1. Introduction
The advent of modern natural language processing (NLP) techniques, driven by advances in machine learning (ML), has significantly enhanced our ability to analyze and interpret complex text data [
1,
2,
3,
4]. These improvements are partly attributed to methods that encode and manipulate text data using latent representations. Those methods embed text into high-dimensional vector spaces that capture the underlying semantics and structure of language. The medical field has witnessed a significant transformation in recent years with the emergence and popularization of various advanced techniques, including Large Language Models (LLMs) [
5,
6].
One of the most critical areas where these advances have made a substantial impact is the integration and analysis of Electronic Health Records (EHR). With its rich and varied information, EHR data offer unique opportunities for developing and validating ML models that can improve diagnostic accuracy, predict patient outcomes, and personalize treatment plans [
7,
8,
9]. For instance, Hashir and Sawhney [
9] demonstrate the potential of unstructured clinical notes in enhancing mortality prediction, while Alghamdi and Mostafa [
8] show that integrating advanced NLP techniques with detailed EHR data significantly improves medication management predictions. In healthcare informatics, converting unstructured clinical data into structured formats like Fast Healthcare Interoperability Resources (FHIR) is crucial for enhancing interoperability and improving patient care. FHIR is a standard for exchanging healthcare information electronically, facilitating the integration of disparate healthcare systems. However, a gap remains between FHIR Implementation Guides (IGs) and the process of building actual services, as IGs provide rules without specifying concrete methods, procedures, or tools [
10]. Thus, stakeholders may feel it nontrivial to participate in the ecosystem, highlighting the need for more actionable practice guidelines to promote FHIR’s fast adoption.
Several studies have highlighted the complexities and importance of this task. For instance, Shah [
11] underscore the significance of FHIR in achieving interoperability between different healthcare systems, demonstrating its potential to improve healthcare delivery and patient outcomes. Hong et al. [
12] discuss the intricacies of mapping unstructured data to the highly structured and standardized FHIR format, emphasizing the need for sophisticated techniques and tools. Their work demonstrates the feasibility of a scalable clinical data normalization pipeline known as NLP2FHIR, which integrates NLP with FHIR to model unstructured EHR data. However, the NLP2FHIR technique currently relies on transformation scripts for mapping and content normalization, which limits its standardization and automation capabilities.
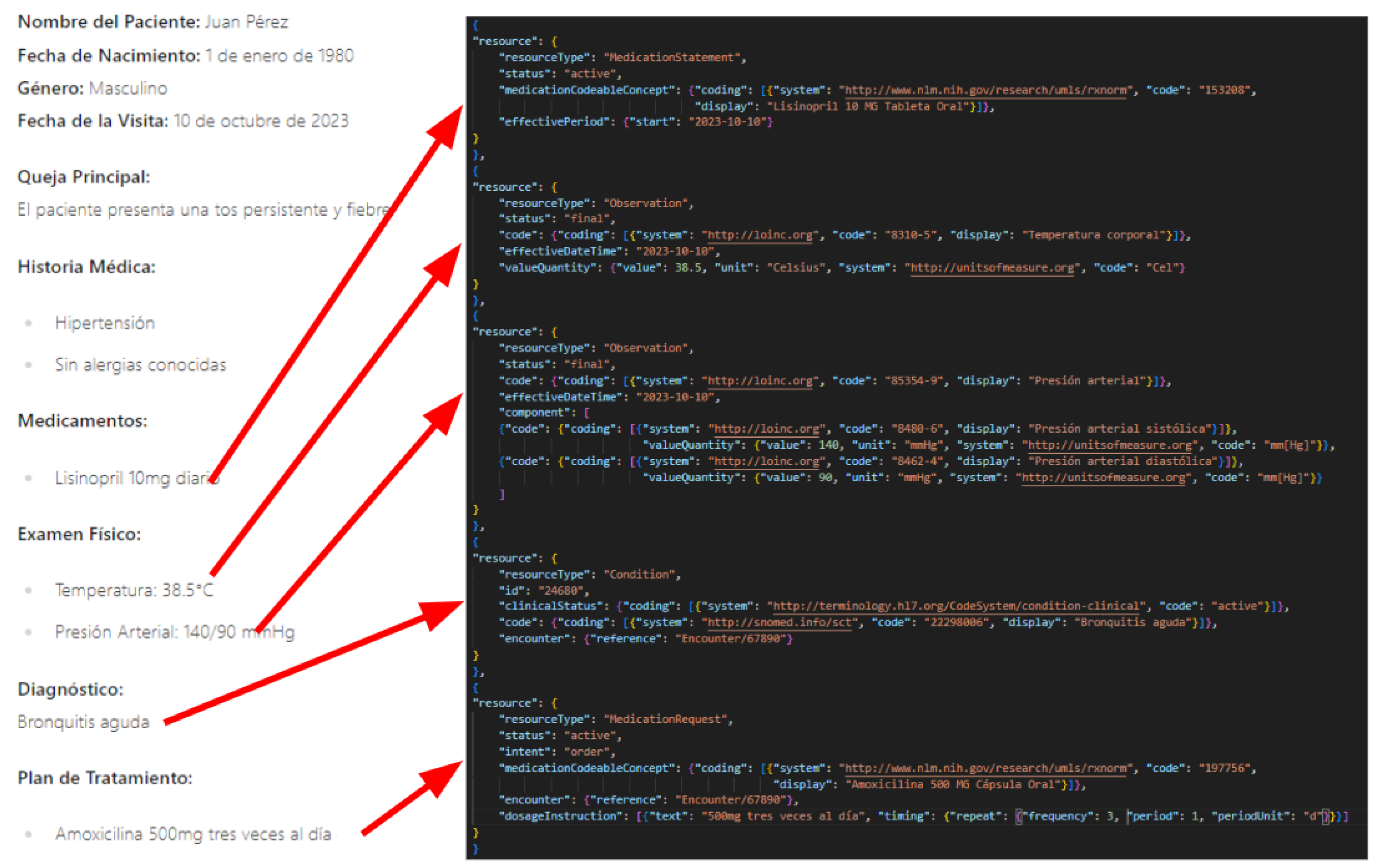
Despite advances in NLP, converting unstructured clinical data, such as clinical reports, into structured formats like FHIR remains a challenging task. The importance of this conversion lies in its potential to enable seamless data sharing, enhance clinical decision support, and facilitate research and public health initiatives [
