560 | TRINITY_DN1894_c0_g2 | UR | P. sativum | XP_050871881.1 | GTPase ERA-like, chloroplastic | AT5G66470 |
| PSF_2010/11 | CHR4 | AX_416773947 | 4g123400 | TRINITY_DN40735_c0_g1 | DR | Vicia faba | CAI8613174.1 | unnamed protein product [Vicia faba] | NA |
| PSF_2010/11 | CHR4 | AX_416777102 | 4g127640 | TRINITY_DN27369_c0_g1 | UR | P. sativum | XP_050871669.1 | cytochrome P450 81E8-like | AT4G37370 |
| PSF_2010/11 | CHR4 | AX_416767177 | 4g127880 | TRINITY_DN2843_c0_g1 | UR | Glycine max | XP_003527222.1 | UDP-glycosyltransferase 79B3 | AT4G27570 |
| PSF_2010/11 | CHR4 | AX_181488376 | 4g128040 | TRINITY_DN41664_c0_g1 | DR | P. sativum | XP_050871664.1 | metal transporter Nramp3-like | AT1G47240 |
| PSF_2010/11 | CHR4 | AX_181497023 | 4g131840 | TRINITY_DN704_c0_g1 | UR | P. sativum | XP_050871597.1 | probable xyloglucan endotransglucosylase/hydrolase protein 6 | AT5G65730 |
| PAPL | CHR4 | AX_416742799 | 4g228400 | TRINITY_DN60908_c0_g1 | DR | P. sativum | XP_050920799.1 | temperature-induced lipocalin-1-like [Pisum sativum] | AT5G58070 |
| SSC_2009/10 + OL/FL | CHR6 | AX_416737553 | 6g112200 | TRINITY_DN22338_c0_g1 | DR | P. sativum | XP_050901848.1 | stress-related protein-like | AT3G05500 |
| SSF_2012/13 + TOTALS + NORMALQ + NORMAL% + RATIO_PSIZE | CHR6 | AX_416792345 | 6g120160 | TRINITY_DN46243_c0_g1 | DR | P. sativum | XP_050900348.1 | la-related protein 6C (LARP6c) isoform X1 | AT3G19090 |
| SSF_2012/13 + TOTALS + NORMALQ + NORMAL% + RATIO_PSIZE | CHR6 | AX_416792345 | 6g120160 | TRINITY_DN61454_c0_g1 | DR | P. sativum | XP_050900348.1 | la-related protein 6C (LARP6c) isoform X1 | AT3G19090 |
| NORMALQ | CHR6 | AX_416723935 | 6g122960 | TRINITY_DN35465_c0_g1 | DR | P. sativum | XP_050898899.1 | U-box domain-containing protein 26-like | AT1G49780 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





