1. Introduction
Cancer is a primary cause of mortality globally. It was responsible for close to 10 million deaths in 2020 [
1]. This disease presents a myriad of unique pathologies [
2], adding a layer of complexity to its diagnosis, research, and the improvement of care. The exploration of a specific pathology often demands the identification of a significant number of patients that typically exceeds the count available at a single healthcare facility [
3]. Therefore, data integration and interoperability across institutions becomes essential to enhance cancer research and care.
Patient identification is a critical yet labor-intensive process [
4]. First step often require manual review and interpretation of electronic health record (EHR) data, a process that is both slow and financially taxing. Despite this, it remains a necessary step, considering that approximately 80% of the pertinent clinical information is encapsulated within the text of health records [
5]. To tackle these challenges, the contribution of natural language processing (NLP) techniques is a key asset to help physicians and data experts identifying potential candidates for research projects.
Beyond being able to search available medical information, data must be structured and standardized to ensure their secondary reuse in research in accordance with the FAIR principles [
6]. An initiative launched by eleven comprehensive cancer centres from the Unicancer network sought to enhance and expedite data sharing in oncology and has been involved in the creation of the OSIRIS model [
7]. This common data model comprises a minimal set of clinical and genomic data, specific to oncology research, serving as the cornerstone of a larger initiative aimed at accelerating cancer research by simplifying the creation of cohorts of cancer patients with similar characteristics.
To navigate regulatory hurdles and reduce data flows, the initiative implemented a federated technical architecture, avoiding the need for a single centralised data warehouse. However, this federated network requires a high level of harmonisation among centres to supply the common model. To this end, the French federation of comprehensive cancer centres (UNICANCER) developed Consore, a unique network equipped with a powerful federated search engine. This tool enables the efficient and reliable identification of patient cohorts by digging into the electronic health records of millions of patients across eleven French comprehensive cancer centres.
The project addresses four major challenges: (i) the aggregation of tremendous amount of heterogeneous data; (ii) the semantic analysis of electronic health records, data standardisation, and modelling of the cancer disease; (iii) the technical implementation of a solution facilitating fast data querying at a national level; (iv) the development of ready-to-use services for clinicians and researchers. This paper presents an in-depth examination of each aspect of these challenges and their respective evaluations.
2. Materials and Methods
Overseen by UNICANCER, the federation of 18 French comprehensive cancer centres, the Consore project incorporates a multidisciplinary team of oncologists, bioinformaticians, project managers, data engineers and Information Technology (IT) engineers. The implementation of Consore was authorised in November 2016 (N°2016-331) by the French regulatory authorities (Commission Nationale de l’Informatique et des Libertés, CNIL). Despite this approval, it remains essential to inform patients individually and collectively about the potential reuse of their health data for cancer research in compliance with European and French regulations.
Data Aggregation
Consore addresses various types of data coming from numerous disparate sources. This includes both structured and unstructured information in electronic medical records, demographic and administrative data, medical activity data derived from France’s nationwide diagnostic-related group (DRG)-based information system - hospital discharge data from the programme for medicalising information systems (PMSI), biobanking data, tumor characteristics, lab results, pharmaceutical data and molecular alteration information.
Medical Concept Inference
Medical reports provide a treasure trove of data for Consore. However, harnessing this data presents a significant challenge due to the voluminous mass of raw unstructured data they contain. Given the content is natural language text, it poses a formidable task for AI to process. To surmount this challenge, Consore is equipped with a specialised web service for natural language processing (NLP) tasks. It receives medical reports, processes them via a spaCy NLP pipeline, and subsequently produces structured documents. This NLP pipeline carries out a series of operations on each report to extract as many medical concepts as possible. These operations include anonymizing personal information for regulatory reasons and text cleaning tasks such as stopwords removal, tokenization, and lemmatization.
