Submitted:
25 November 2023
Posted:
27 November 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Protein Preparation
2.2. Ligand preparation
2.3. Molecular docking
2.4. MM-GBSA calculations
2.5. Molecular dynamics studies
2.6. Entropy calculation for molecular dynamics trajectories
3. Results and Discussion
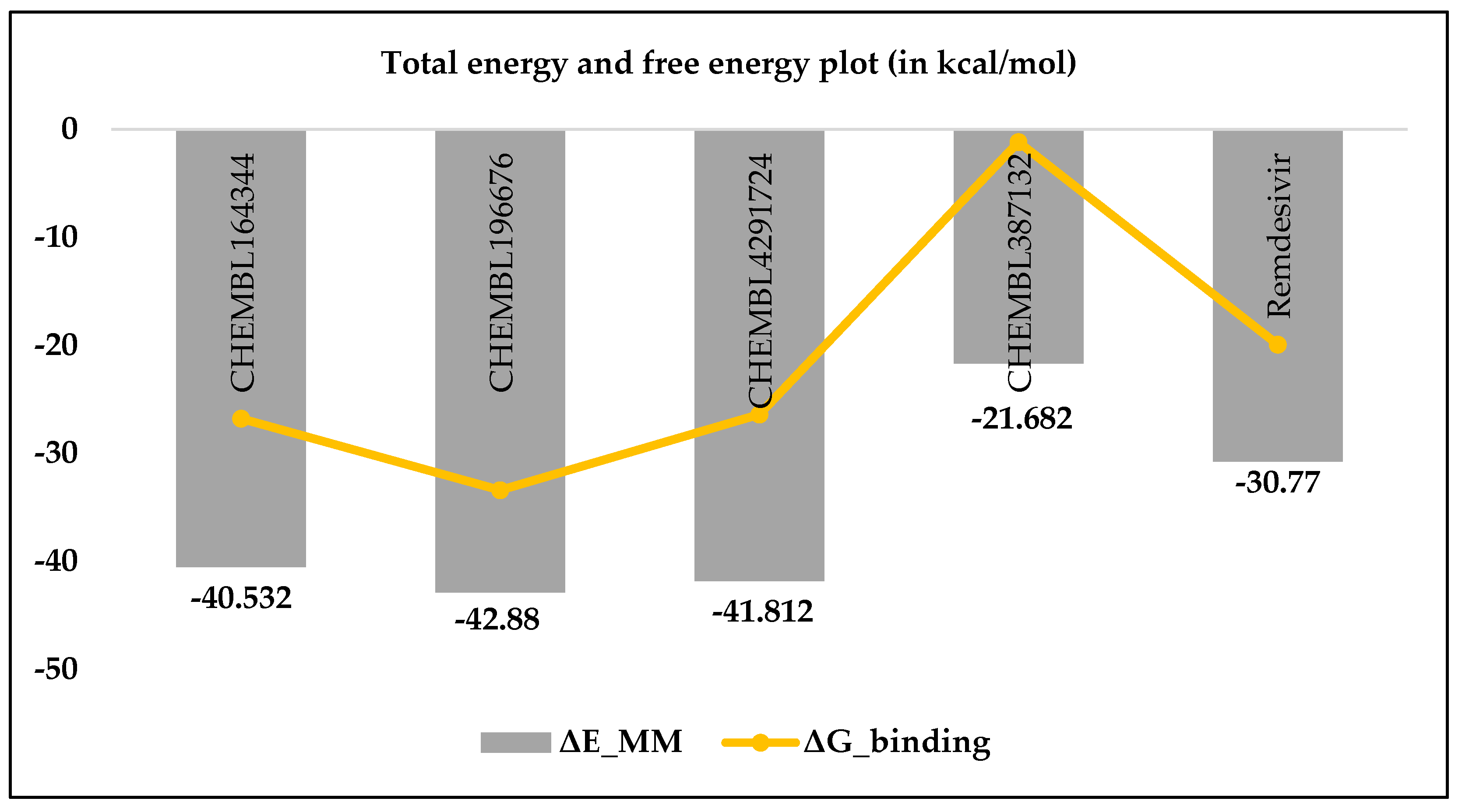
3.1. MM-GBSA studies
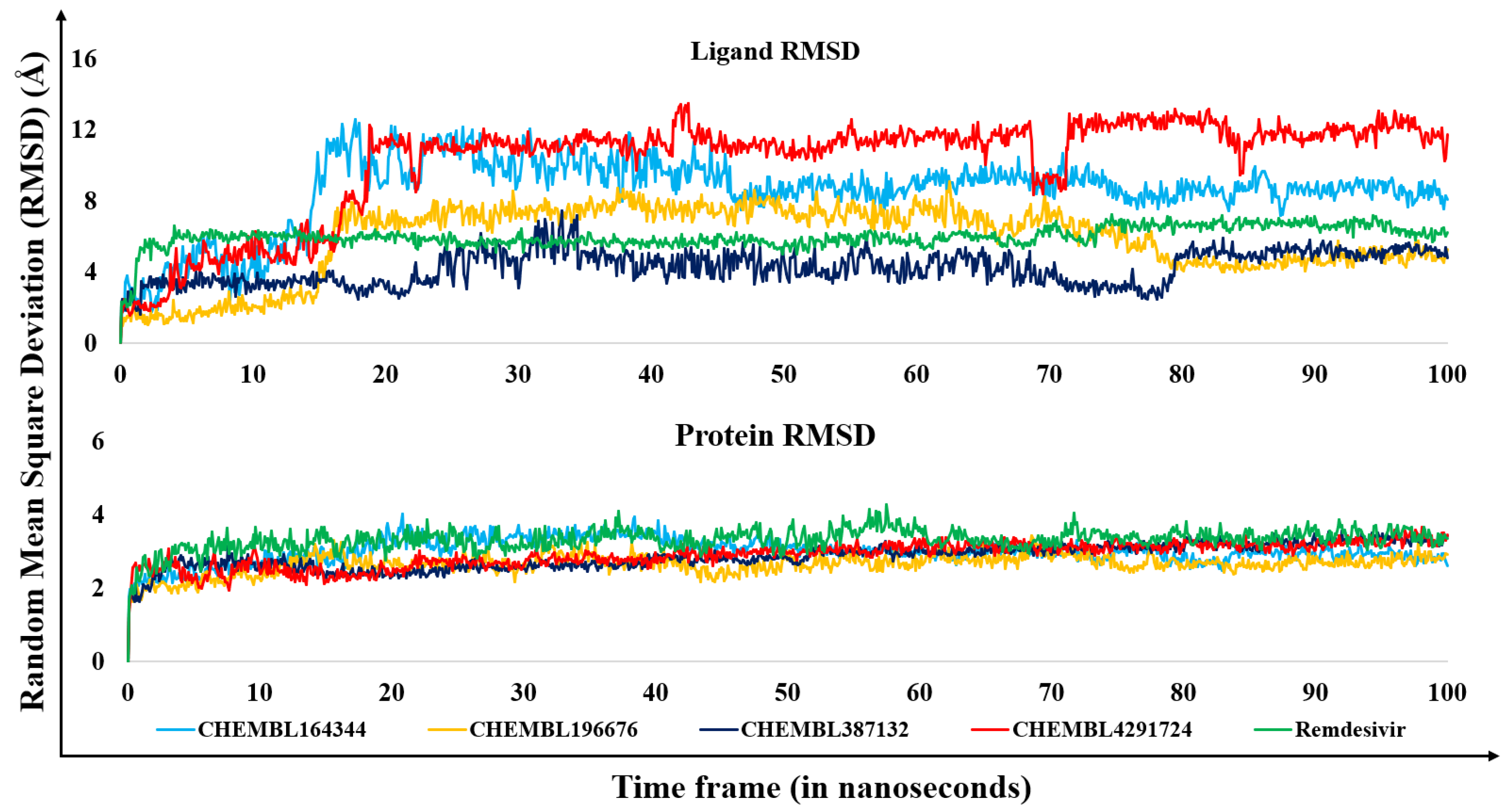
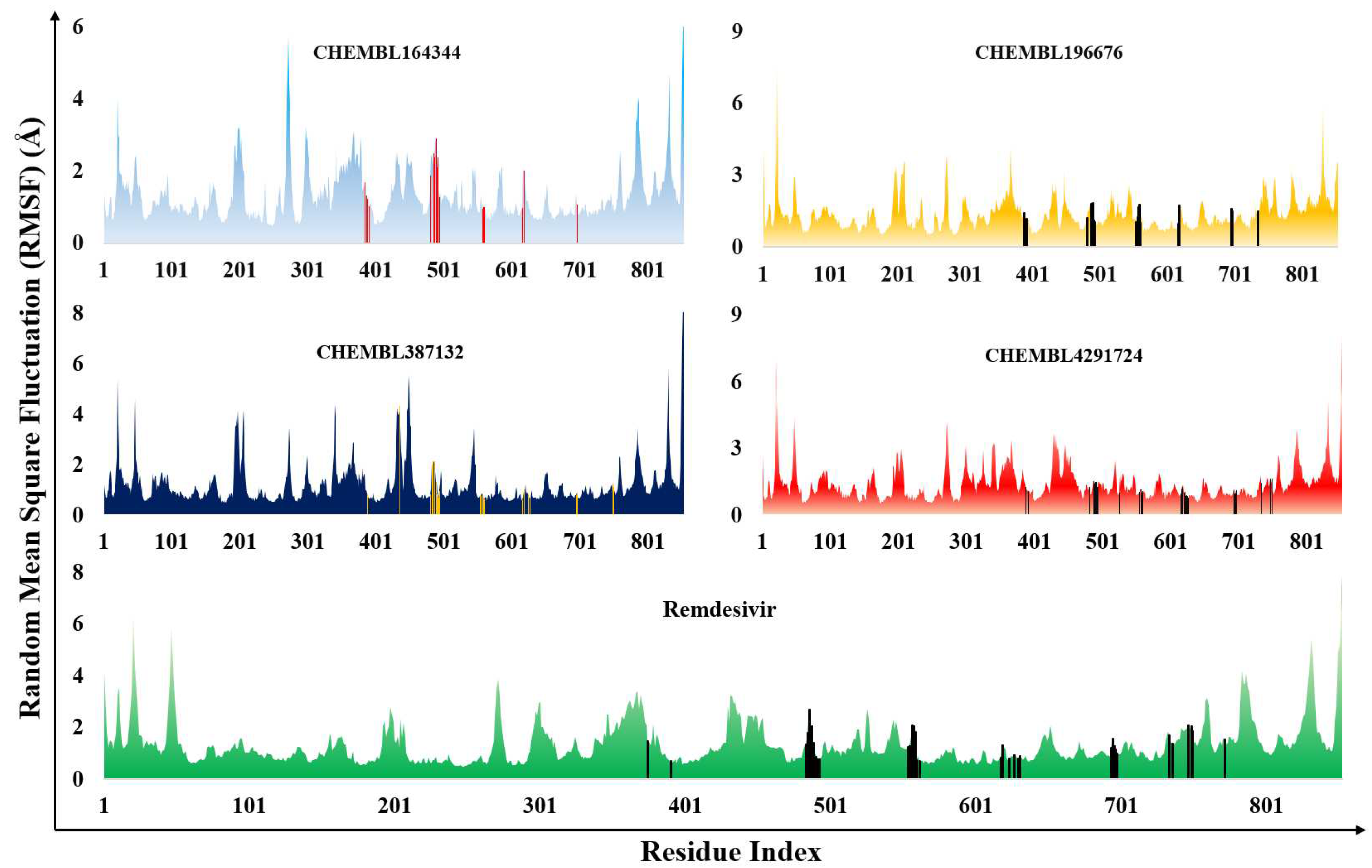
3.2. Molecular dynamics results
3.3. Estimation of entropic contribution by gmx_MMPBSA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mesel-Lemoine, M.; Millet, J.; Vidalain, P.-O.; Law, H.; Vabret, A.; Lorin, V.; Escriou, N.; Albert, M.L.; Nal, B.; Tangy, F. A Human Coronavirus Responsible for the Common Cold Massively Kills Dendritic Cells but Not Monocytes. J. Virol. 2012, 86, 7577–7587. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A Brief Summary and Comparison of Severe Acute Respiratory Infections Caused by Three Highly Pathogenic Human Coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. Available online: https://covid19.who.int/ (accessed on 2 November 2023).
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; Mccomsey, G.A.; Mccorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-Acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 1–24. [Google Scholar] [CrossRef]
- Office for National Statistics (ONS) Prevalence of Ongoing Symptoms Following Coronavirus (COVID-19) Infection in the UK: 2 February 2023. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/prevalenceofongoingsymptomsfollowingcoronaviruscovid19infectionintheuk/2february2023 (accessed on 6 November 2023).