13,
14]. For instance, consider the artificial example shown in
Figure 1, which illustrates the conversion of information from a clinical report (on the left) to a FHIR bundle (on the right). This example underscores the well-defined structure of FHIR and highlights the complexity involved in converting even simple elements from a clinical report into this structured format. This challenge underscores the need for advanced techniques, such as those explored in our study, to facilitate this conversion process effectively.
The primary objective of this research is to evaluate the effectiveness of various LLMs in converting unstructured clinical reports in Spanish into structured FHIR bundles. Different prompting techniques are employed, including providing examples of clinical reports and their corresponding FHIR bundles. The impact of iterative correction on the quality of generated FHIR bundles is also explored, with the hypothesis that multiple correction cycles may yield more accurate results compared to a single generation attempt.
The methodology includes gathering diverse clinical reports, designing prompting techniques, investigating the impact of temperature and iterative correction, and evaluating the precision, hallucination rate, and resource mapping accuracy of the generated FHIR bundles. Results indicate that providing well-defined examples within prompts and employing a two-step conversion approach are particularly effective strategies for accurate and reliable FHIR conversion. This study aims to demonstrate the potential of LLMs in automating the conversion of unstructured clinical data into structured formats, thereby enhancing data interoperability and improving healthcare outcomes.
The remainder of this paper is organized as follows.
Section 2 presents the preliminaries, defining what is a large language model and FHIR resources.
Section 3 provides an overview of related work on the use of LLMs in medical data and their performance in data conversion.
Section 4 describes the datasets, LLMs, prompt techniques, and temperature levels used in our study.
Section 5 presents our methodology for evaluating the conversion made by the LLMs.
Section 6 reports the results of our study while
Section 7 discusses the implications of our findings for the use of LLMs in healthcare. Finally,
Section 8 concludes the paper and outlines directions for future research.
2. Preliminaries
This section provides an overview of the key concepts and technologies used in this study. This ensures that readers from both healthcare and natural language processing (NLP) domains are familiar with the main concepts from both fields. We discuss tokenization and embeddings, which are fundamental to NLP. Then, we present Large Language Models (LLMs), which are central to our approach and cover advanced techniques such as Mixture of Experts (MoE) and the fine-tuning of LLMs to specific domains. We also discuss the impact of prompts and temperature settings on their performance. Additionally, we introduce Fast Healthcare Interoperability Resources (FHIR), a standard for exchanging healthcare information electronically.