The web service extracts several concepts from the reports using NER models, and structures them through entity linking tasks. The latter are operated according to the same standards and classifications used for structured data sources in Consore, such as:
The “Classification Commune des Actes Médicaux” (CCAM), as the French classification for medical procedures [
8];
The 10th revision of the International Classification of Diseases (ICD-10) [
9] and the 3rd edition of the ICD for Oncology (ICD-O-3) [
10]
Another issue we address using Consore’s NLP web service is redundancy and lack of precision. Indeed, for each patient, hundreds of reports are processed; most contain a redundant history section that is rarely updated. Moreover, in a single report, we tend to detect various occurrences of the same tumor, yet with different details: sometimes it is accompanied by the tumor location, its morphology, the associated biomarkers, the administered treatment, etc. These remain unitary concepts of different categories. Detecting them and representing them in a structured form does not solve all the challenges.
In fact, not only do we detect single concepts, but we also need to infer the links between them. For instance, there is high interest in data structuring to find the dates linked to a concept in order to locate it in time. In addition to this example, we also tackle linking a treatment response to the involved treatment or recognising whether a tumor remains in its first stages or if a metastasis is diagnosed.
Pivot Model for Diverse Data Sources
Consore handles a variety of data from several different sources that do not have a shared structure. Thus, a pivot model was designed in compliance with the OSIRIS model, capable of storing information from these diverse sources. This model addresses several hurdles such as partial data, conceptual links, volumetry and redundancy, and data sourcing.