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. acta. Mol. basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and Functions of Coronavirus Replication–Transcription Complexes and Their Relevance for SARS-CoV-2 Drug Design. Nat. Rev. Mol. Cell Biol. 2021, 23, 21–39. [Google Scholar] [CrossRef]
- Gangadharan, S.; Ambrose, J.M.; Rajajagadeesan, A.; Kullappan, M.; Patil, S.; Gandhamaneni, S.H.; Veeraraghavan, V.P.; Nakkella, A.K.; Agarwal, A.; Jayaraman, S.; et al. Repurposing of Potential Antiviral Drugs against RNA-Dependent RNA Polymerase of SARS-CoV-2 by Computational Approach. J. Infect. Public Health 2022, 15, 1180–1191. [Google Scholar] [CrossRef]
- Sivaraman, H.; Er, S.Y.; Choong, Y.K.; Gavor, E.; Sivaraman, J. Structural Basis of SARS-CoV-2- and SARS-CoV-Receptor Binding and Small-Molecule Blockers as Potential Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 465–493. [Google Scholar] [CrossRef]
- Bertolin, A.P.; Weissmann, F.; Zeng, J.; Posse, V.; Milligan, J.C.; Canal, B.; Ulferts, R.; Wu, M.; Drury, L.S.; Howell, M.; et al. Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of Nsp12/7/8 RNA-Dependent RNA Polymerase. Biochem. J. 2021, 478, 2425–2443. [Google Scholar] [CrossRef]
- Baby, K.; Maity, S.; Mehta, C.H.; Suresh, A.; Nayak, U.Y.; Nayak, Y. Targeting SARS-CoV-2 RNA-Dependent RNA Polymerase: An in Silico Drug Repurposing for COVID-19. F1000Research 2020, 9, 1166. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef]
- Ahmad, J.; Ikram, S.; Ahmad, F.; Rehman, I.U.; Mushtaq, M. SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) – A Drug Repurposing Study. Heliyon 2020, 6, e04502. [Google Scholar] [CrossRef]
- El Sohaimy, S.; Abdo, N.; Shehata, M.; Moheyeldin, O. Inhibition of COVID-19 RNA-Dependent RNA Polymerase by Natural Bioactive Compounds: Molecular Docking Analysis. Egypt. J. Chem. 2021, 64, 1989–2001. [Google Scholar] [CrossRef]
- McDonald, E.G.; Lee, T.C. Nirmatrelvir-Ritonavir for COVID-19. CMAJ 2022, 194, E218. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA) FDA Approves First Oral Antiviral for Treatment of COVID-19 in Adults. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-oral-antiviral-treatment-covid-19-adults (accessed on 6 November 2023).
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Manabe, T.; Kambayashi, D.; Akatsu, H.; Kudo, K. Favipiravir for the Treatment of Patients with COVID-19: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 489. [Google Scholar] [CrossRef]
- McAuley, A.J.; Jansen van Vuren, P.; Mohammed, M.-U.-R.; Faheem; Goldie, S.; Riddell, S.; Gödde, N.J.; Styles, I.K.; Bruce, M.P.; Chahal, S.; et al. Use of Human Lung Tissue Models for Screening of Drugs against SARS-CoV-2 Infection. Viruses 2022, 14, 2417. [Google Scholar] [CrossRef]
- Reis, G.; dos Santos Moreira-Silva, E.A.; Silva, D.C.M.; Thabane, L.; Milagres, A.C.; Ferreira, T.S.; dos Santos, C.V.Q.; de Souza Campos, V.H.; Nogueira, A.M.R.; de Almeida, A.P.F.G.; et al. Effect of Early Treatment with Fluvoxamine on Risk of Emergency Care and Hospitalisation among Patients with COVID-19: The TOGETHER Randomised, Platform Clinical Trial. Lancet Glob. Heal. 2022, 10, e42–e51. [Google Scholar] [CrossRef]