2.1. Tokenization & Embeddings
Tokenization is a crucial preprocessing step in NLP that converts text into tokens, which are smaller units of meaning. In the context of LLMs, tokenization is essential for transforming clinical reports into a format that the model can understand and process. Effective tokenization ensures that the context and semantic meaning of the clinical data are preserved. This is particularly important in healthcare, where the accuracy of the converted data can directly impact patient outcomes. Proper tokenization allows the LLM to process the text more efficiently, reducing computational resources and time. Consistent tokenization ensures that similar clinical terms are represented uniformly, which is crucial for maintaining the integrity of the converted data. The process involves breaking down the text into individual words or subwords, which are then converted into numerical representations called embeddings.
Embeddings capture the semantic meaning of the text, allowing the LLM to understand the context and relationships between different parts of the clinical report. In LLMs, embeddings are generated through a process that maps tokens to high-dimensional vectors. These vectors are used by the model to understand and generate text. There are several types of embeddings, including word embeddings, subword embeddings, and contextual embeddings. Word embeddings represent individual words in a continuous vector space, capturing semantic and syntactic similarities. Techniques like Word2Vec [
15] and GloVe [
16] are commonly used to create word embeddings. Subword embeddings break words into smaller units, such as morphemes, to handle out-of-vocabulary words and rare terms more effectively. Byte Pair Encoding (BPE) and WordPiece are popular methods for subword tokenization. Contextual embeddings consider the context in which a word appears, allowing the same word to have different embeddings based on its usage. Models like BERT [
1] and RoBERTa [
2] use contextual embeddings to capture nuanced meanings.
2.2. Large Language Models
Large Language Models are a class of artificial intelligence models designed to understand and generate human language. These models are trained on vast amounts of text data and can perform a wide range of natural language processing tasks, such as text generation, translation, summarization, and question answering [
17,
18,
19,
20]. LLMs leverage deep learning techniques, particularly transformer architectures [
4], to capture complex linguistic patterns and contextual information.
Key characteristics of LLMs include scalability, versatility, and the ability to improve performance through prompt engineering. Scalability allows LLMs to handle large volumes of text data and generate coherent and contextually relevant responses. Versatility enables their application across various domains, including healthcare, finance, and customer service, to automate and enhance text-based tasks. Prompt engineering involves crafting specific input prompts to guide the model’s output, significantly improving performance. Advanced LLMs are designed to be computationally efficient, allowing them to handle complex tasks with high performance and minimal resource usage.
Mixture of Experts (MoE) is an advanced technique used to enhance the capabilities of LLMs by leveraging a combination of specialized sub-models, or “experts,” each trained to handle specific aspects of a task. In an MoE architecture, the model dynamically selects the most appropriate experts to process different parts of the input, allowing for more efficient and effective handling of complex tasks. This approach has been shown to improve the performance and scalability of LLMs, especially in scenarios requiring specialized knowledge or fine-grained processing [
21,
22]. The MoE approach allows for the integration of a large number of parameters, making it well-suited for handling complex and diverse datasets.
Fine-tuning is a process used to adapt pre-trained LLMs to specific domains or tasks by further training the model on domain-specific data. This approach allows the model to learn the unique linguistic patterns, terminology, and contextual nuances of the target domain, enhancing its performance and accuracy. Fine-tuning has been successfully applied in various domains, including healthcare, where models are fine-tuned using specialized clinical data to improve their understanding and generation of medical information [
23,
24]. By adapting the model to the unique characteristics of the target domain, fine-tuning can significantly improve its accuracy and reliability. Fine-tuned models are better equipped to understand and generate contextually relevant responses, making them more effective in specialized applications.
In the context of healthcare, LLMs have shown promise in tasks such as clinical note summarization, medical information extraction, and diagnostic support [
24,
25,
26]. However, their effectiveness in converting unstructured clinical data into structured formats, such as FHIR, remains an active area of research.