- -
Partial data: we often deal with information about the morphology of a tumor, but we have nothing about the initial diagnostic or its location; or treatment responses without the involved treatment.
- -
Concept links: the pivot model must preserve, when applicable, the links between the detected concepts (e.g. the link between a metastasis and the primary tumor)
- -
Volumetry and redundancy: Consore might detect or receive thousands of concepts, often redundant, for a single patient.
- -
Data sourcing: for each data item, we need to identify its source and the date it was recorded in the information system.
- -
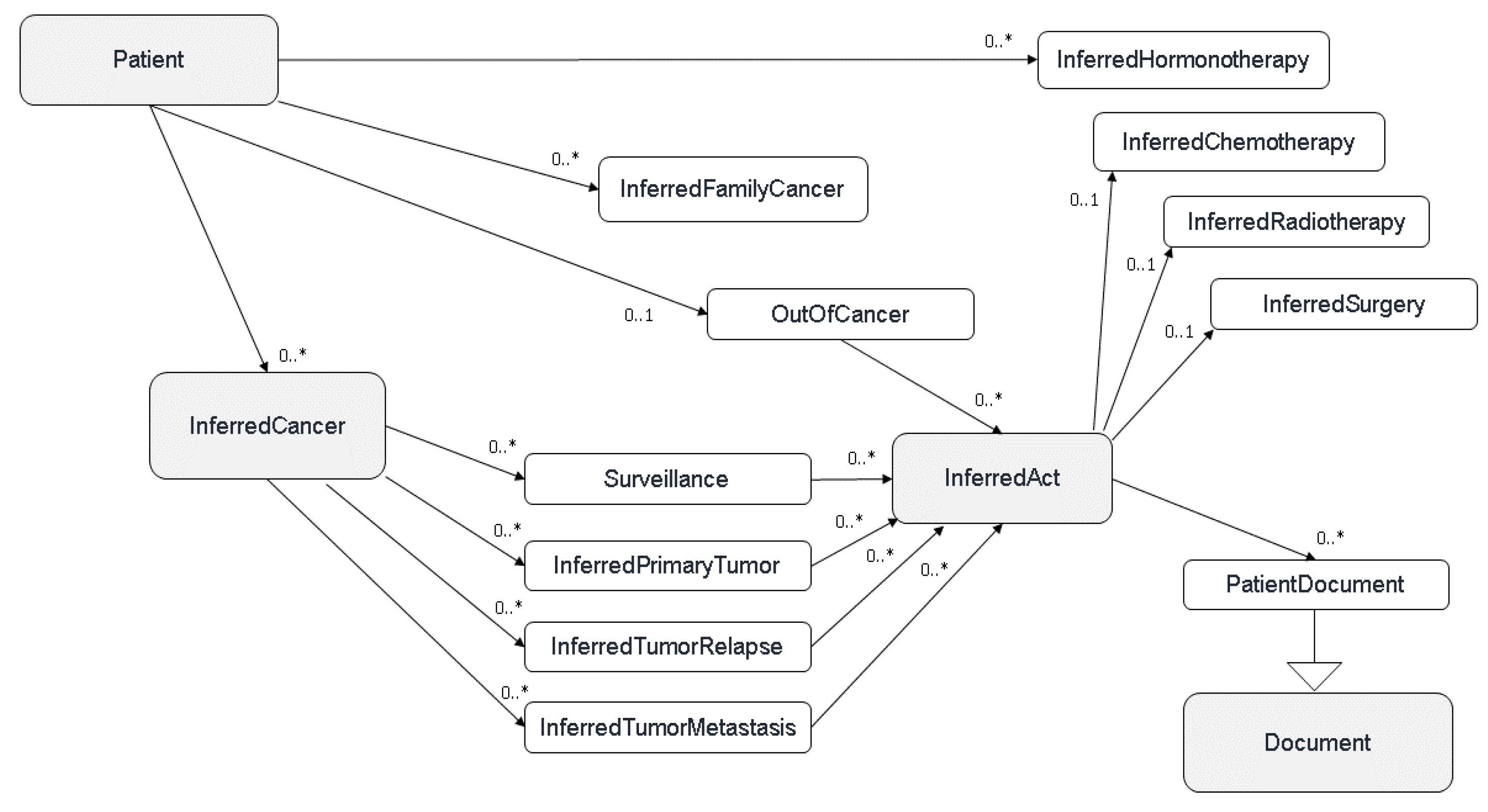
In order to organise and classify all the identified concepts, we developed a common model defining the cancer disease based on several main hierarchical classes (or layers): Cancers (all cancer recurrences for a given patient), Tumor events (primary tumor, local or metastatic relapse), Acts (treatments and/or analysis), and Documents (all the documents of a patient or available biological samples) (
Figure 1).
Upon completion of documents processing through various pipelines, each data source possessing its own, Consore carries out its primary function: data inference and structuring. The upcoming sections will delve into metastasis structuring as an example of data structuring within Consore.
3. Results
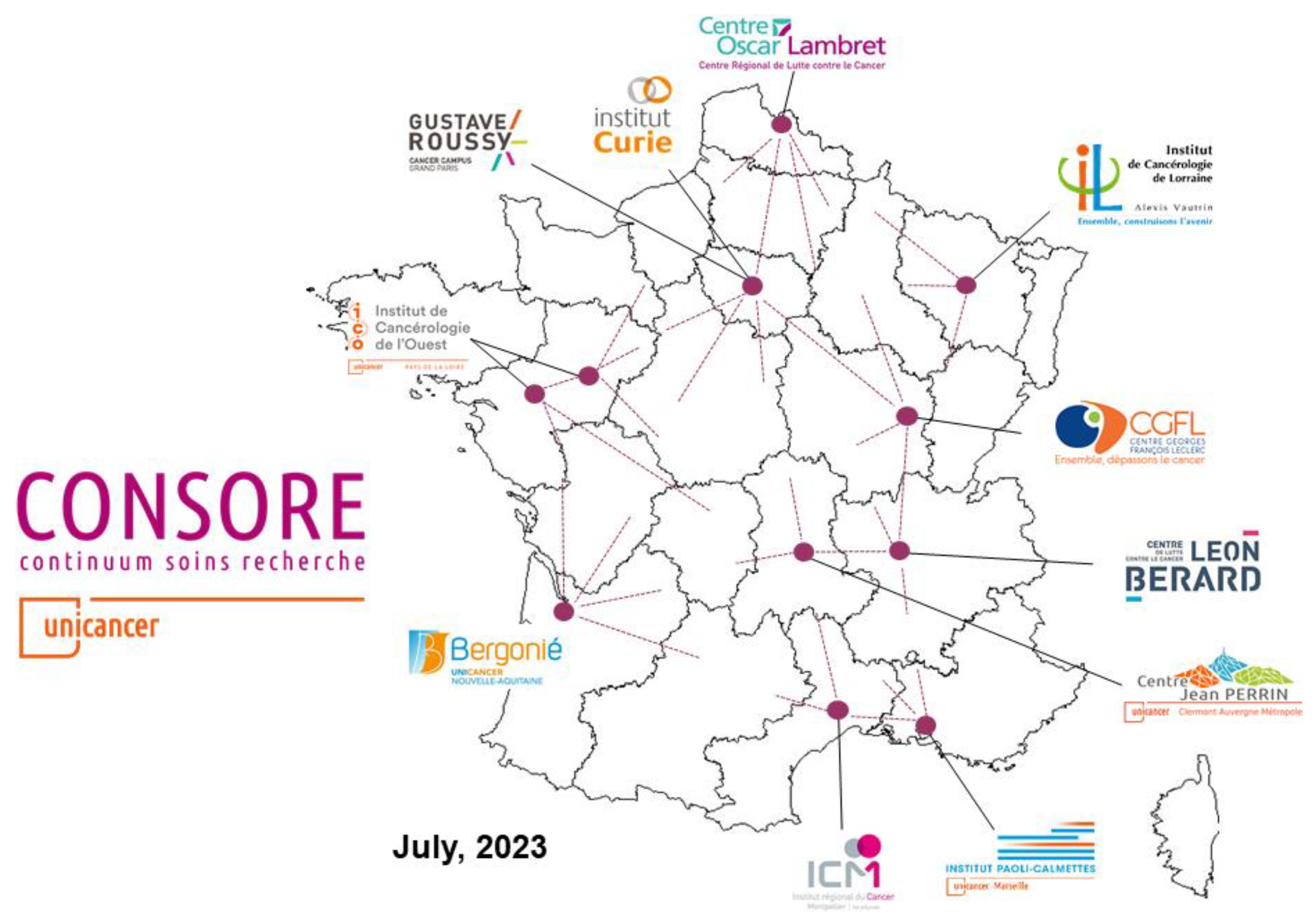
Consore is a software deployed within eleven major cancer centres in France (
Figure 3).
The following table summarises the number of patients taken cared of and whose data are included in Consore. This is to help represent the volume of data our software deals with.