- Butler, C.C.; Hobbs, F.D.R.; Gbinigie, O.A.; Rahman, N.M.; Hayward, G.; Richards, D.B.; Dorward, J.; Lowe, D.M.; Standing, J.F.; Breuer, J.; et al. Molnupiravir plus Usual Care versus Usual Care Alone as Early Treatment for Adults with COVID-19 at Increased Risk of Adverse Outcomes (PANORAMIC): An Open-Label, Platform-Adaptive Randomised Controlled Trial. Lancet 2023, 401, 281–293. [Google Scholar] [CrossRef]
- Wise, J. Covid-19: Molnupiravir Does Not Cut Hospital Admissions or Deaths in Vaccinated People at High Risk, Trial Finds. BMJ 2022, o3055. [Google Scholar] [CrossRef]
- Kozlov, M. Merck’s COVID Pill Loses Its Lustre: What That Means for the Pandemic. Nature 2021. [Google Scholar] [CrossRef]
- Batool, S.; Vuthaluru, K.; Hassan, A.; Bseiso, O.; Tehseen, Z.; Pizzorno, G.; Rodriguez Reyes, Y.; Saleem, F. Efficacy and Safety of Favipiravir in Treating COVID-19 Patients: A Meta-Analysis of Randomized Control Trials. Cureus 2023, 15, e33676. [Google Scholar] [CrossRef]
- Bosaeed, M.; Alharbi, A.; Mahmoud, E.; Alrehily, S.; Bahlaq, M.; Gaifer, Z.; Alturkistani, H.; Alhagan, K.; Alshahrani, S.; Tolbah, A.; et al. Efficacy of Favipiravir in Adults with Mild COVID-19: A Randomized, Double-Blind, Multicentre, Placebo-Controlled Clinical Trial. Clin. Microbiol. Infect. 2022, 28, 602–608. [Google Scholar] [CrossRef]
- Shah, P.L.; Orton, C.M.; Grinsztejn, B.; Donaldson, G.C.; Crabtree Ramírez, B.; Tonkin, J.; Santos, B.R.; Cardoso, S.W.; Ritchie, A.I.; Conway, F.; et al. Favipiravir in Patients Hospitalised with COVID-19 (PIONEER Trial): A Multicentre, Open-Label, Phase 3, Randomised Controlled Trial of Early Intervention versus Standard Care. Lancet Respir. Med. 2023, 11, 415–424. [Google Scholar] [CrossRef]
- Siripongboonsitti, T.; Ungtrakul, T.; Tawinprai, K.; Nimmol, T.; Buttakosa, M.; Sornsamdang, G.; Jarrusrojwuttikul, T.; Silapant, P.; Mahanonda, N. Efficacy of Combination Therapy of Fluvoxamine and Favipiravir vs Favipiravir Monotherapy to Prevent Severe COVID-19 among Mild to Moderate COVID-19 Patients: Open-Label Randomized Controlled Trial (EFFaCo Study). Int. J. Infect. Dis. 2023, 134, 211–219. [Google Scholar] [CrossRef]
- Lenze, E.J.; Mattar, C.; Zorumski, C.F.; Stevens, A.; Schweiger, J.; Nicol, G.E.; Miller, J.P.; Yang, L.; Yingling, M.; Avidan, M.S.; et al. Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients With Symptomatic COVID-19. JAMA 2020, 324, 2292. [Google Scholar] [CrossRef] [PubMed]
- Calusic, M.; Marcec, R.; Luksa, L.; Jurkovic, I.; Kovac, N.; Mihaljevic, S.; Likic, R. Safety and Efficacy of Fluvoxamine in COVID-19 ICU Patients: An Open Label, Prospective Cohort Trial with Matched Controls. Br. J. Clin. Pharmacol. 2022, 88, 2065–2073. [Google Scholar] [CrossRef]
- Muzaffar-Ur-Rehman, M.; Suryakant, C.K.; Chandu, A.; Kumar, B.K.; Joshi, R.P.; Jadav, S.R.; Sankaranarayanan, M.; Vasan, S.S. Molecular Docking and Dynamics Identify Potential Drugs to Be Repurposed as SARS-CoV-2 Inhibitors. J. Comput. Biophys. Chem. 2023, 1–23. [Google Scholar] [CrossRef]
- Zamzami, M.A. Molecular Docking, Molecular Dynamics Simulation and MM-GBSA Studies of the Activity of Glycyrrhizin Relevant Substructures on SARS-CoV-2 RNA-Dependent-RNA Polymerase. J. Biomol. Struct. Dyn. 2023, 41, 1846–1858. [Google Scholar] [CrossRef]