2.3. Impact of Prompts and Temperature
LLMs offer promising avenues for automating many diverse complex tasks, including converting unstructured clinical reports into standardized FHIR bundles. However, the effectiveness of these models is significantly influenced by two key factors: the design of input prompts and the temperature setting.
Prompts: The way information is presented to an LLM through prompts plays a crucial role in shaping its output. Different prompting strategies can guide the model’s reasoning and influence its ability to accurately extract information and map it to FHIR resources. Broadly, these strategies can be categorized by the level of guidance and examples provided.
A basic prompting approach involves providing a direct instruction to the LLM, such as “Convert this clinical report to a FHIR bundle.” While simple to implement, basic prompting often relies heavily on the model’s pre-existing knowledge and may not provide sufficient context for complex conversion tasks like FHIR.
More sophisticated, or advanced, prompting techniques aim to enhance the model’s performance by providing additional context, structure, or examples. These techniques can include providing explicit instructions on the desired output format, which helps the model understand the specific requirements of FHIR bundles. They can also involve incorporating examples of correctly formatted FHIR resources, which allows the model to learn from examples and improve its ability to generate valid bundles. Finally, these advanced techniques can guide the model’s reasoning process through step-by-step instructions, which helps the model break down the complex conversion task into smaller, more manageable steps.
Temperature Control: Temperature is a hyperparameter that controls the randomness or “creativity” of the LLM’s output. A lower temperature setting results in more deterministic and focused responses, prioritizing accuracy and consistency. This is particularly important for tasks like FHIR conversion, where adherence to strict standards is crucial. Conversely, a higher temperature setting introduces more randomness, leading to more diverse and potentially creative outputs, but at the cost of consistency and potentially impacting the validity of the generated FHIR bundles. Therefore, finding an appropriate temperature is essential for balancing the need for accuracy with the potential benefits of exploring diverse conversion strategies.
The choice of prompting strategy and temperature value has a direct impact on the quality, validity, and completeness of the generated FHIR bundles. By analyzing the interaction between prompts and temperature settings, this study sheds light on the optimal configurations for leveraging LLMs in converting unstructured clinical data to structured FHIR formats, particularly for Spanish clinical cases from different specialties such as skin care and mammography.
2.4. Fast Healthcare Interoperability Resources
Fast Healthcare Interoperability Resources (FHIR) is a standard for exchanging healthcare information electronically. It aims to facilitate the interoperability of healthcare systems by providing a consistent and flexible framework for representing clinical data.
Key components of FHIR include:
Resources: FHIR defines a set of resources that represent specific healthcare concepts, such as patients, observations, medications, and diagnoses. Each resource has a well-defined structure and can be combined to form complex clinical records. In our experiments, we restricted the resources types that the LLMs might use to convert input text to
Condition, Observation, MedicationStatement, Procedure, AllergyIntolerance, Encounter, Immunization, and EpisodeOfCare. We detail why we selected this curated set in
Appendix A.1.
Interoperability: FHIR supports various data exchange formats, including JSON and XML, making it compatible with a wide range of healthcare systems and technologies.
Extensibility: FHIR is designed to be extensible, allowing for the addition of new resources and the customization of existing ones to meet specific healthcare needs.
Standardization: FHIR adheres to strict standards and protocols, ensuring consistency and reliability in data exchange across different healthcare systems.
The adoption of FHIR has the potential to significantly improve healthcare outcomes by enabling seamless data sharing, enhancing clinical decision support, and facilitating research and public health initiatives. However, converting unstructured clinical data into the structured FHIR format is a complex task that requires sophisticated techniques and tools.
In our implementation, we utilized the HAPI FHIR library, an open-source Java implementation of the FHIR specification. HAPI FHIR defines model classes for every resource type and data type specified by FHIR, ensuring robust data validation and facilitating the serialization of clinical document elements into standard FHIR JSON representations.
In this study, we explore the use of LLMs to automate the conversion of unstructured clinical data into the FHIR format, with a focus on improving the accuracy and reliability of the generated FHIR bundles.
3. Related Works
This section provides an overview of the current research landscape related to the application of LLMs in healthcare, the role of Electronic Health Records (EHRs) and the FHIR standard, and methods for converting unstructured data into structured formats. We highlight key studies and findings that underscore the potential and challenges of these technologies in healthcare settings.