The difference between the enumeration of patients and those with at least a cancer disease is due to the inclusion into Consore of data of patients who have benign tumors or those who came into a centre for clinical tests that concluded an absence of cancerous disease.
Table 1.
Overview of data volumes in the different Consore data warehouses (*February 2023;** August 2023).
Table 1.
Overview of data volumes in the different Consore data warehouses (*February 2023;** August 2023).
| French Cancer Centres |
Nb of patients |
Nb of patients with at least one cancer |
Nb of patients with a metastatic relapse |
Nb of medical records |
| Institut Curie* |
572 421 |
280 924 |
95 025 |
13 431 874 |
| Centre Léon Bérard* |
359 634 |
207 657 |
85 210 |
18 711 561 |
| Institut Paoli-Calmettes* |
347 415 |
136 500 |
43 767 |
4 464 580 |
| Gustave Roussy* |
399 665 |
237 132 |
96 074 |
12 856 023 |
| Institut de Cancérologie de l’Ouest |
N/A (deployment in progress) |
N/A |
N/A |
|
| Centre Oscar Lambret* |
182 436 |
118 506 |
57 784 |
5 865 404 |
| Institut du Cancer de Montpellier* |
176 257 |
79 601 |
34 138 |
3 401 825 |
| Centre Georges-François Leclerc* |
282 948 |
79 592 |
36 635 |
3 207 721 |
| Centre Jean Perrin* |
397 179 |
124 080 |
44 548 |
2 776 005 |
| Institut Bergonié* |
285 129 |
153 589 |
52 290 |
3 806 476 |
| Institut de Cancérologie de Lorraine** |
247869 |
63350 |
19105 |
1 096 485 |
Cancer of unknown primary
Cancers of unknown primary (CUP) are metastatic cancers for which a diagnostic work-up fails to identify the site of origin at the time of diagnosis and account for < 5% of all cancers. It is a rare disease and often poorly documented in the EHR and for which the diagnosis is made by default, that is to say when all the primary cancer sites have been eliminated which can sometimes take several months.