- Brunt, D.; Lakernick, P.M.; Wu, C. Discovering New Potential Inhibitors to SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Using High Throughput Virtual Screening and Molecular Dynamics Simulations. Sci. Rep. 2022, 12, 19986. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lu, W.; Jiang, H.; Yang, C.; Dong, X. Molecular Docking and Dynamics of Phytochemicals From Chinese Herbs With SARS-CoV-2 RdRp. Nat. Prod. Commun. 2022, 17, 1934578X2211056. [Google Scholar] [CrossRef]
- Askari, F.S.; Ebrahimi, M.; Parhiz, J.; Hassanpour, M.; Mohebbi, A.; Mirshafiey, A. Digging for the Discovery of SARS-CoV-2 Nsp12 Inhibitors: A Pharmacophore-Based and Molecular Dynamics Simulation Study. Future Virol. 2022, 17, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Uengwetwanit, T.; Chutiwitoonchai, N.; Wichapong, K.; Karoonuthaisiri, N. Identification of Novel SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Inhibitors: From in Silico Screening to Experimentally Validated Inhibitory Activity. Comput. Struct. Biotechnol. J. 2022, 20, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Chinnamadhu, A.; Ramakrishnan, J.; Suresh, S.; Ramadurai, P.; Poomani, K. Dynamics and Binding Affinity of Nucleoside and Non-Nucleoside Inhibitors with RdRp of SARS-CoV-2: A Molecular Screening, Docking, and Molecular Dynamics Simulation Study. J. Biomol. Struct. Dyn. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, F.A.; Alkarim, S.A.; Hawsawi, Y.M.; Abdulaal, W.H.; Albiheyri, R.; Kurdi, B.; Alguridi, H.; El-Magd, M.A. 25 (S)-Hydroxycholesterol Acts as a Possible Dual Enzymatic Inhibitor of SARS-CoV-2 M pro and RdRp–: An Insight from Molecular Docking and Dynamics Simulation Approaches. J. Biomol. Struct. Dyn. 2023, 41, 4744–4755. [Google Scholar] [CrossRef] [PubMed]
- Alexpandi, R.; De Mesquita, J.F.; Pandian, S.K.; Ravi, A.V. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in Silico Analysis. Front. Microbiol. 2020, 11, 1796. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.P.; Singh, A.K.; Bansal, T.; Yadav, A.; Prajapati, K.S.; Shuaib, M.; Kumar, S. Identification of Natural Inhibitors Against SARS-CoV-2 Drugable Targets Using Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Approach. Front. Cell. Infect. Microbiol. 2021, 11, 730288. [Google Scholar] [CrossRef]
- Shady, N.H.; Hayallah, A.M.; Mohamed, M.F.A.; Ghoneim, M.M.; Chilingaryan, G.; Al-Sanea, M.M.; Fouad, M.A.; Kamel, M.S.; Abdelmohsen, U.R. Targeting 3CLpro and SARS-CoV-2 RdRp by Amphimedon Sp. Metabolites: A Computational Study. Molecules 2021, 26, 3775. [Google Scholar] [CrossRef]
- Gajjar, N.D.; Dhameliya, T.M.; Shah, G.B. In Search of RdRp and Mpro Inhibitors against SARS CoV-2: Molecular Docking, Molecular Dynamic Simulations and ADMET Analysis. J. Mol. Struct. 2021, 1239, 130488. [Google Scholar] [CrossRef]
- Parihar, A.; Sonia, Z.F.; Akter, F.; Ali, M.A.; Hakim, F.T.; Hossain, M.S. Phytochemicals-Based Targeting RdRp and Main Protease of SARS-CoV-2 Using Docking and Steered Molecular Dynamic Simulation: A Promising Therapeutic Approach for Tackling COVID-19. Comput. Biol. Med. 2022, 145, 105468. [Google Scholar] [CrossRef]