3.1. Large Language Models in Healthcare
LLMs have been increasingly applied in healthcare for various tasks, including text summarization and information extraction [
26]. Recent studies have demonstrated their potential in clinical text summarization, radiology report analysis, and extracting structured information from electronic health records [
24,
25,
26]. Prompt engineering has emerged as a critical technique to enhance the performance of LLMs in these tasks. These studies are particularly relevant to our research as they highlight the effectiveness of prompt engineering and the potential of fine-tuned LLMs in healthcare applications.
For instance, Wei et al. [
27] introduced chain-of-thought prompting, which improved LLM performance on multi-step reasoning problems. Additionally, Kojima et al. [
28] demonstrated that zero-shot chain-of-thought prompting can enhance LLM capabilities without task-specific examples. These findings underscore the importance of prompt engineering in maximizing the potential of LLMs for diverse applications. Building on these insights, our study evaluates different prompting techniques to improve the accuracy and efficiency of converting unstructured clinical data into structured formats.
The aim of this adapted Delphi study [
29] from 2024, was to collect researchers’ opinions on how LLMs might influence health care and on the strengths, weaknesses, opportunities, and threats of LLM use in health care. Participants offered several use cases, including supporting clinical tasks, documentation tasks, and medical research and education, and agreed that LLM-based systems will act as health assistants for patient education. The agreed-upon benefits included increased efficiency in data handling and extraction, improved automation of processes, improved quality of health care services and overall health outcomes, provision of personalized care, accelerated diagnosis and treatment processes, and improved interaction between patients and health care professionals.
In their study, Dalmaz et al. [
30] leverage clinician feedback to learn from their preferences to tailor the model’s outputs, ensuring they are aligned with the needs of clinical practice. Their experiments demonstrate that this approach improves upon the performance of the model post-supervised fine tuning. Their findings underscore the potential of integrating direct clinician input into LLM training processes, paving the way for more accurate, relevant, and accessible tools for clinical text summarization. These insights are particularly relevant to our study as we evaluate the performance of a fine-tuned LLM in healthcare contexts.
3.2. Electronic Health Records and FHIR
EHR systems are essential for modern healthcare, and the FHIR standard plays a crucial role in ensuring interoperability. Studies have shown that EHRs improve patient care by enhancing communication between clinicians, streamlining clinical activities, and providing comprehensive patient information at the point of care. A study by Koo et al. [
31] showed EHR-based sign-outs reduced medical errors by 30% compared to verbal hand-offs between hospital providers while a review found EHR enhanced communication between providers reduced hospital readmissions by 20-30% [
32].
The FHIR standard has emerged as a critical component in achieving interoperability between different healthcare systems. Bender and Sartipi [
13] introduced FHIR as a solution to address the limitations of previous standards, highlighting its potential to facilitate seamless data exchange. Mandel et al. [
14] further demonstrated FHIR’s effectiveness in enabling third-party apps to integrate with EHRs, enhancing functionality and data accessibility. Implementation of EHR systems with FHIR capabilities has shown promising results in improving healthcare delivery [
33]. For instance, Ayaz et al. [
33] developed a data analytic framework that supports clinical statistics and analysis by leveraging FHIR, showcasing the practical applications and benefits of FHIR in real-world healthcare settings. These studies collectively emphasize the crucial role of EHR systems and the FHIR standard in modernizing healthcare delivery, improving patient outcomes, and ensuring efficient, interoperable health information exchange.
3.3. Data Conversion and Structuring
Several studies have demonstrated the effectiveness of Machine Learning and NLP techniques in extracting structured information from scientific text. For example, a recent paper [
34] demonstrated the effectiveness of fine-tuned LLMs in extracting structured information from scientific text. Their findings include fine-tuning LLMs can extract complex scientific knowledge and format it as JSON objects. The method works well with only a few hundred training examples. Models showed the ability to automatically correct errors and normalize common entity patterns. In [
35], the authors tested various LLMs including GPT-3, InstructGPT, and PaLM on tasks like question answering, summarization, and code generation. For each task, LLMs were asked to generate responses in three formats: free-form text, structured formats (e.g., tables, lists, code), and a mix of both. They found that LLMs consistently performed better on free-form text generation compared to structured formats. Moreover, this study revealed that LLMs have inherent biases towards producing natural language text and struggle more with adhering to the constraints of structured data formats. Finally, stricter format constraints resulted in greater performance degradation in the LLMs’ reasoning abilities.