Results at the “Centre Léon Bérard” (CLB)
After analysis of an "institutional" database and the Consore query, we have a total of 145 CUP at the “Centre Leon Berard”. The selection criteria used in Consore were as follows: patients initially diagnosed with de novo metastatic cancer and for whom the mention “unknown primary” was present in the electronic health record since January 1, 2010. The difficulty of this query lies in the fact that it is based both on a textual search but also on data structured by Consore (here the dates of diagnosis and the metastatic stage). Consore offers 2577 patients but with 1 patient from the institutional database not found by Consore. Focusing only on cancer diagnoses between January 1, 2019 and June 30, 2021, we manually reviewed 121 electronic medical records. Concerning these 121 patients, 69 had a CUP while 52 patients finally had a cancer whose primary was identified. The large number of false positives is linked to the period of diagnostic uncertainty which can last several months with cancers of unknown primary (the diagnosis of cancer of unknown primary being ultimately excluded when a primary tumor is identified). So over this period, Consore finds 69 additional diagnoses but detects at the same time 52 false positives.
-
To assess the performance, recall, precision [
11] and F1 score were calculated as followed [
12]:
- ○
Recall = 99%;
- ○
Precision = 57%;
- ○
F1-score = 0,66.
- ○
calculation of inverse recall is not possible because manual control of all EMRs to identify true negatives is not possible.
Results at the “Institut Curie” (IC)
Based on the Consore query first designed at the “Centre Léon Bérard”, we identified a selection of 133 CUP at Institut Curie. The primary comparison with the institutional TransCUPtomics [
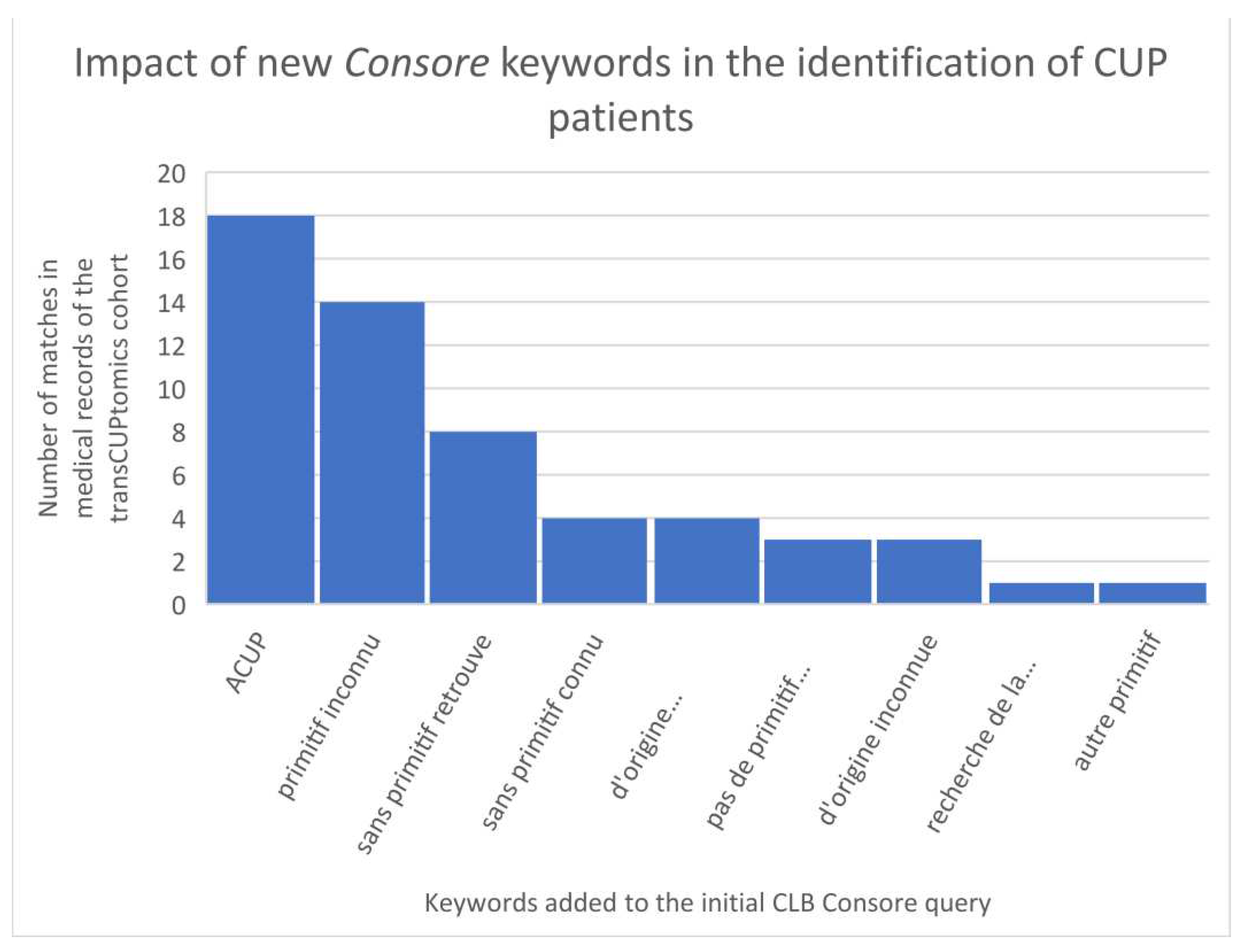
13] study (48 CUP patients) showed an identification of only 8 patients (17%). This poor result is mainly due to the initial criteria: in the TransCUPtomics study, 15 patients were diagnosed before 2010 and 33 patients had a metastatic relapse after the initial diagnosis. However, there was also a lack of keywords to properly identified CUP patients in