- M A Kawsar, S.; Hosen, M.A.; Ahmad, S.; El Bakri, Y.; Laaroussi, H.; Ben Hadda, T.; Almalki, F.A.; Ozeki, Y.; Goumri-Said, S. Potential SARS-CoV-2 RdRp Inhibitors of Cytidine Derivatives: Molecular Docking, Molecular Dynamic Simulations, ADMET, and POM Analyses for the Identification of Pharmacophore Sites. PLoS One 2022, 17, e0273256. [Google Scholar] [CrossRef]
- Veerasamy, R.; Karunakaran, R. Molecular Docking Unveils the Potential of Andrographolide Derivatives against COVID-19: An in Silico Approach. J. Genet. Eng. Biotechnol. 2022, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, S.; Ambrose, J.M.; Rajajagadeesan, A.; Kullappan, M.; Patil, S.; Gandhamaneni, S.H.; Veeraraghavan, V.P.; Nakkella, A.K.; Agarwal, A.; Jayaraman, S.; et al. Repurposing of Potential Antiviral Drugs against RNA-Dependent RNA Polymerase of SARS-CoV-2 by Computational Approach. J. Infect. Public Health 2022, 15, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.D.; Jeon, S.; Kim, S.; Lee, S.Y. Drugs Repurposed for COVID-19 by Virtual Screening of 6,218 Drugs and Cell-Based Assay. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2024302118. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, M.; Chen, W.; Xiao, D.; Wang, C. Computational Molecular Docking and Virtual Screening Revealed Promising SARS-CoV-2 Drugs. Precis. Clin. Med. 2021, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- El Hassab, M.A.; Hemeda, L.R.; Elsayed, Z.M.; Al-Rashood, S.T.; Abdel-Hamid Amin, M.K.; Abdel-Aziz, H.A.; Eldehna, W.M. Computational Prediction of the Potential Target of SARS-CoV-2 Inhibitor Plitidepsin via Molecular Docking, Dynamic Simulations and MM-PBSA Calculations. Chem. Biodivers. 2022, 19, e202100719. [Google Scholar] [CrossRef] [PubMed]
- Vesga, L.C.; Ruiz-Hernández, C.A.; Alvarez-Jacome, J.J.; Duque, J.E.; Rincon-Orozco, B.; Mendez-Sanchez, S.C. Repurposing of Four Drugs as Anti-SARS-CoV-2 Agents and Their Interactions with Protein Targets. Sci. Pharm. 2022, 90, 24. [Google Scholar] [CrossRef]
- Elfiky, A.A. Dual Targeting of RdRps of SARS-CoV-2 and the Mucormycosis-Causing Fungus: An in Silico Perspective. Future Microbiol. 2022, 17, 755–762. [Google Scholar] [CrossRef]
- Mohammed, A.O.; Abo-Idrees, M.I.; Makki, A.A.; Ibraheem, W.; Alzain, A.A. Drug Repurposing against Main Protease and RNA-Dependent RNA Polymerase of SARS-CoV-2 Using Molecular Docking, MM-GBSA Calculations and Molecular Dynamics. Struct. Chem. 2022, 33, 1553–1567. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Oselladore, E.; Zagotto, G.; Memo, M.; Gianoncelli, A. A Computational Approach to Drug Repurposing against SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). J. Biomol. Struct. Dyn. 2022, 40, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; Wang, Y.; Dreischulte, T.; Barreiro, O.; Gonzalez, R.J.; Hanč, P.; Matysiak, C.; Neely, H.R.; Rottenkolber, M.; Haskell, T.; et al. Association between Bisphosphonate Use and COVID-19 Related Outcomes. Elife 2023, 12, e79548. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Svetlov, D.; Artsimovitch, I. NMPylation and De-NMPylation of SARS-CoV-2 Nsp9 by the NiRAN Domain. Nucleic Acids Res. 2021, 49, 8822–8835. [Google Scholar] [CrossRef] [PubMed]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of Action and Role in Clinical Practice. Mayo Clin. Proc. 2008, 83, 1032–1045. [Google Scholar] [CrossRef] [PubMed]
- Kunzmann, V.; Bauer, E.; Feurle, J.; Tony Florian Weißinger, H.-P.; Wilhelm, M.; Tony, Florian Weißinger, H.-P.; Wilhelm, M. Stimulation of Γδ T Cells by Aminobisphosphonates and Induction of Antiplasma Cell Activity in Multiple Myeloma. Blood 2000, 96, 384–392. [CrossRef]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL Web Services: Streamlining Access to Drug Discovery Data and Utilities. Nucleic Acids Res. 2015, 43, W612–20. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PK( a ) Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided. Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Koulgi, S.; Jani, V.; Uppuladinne, M.V.N.; Sonavane, U.; Joshi, R. Remdesivir-Bound and Ligand-Free Simulations Reveal the Probable Mechanism of Inhibiting the RNA Dependent RNA Polymerase of Severe Acute Respiratory Syndrome Coronavirus 2. RSC Adv. 2020, 10, 26792–26803. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Liu, X.; Zhang, J.Z.H. Interaction Entropy: A New Paradigm for Highly Efficient and Reliable Computation of Protein-Ligand Binding Free Energy. J. Am. Chem. Soc. 2016, 138, 5722–5728. [Google Scholar] [CrossRef]
- Shirts, M.R.; Klein, C.; Swails, J.M.; Yin, J.; Gilson, M.K.; Mobley, D.L.; Case, D.A.; Zhong, E.D. Lessons Learned from Comparing Molecular Dynamics Engines on the SAMPL5 Dataset. J. Comput. Aided. Mol. Des. 2017, 31, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ekberg, V.; Ryde, U. On the Use of Interaction Entropy and Related Methods to Estimate Binding Entropies. J. Chem. Theory Comput. 2021, 17, 5379–5391. [Google Scholar] [CrossRef]
- Dutta, K.; Shityakov, S.; Morozova, O.; Khalifa, I.; Zhang, J.; Zhu, W.; Panda, A.; Ghosh, C. Beclabuvir Can Inhibit the RNA-Dependent RNA Polymerase of Newly Emerged Novel Coronavirus (SARS-CoV-2). Preprint 2020. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson−Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, M.J.; Russ, G.R. Principles of Drug Therapy, Dosing, and Prescribing in Chronic Kidney Disease and Renal Replacement Therapy. In Comprehensive Clinical Nephrology; Elsevier, 2010; pp. 871–893. ISBN 9780323077668. [Google Scholar]
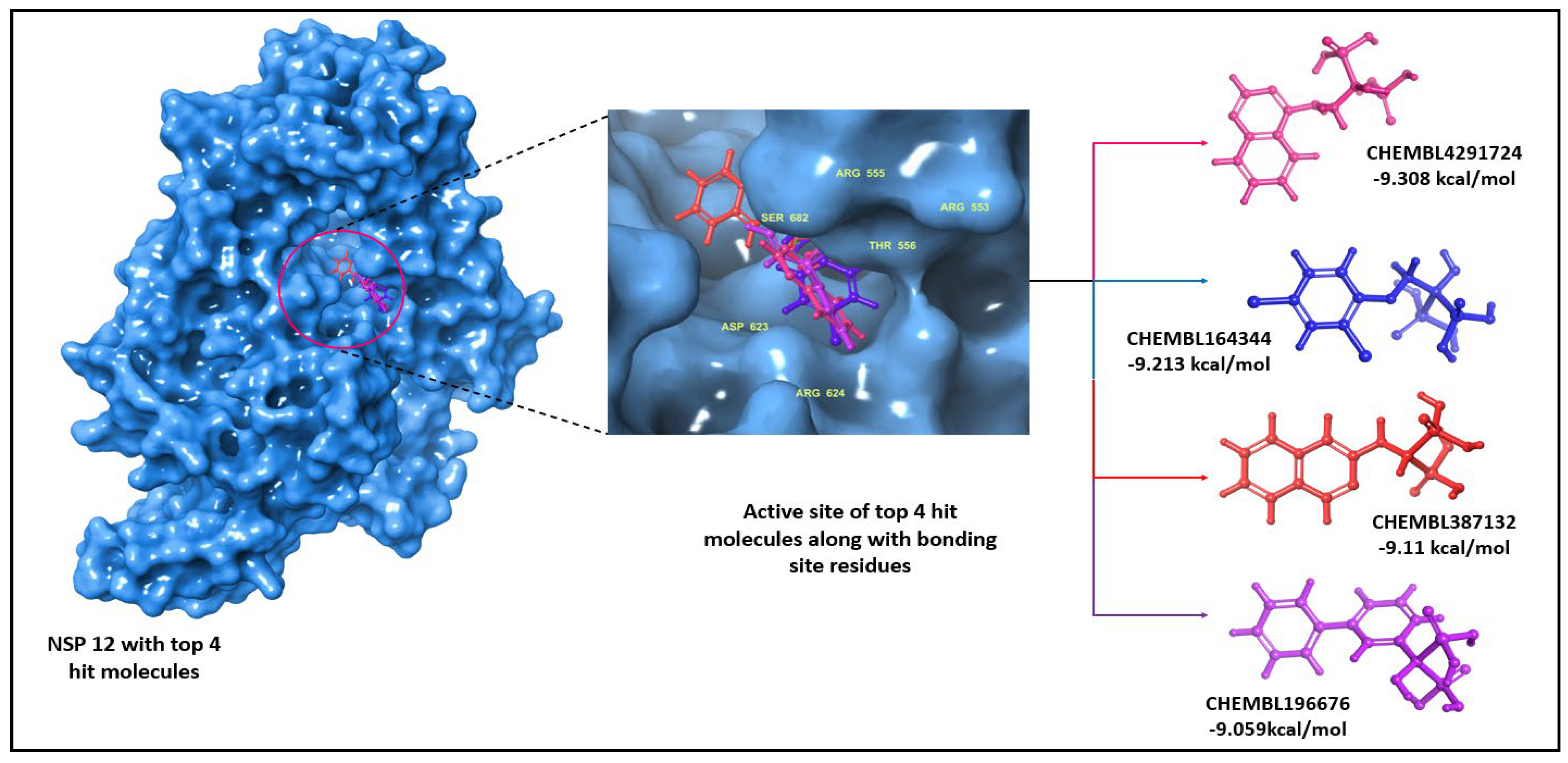
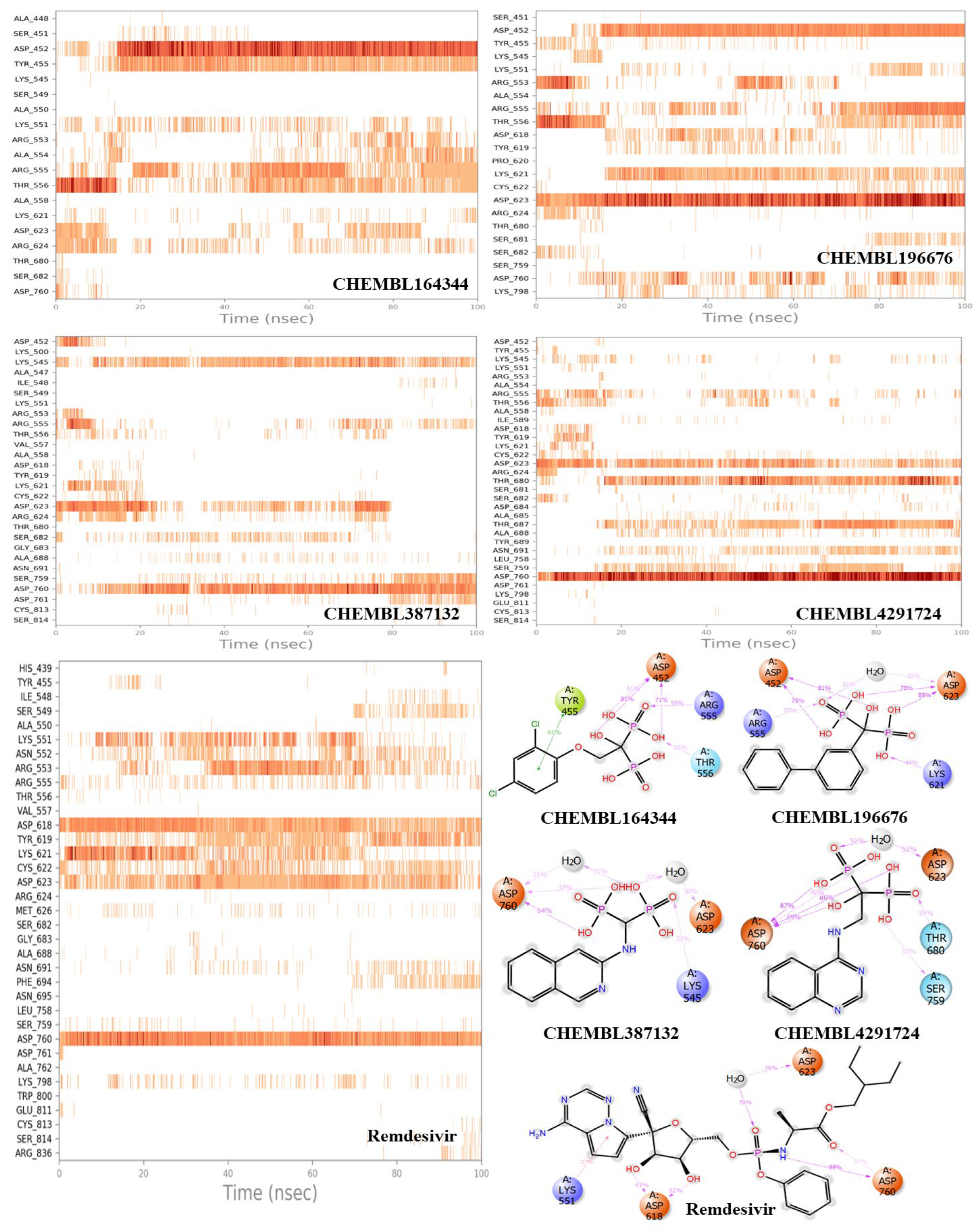
| S. No | ChEMBL ID | Structure | Docking score kcal/mol | S. No | ChEMBL ID | Structure | Docking score kcal/mol |
|---|---|---|---|---|---|---|---|
| 1 | CHEMBL1213265 | 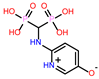 |
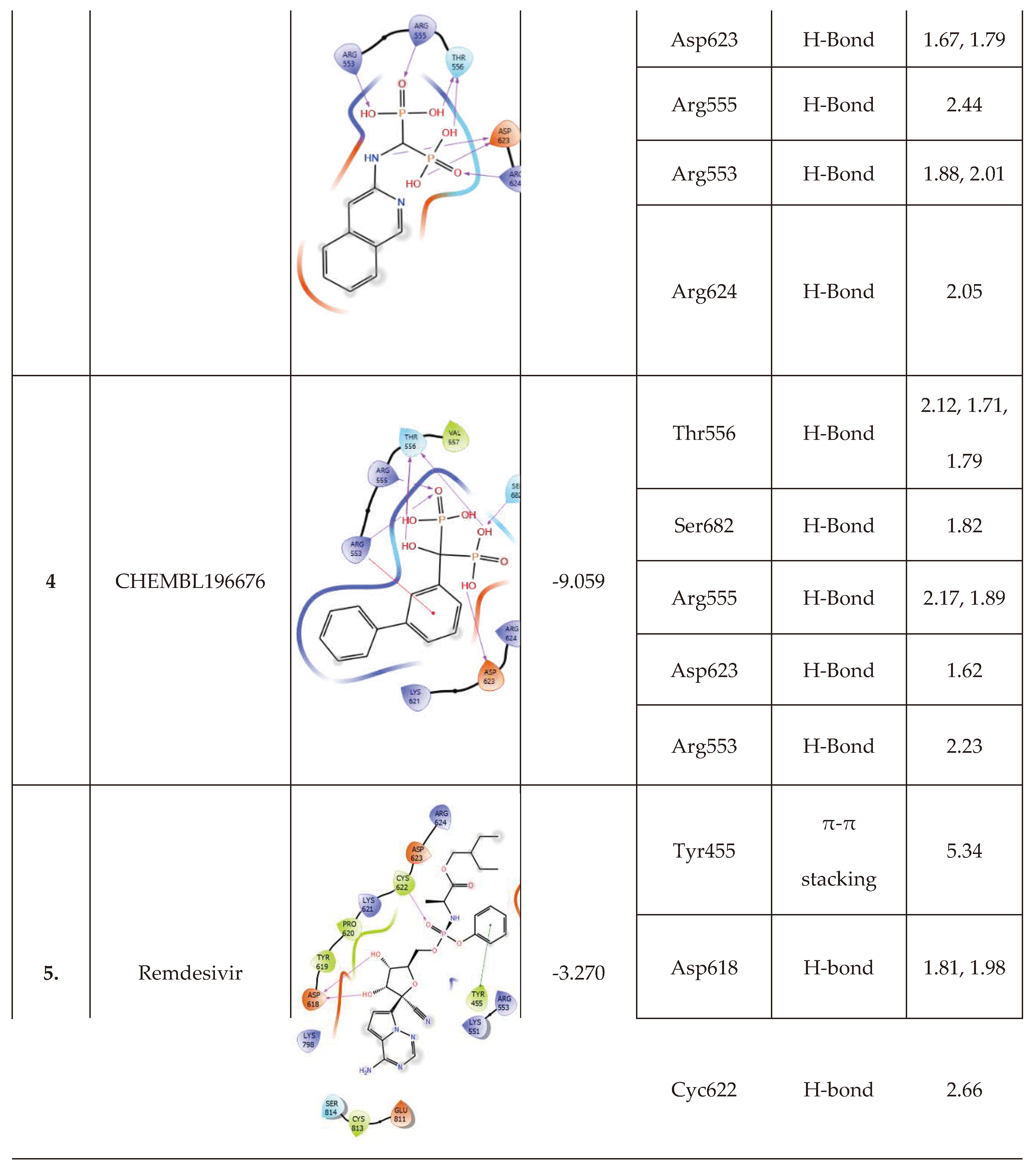
-10.235 | 8 | CHEMBL164344 | 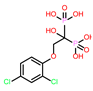 |
-9.213 |
| 2 | CHEMBL608526 | 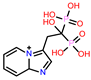 |
-9.706 | 9 | CHEMBL300361 | 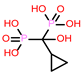 |
-9.151 |
| 3 | CHEMBL319144 | 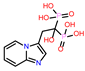 |
-9.657 | 10 | CHEMBL4289996 | 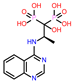 |
-9.119 |
| 4 | CHEMBL4802971 | 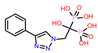 |
-9.355 | 11 | CHEMBL387132 | 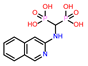 |
-9.11 |
| 5 | CHEMBL98211 | 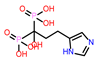 |
-9.347 | 12 | CHEMBL196676 | 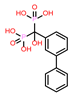 |
-9.059 |
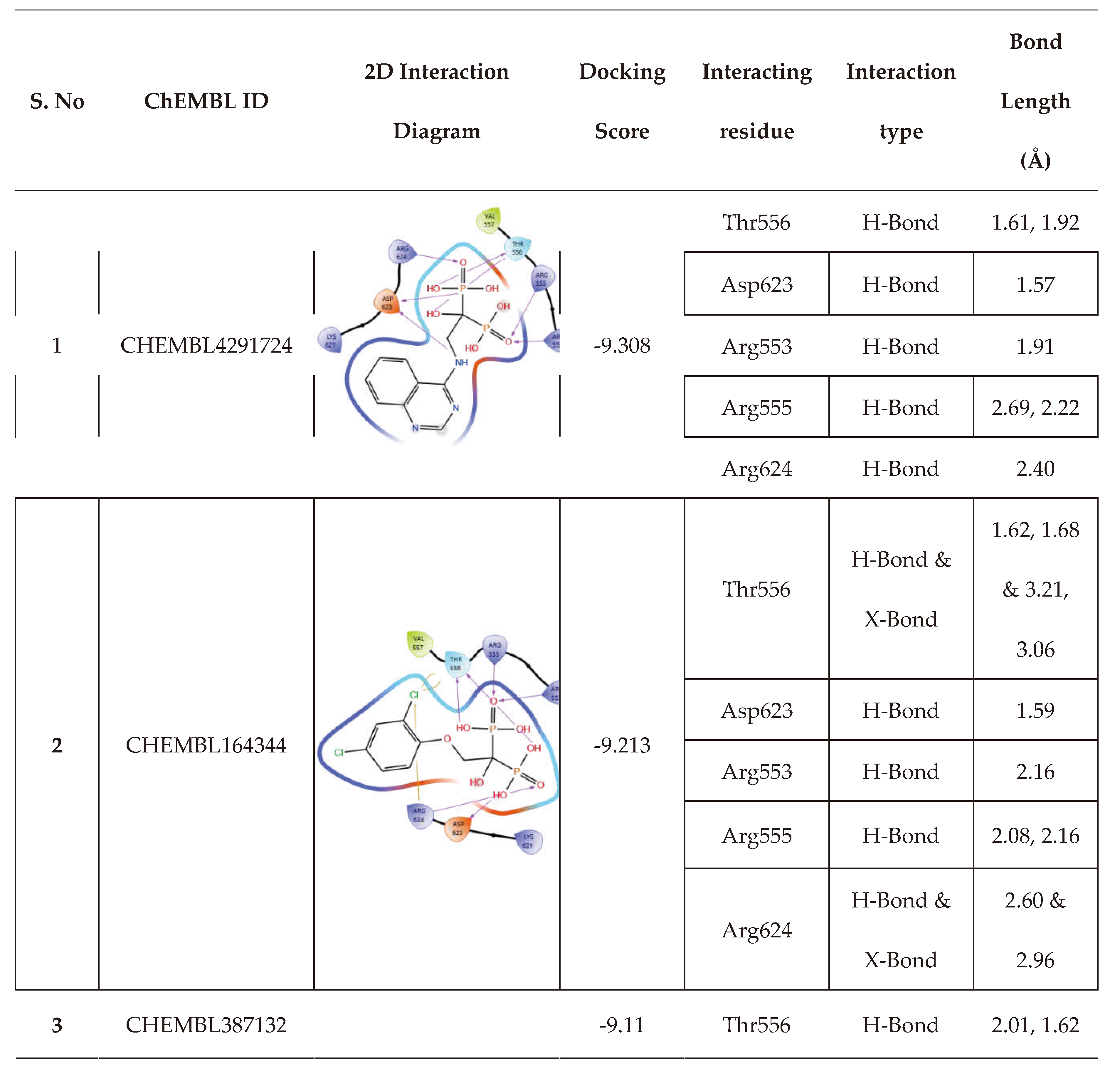
| 6 | CHEMBL4291724 | 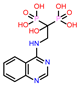 |
-9.308 | 13 | CHEMBL4569308 | 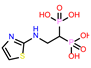 |