Consore. In a second step, we have optimized the Consore query by adding the following list of French keywords: “ACUP”, “primitif inconnu”, “de primitif inconnu”, “sans primitif retrouve”, “sans primitif connu”, “d’origine indéterminée”, “pas de primitif retrouve”, “d’origine inconnue”, “recherche de la tumeur primitive”, “autre primitif” (
Figure 4).
With this new query, we identified 2871 CUP patients on the overall database where we retrieved 45 of the 48 TransCUPtomics patients (94%): 2 patients remained not found because of a lack of EHRs (less than 5 documents available), 1 patient had a CUP status unclear with minimal description in the EHR. In order to assess the positive predictive value (PPV), we reviewed a sample of 119 patients diagnosed between January 1, 2019 and June 30, 2021. Concerning these 119 patients, 58 had a CUP while 26 patients finally had a cancer whose primary was identified (false positives linked to the period of diagnostic uncertainty). So over this period, Consore finds 58 additional diagnoses but detects at the same time 61 false positives.
4. Discussion
Considering the growing impact of RWD for clinical practice and research, the development of digital solutions to gather and use such data becomes essential. Beyond the description of this innovative tool, our work underlines the major role of collaborative efforts in constructing a federated technical architecture and agreeing on a unified data model.
With the creation of the OSIRIS model, French cancer centres have laid down a foundation of a minimal data set, which serves as an essential starting point for any collaborative cancer research [
7]. Consore, to date, has facilitated numerous research projects on large cohorts of cancer patients, including timely studies on topics such as immunotherapy or COVID-19 [
14]. The tool’s role in RWD research has been bolstered through collaboration with the Health Data Hub [
15], through a project initiative aimed at cross-referencing Consore-provided data with data from the national health data system [
16]. A dialogue has also been initiated to foster communication between this data model and international initiatives, such as OMOP [
17].