-9.02 |
| 7 | CHEMBL301247 | 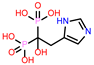 |
-9.219 | 14 | CHEMBL338622 | 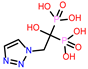 |
-9.014 |
| 15 | Remdesivir(Reference drug) |  |
-3.270 | ||||
| S. No | Category | Drugs | MM-GBSA dG Bind (Kcal/mol) |
|---|---|---|---|
| 1. | High binding energy | CHEMBL1213265 | -7.74 |
| CHEMBL338622 | -24.14 | ||
| CHEMBL301247 | -24.22 | ||
| CHEMBL4289996 | -24.81 | ||
| CHEMBL98211 | -26.04 | ||
| CHEMBL300361 | -26.68 | ||
| 2. | Moderate binding energy | CHEMBL608526 | -33.42 |
| CHEMBL319144 | -35.2 | ||
| CHEMBL4802971 | -36.77 | ||
| CHEMBL4569308 | -40.94 | ||
| 3. | Low binding energy | CHEMBL4291724 | -41.51 |
| CHEMBL387132 | -43.28 | ||
| CHEMBL196676 | -44.14 | ||
| CHEMBL164344 | -46.65 | ||
| 4. | Reference ligand | Remdesivir | -40.32 |
|
| Compounds | I.E. | Total energy contributions (ΔEMM) | ΔGbinding | ||||
|---|---|---|---|---|---|---|---|
| ΔEVDW | ΔEEL | ΔEPB | ΔENPOLAR | ΔEMM = ∑ ΔE | |||
| CHEMBL164344 | 13.76 | -22.77 | -93.71 | 79.14 | -3.19 | -40.53 | -26.77 |
| CHEMBL196676 | 9.48 | -13.06 | -90.55 | 63.70 | -2.98 | -42.88 | -33.41 |
| CHEMBL4291724 | 15.43 | -13.47 | -123.47 | 97.86 | -2.73 | -41.81 | -26.38 |
| CHEMBL387132 | 20.52 | -20.15 | -87.58 | 88.93 | -2.88 | -21.68 | -1.16 |
| Remdesivir | 10.86 | -52.27 | -64.85 | 92.31 | -5.95 | -30.77 | -19.91 |
| Parameters | Hit molecules | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHEMBL → | 4291724 | 164344 | 387132 | 196676 | Remdesivir | ||||||
| Absorption | WS (log mol/L) | -2.47 | -2.152 | -2.046 | -3.677 | -3.07 | |||||
| CP (log Papp in 10-6 cm/s) | -0.438 | -0.461 | 0.093 | 1.245 | 0.635 | ||||||
| IA (% Absorbed) | 42.008 | 16.774 | 71.627 | 38.34 | 71.109 | ||||||
| S.P. (log Kp) | -2.735 | -2.743 | -2.759 | -2.735 | -2.735 | ||||||
| P-glyco protein | Substrate | No | No | No | No | Yes | |||||
| Inhibitor | I | No | No | No | No | Yes | |||||
| II | No | No | No | No | No | ||||||
| Distribution | V.D. ss (log L/kg) | -0.768 | -0.558 | -0.814 | 0.578 | 0.307 | |||||
| FU (Fu) | 0.331 | 0.469 | 0.516 | 0.028 | 0.005 | ||||||
| BBB (log BB) | -2.302 | -2.284 | -1.86 | -1.908 | -2.056 | ||||||
| CNS (log P.S.) | -4.756 | -3.881 | -4.034 | -3.879 | -4.675 | ||||||
| Metabolism | CYP action | Substrate | 2D6 | No | No | No | No | No | |||
| 3A4 | No | No | No | No | Yes | ||||||
| Inhibition against 1A2, 2C19, 2C9, 2D6, 3A4 | No for all four molecules | ||||||||||
| Excretion | T.C. (log ml/min/kg) | 0.146 | 0.032 | -0.025 | -0.119 | 0.198 | |||||
| ROC | No | No | No | No | No | ||||||
| Toxicity | AMES | No | No | No | Yes | No | |||||
| MTD (log mg/kg/day) | 0.841 | 0.445 | 0.689 | 0.574 | 0.15 | ||||||
| hERG I inhibitor | No | No | No | No | No | ||||||
| hERG II inhibitor | No | No | No | Yes | Yes | ||||||
| Rat oral toxicity | Acute (LD50) (mol/kg) | 2.67 | 2.613 | 1.886 | 3.117 | 2.043 | |||||
| Chronic (LOAEL) (Log mg/kg_bw/day) |
3.195 | 3.597 | 3.052 | 3.404 | 1.639 | ||||||
| HT | Yes | No | Yes | No | Yes | ||||||
| SS | No | No | No | No | No | ||||||
| TT (log ug/L) | 0.285 | 0.285 | 0.288 | 0.285 | 0.285 | ||||||
| MT (log mM) | 2.992 | 1.686 | 2.938 | 0.427 | 0.291 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