Numerous French and international consortium are aiming to structure information within medical records to facilitate their reuse for research purposes. Examples include Dr Warehouse [
18] and the EHOP warehouse [
19] in France, and CancerLinq [
20] and FLATIRON [
21] in the United States. In this ecosystem, Consore stands out as one of the few solutions dedicated to oncology with a federated architecture, employing natural language processing.
However, this project isn’t without limitations. While the federated architecture is a strength, it brings also some issues. Each cancer centre maintains its own EHR, which contains heteregeneous data sources in different formats and varying data quality levels. Although the OSIRIS model allows data format standardisation, quality variability remains a challenge that must be acknowledged, quantified, and considered in the analyses and outcomes of multicentre projects. The question of completeness and quality of the data source as well as the results persist for each research work.
Another identified limitation deals with the common data model, which relies on the primary tumor identification. While suitable for most solid tumors, such as breast or lung cancer, it presents more challenges and therefore errors, for haematological cancers, sarcomas, and skin cancers. For these types of neoplastic diseases, the histological type is indeed more critical than the primary tumor location. Efforts are underway to improve these results.
As an expert tool, Consore is dedicated to oncology and its NLP algorithms have been exclusively trained on French language-based medical reports. A wider dissemination of Consore, at an international level or for other chronic disease (such as chronic obstructive pulmonary disease or chronic renal failure) would require substantial modifications such as the development of an English version or significant data model transformations.
The previously highlighted limitations suggest several potential areas for future enhancement. Special emphasis will be placed on improving the comprehensiveness and quality of the results, as well as bridging the data model into the OMOP-CDM format (Observational Medical Outcomes Partnership - Common Data Model) [
16] to facilitate international engagement. The pivotal aspect, thus far, revolves around the update on data quality. Despite our awareness of the inherent constraints of Real-World Data (RWD), we have established a dedicated working group to systematically assess this quality and to enhance the overall robustness of the results.
5. Conclusions
Consore stands as a powerful tool dedicated to oncology and capable of modelling patients’ neoplastic histories by structuring data from EHRs. Despite the discussed limitations, Consore has already demonstrated its potential in accelerating the identification of patient cohorts and the implementation of research projects. With its federated architecture and the use of a common data model, Consore is playing a key role in the development of multicentric projects in the field of oncology at a national level. The next steps will aim to enhance interoperability between Consore and other international networks as well as the development of next generation NLP algorithms based on large language models (LLM) [
22].
Authors Contributions
Writing – original draft, Julien Guérin, Amine Nahid, Frédérik Joly and Pierre-Etienne Heudel; Writing – review & editing, Louis Tassy, Marc Deloger, François Bocquet, Simon Thézenas, Emmanuel Desandes, Marie-Cécile Le Deley, Xavier DURANDO, Anne Jaffré, Ikram Es Saad, Hugo Crochet, Marie Le Morvan, François Lion, Judith Raimbourg, Oussama Khay, Franck Craynest, Alexia Giro, Yec’han Laizet, Aurélie Bertaut and Alain Livartowski.
Acknowledgments
This project received special support from grant ANR-10-EQPX-03 (Equipex), Inca-DGOS-4654 (SiRIC), INCa-DGOS-Inserm_12554 (SiRIC). We thank the ICGEx team (Institut Curie) and the SiRIC-Curie program for their commitment.
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M. Global Cancer Observatory: Cancer Today. Lyon: International Agency for Research on Cancer. 20 May 2020. Available online: https://gco.iarc.fr/today accessed on May 2023).
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lainé, A.; Hanvic, B.; Ray-Coquard, I. Importance of guidelines and networking for the management of rare gynecological cancers. Curr Opin Oncol. 2021, 33, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Wilke, R.A.; Berg, R.L.; Peissig, P.; Kitchner, T.; Sijercic, B.; McCarty, C.A. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clinical Medicine & Research 2007, 5, 1–7. [Google Scholar] [CrossRef]
- Hersh, WR; Weiner, MG; et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.; Dumontier, M.; Aalbersberg, I.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Guérin, J.; Laizet, Y.; Le Texier, V.; Chanas, L.; Rance, B.; Koeppel, F.; Lion, F.; Gourgou, S.; Martin, A.L.; Tejeda, M.; Toulmonde, M.; Cox, S.; Hess, E.; Rousseau-Tsangaris, M.; Jouhet, V.; Saintigny, P. OSIRIS: A Minimum Data Set for Data Sharing and Interoperability in Oncology. JCO Clin Cancer Inform. 2021, 5, 256–265. [Google Scholar] [CrossRef] [PubMed]
- CCAM. 20 October. Available online: https://sante.gouv.fr/professionnels/gerer-un-etablissement-de-sante-medico-social/financement/financement-des-etablissements-de-sante-10795/financement-des-etablissements-de-sante-glossaire/article/classification-commune-des-actes-medicaux-ccam accessed on October 2023.
- World Health Organization. (2004). ICD-10: international statistical classification of diseases and related health problems: tenth revision, 2nd ed. World Health Organization.
- AFrizt; Percy, C.; Jack, A.; Shanmagaratnam, K.; Sobin, L.; Parkin, D.M.; Whelan, S. International Classification of Diseases for Oncology. Third edition. First Revision, World Health Organization, Geneva, 2013.
- Fraser, Alexander & Daniel Marcu. Measuring Word Alignment Quality for Statistical Machine Translation. Computational Linguistics 2007, 33, 293–303. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J Thoracic Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Vibert, J.; Pierron, G.; Benoist, C.; Gruel, N.; Guillemot, D.; Vincent-Salomon, A.; Le Tourneau, C.; Livartowski, A.; Mariani, O.; Baulande, S.; Bidard, F.C.; Delattre, O.; Waterfall, J.J.; Watson, S. Identification of Tissue of Origin and Guided Therapeutic Applications in Cancers of Unknown Primary Using Deep Learning and RNA Sequencing (TransCUPtomics). J Mol Diagn. 2021, 23, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Heudel, P.; Favier, B.; Solodky, M.L.; et al. Survival and risk of COVID-19 after SARS-COV-2 vaccination in a series of 2391 cancer patients. Eur J Cancer. 2022, 165, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Health data Hub. Available online: https://www.health-data-hub.fr/page/faq-english accessed on May 2023.
- Health data Hub, UNIBASE results. Available online: https://www.health-data-hub.fr/annonce-laureats-unibase accessed on May 2023, French version only.
- OHDSI. Available online: https://www.ohdsi.org/data-standardization/the-common-data-model/ accessed on May 2023.
- Garcelon, N.; Neuraz, A.; Salomon, R.; Faour, H.; Benoit, V.; Delapalme, A.; Munnich, A.; Burgun, A.; Rance, B. A clinician friendly data warehouse oriented toward narrative reports: Dr. Warehouse. J Biomed Inform. 2018, 80, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Madec , J; Bouzillé, G; Riou, C; Van Hille, P; Merour, C; Artigny, ML; Delamarre, D; Raimbert, V; Lemordant, P; Cuggia, M. eHOP Clinical Data Warehouse: From a Prototype to the Creation of an Inter-Regional Clinical Data Centers Network. Stud Health Technol Inform. 2019, 264, 1536–1537. [Google Scholar] [CrossRef] [PubMed]
- CancerLinq. Available online: https://www.cancerlinq.org/ accessed on May 2023.
- Flatiron. Available online: https://flatiron.com/ accessed on May 2023.
- Gilson, A.; Safranek, C.W.; Huang, T.; Socrates, V.; Chi, L.; Taylor, R.A.; Chartash, D. How Does ChatGPT Perform on the United States Medical Licensing Examination? The Implications of Large Language Models for Medical Education and Knowledge Assessment. JMIR Med Educ. 2023, 9, e45312. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).