Despite these advancements, there are relatively few tools and methods specifically designed for converting unstructured clinical data into structured formats, particularly in healthcare. Our study addresses this gap by being the first to rigorously test the capabilities of LLMs in converting unstructured clinical data into the highly structured and complex FHIR format. This approach not only highlights the potential of LLMs in healthcare data conversion but also aims to guide other researchers by providing insights into which LLMs to use, how to prompt the models effectively, and what parameter values to choose for optimal performance.
4. Method
In this section, we first present the documents we used to evaluate the capacities of diverse LLMs in converting unstructured information in Spanish to FHIR. Then, we outline the various models we used to convert the information and evaluate the capacities of each models. Finally, we present technical details including prompt techniques and temperature values.
4.1. Data Collection from Clinical Centers
In this study, we extracted the clinical cases from two different centres; Mammography Clinic, specializing in plastic surgery, and Dermatology Clinic, focusing on skin care, providing a compelling and complementary dataset for this study. Both clinics generate a significant volume of unstructured clinical data that offers a rich opportunity to explore the application of LLMs and FHIR in healthcare.
4.1.1. Mammography Clinic
Breast surgery reports, which constitute the majority of Mammography Clinic’s unstructured data, represent a valuable resource for several reasons. First, mammography is a routine screening procedure with well-established reporting standards, providing a consistent format for data extraction. Second, the detailed descriptions of findings within mammography reports offer a complex and nuanced language that can challenge the capabilities of LLMs to accurately identify and classify entities. By focusing on mammography reports, we can evaluate the performance of LLMs in handling medical terminology, handling negations, and recognizing subtle nuances in clinical language.
4.1.2. Dermatology Clinic
Dermatological clinical reports complement the mammography data by providing a broader range of clinical conditions and presenting different challenges for natural language processing. While both mammography and dermatology reports contain detailed descriptions of findings, dermatology reports often involve more subjective assessments and may include descriptions of complex lesions and medication. This diversity in clinical presentation allows us to assess the degree of generalization of our findings and to identify any limitations of the models when applied to different types of clinical data.
The use of both mammography and dermatological reports offers several advantages. While the specific conditions and findings differ, both types of reports employ similar linguistic structures and conventions. By comparing the performance of LLMs on these two datasets, we can gain insights into the factors that influence the accuracy and efficiency of natural language processing in healthcare.
The combination of mammography and dermatological reports provides a diverse and challenging dataset that can be used to evaluate the potential of LLMs and FHIR in transforming unstructured clinical data into structured, machine-readable formats. By addressing the specific challenges posed by these datasets, we can advance the state of the art in natural language processing for healthcare applications. This comparative analysis allows us to identify commonalities and differences in how LLMs handle various types of clinical language, ultimately contributing to more robust and versatile NLP models for healthcare.
4.2. Models
In this study, we selected three cutting-edge LLMs to assess their proficiency in converting unstructured documents to FHIR. Each model possesses distinct features and capabilities that render them well-suited for this task and merit further exploration. To facilitate reproducibility of our experiments, we leverage the Langchain library
1, which serves as the foundation for the LLMs employed in our evaluation. By using the Langchain library, researchers can replicate our experiments and build upon our findings to advance the state of the art in LLM-based conversion quality. The LLMs evaluated in this study were accessed on 2025/03/18 09:53:30 and are presented in order of increasing transparency, ranging from the more opaque to the most open-source model.
Table 1.
This table includes information on the LLMs evaluated in this study such as the number of parameters, whether the model was fine-tuned, is open source, and the company that developed it.
Estimated count as the exact number of parameters is not publicly disclosed by Google.
* Mixtral 8x22b is a SMoE model with 22 distinct models at 7B, using 39B active parameters out of 141B.
Table 1.
This table includes information on the LLMs evaluated in this study such as the number of parameters, whether the model was fine-tuned, is open source, and the company that developed it.
Estimated count as the exact number of parameters is not publicly disclosed by Google.
* Mixtral 8x22b is a SMoE model with 22 distinct models at 7B, using 39B active parameters out of 141B.
| Model Name |
Parameters |
Fine-Tune |
Open Source |
Company |
| Gemini 1.5 Pro |
540B* |
No |
No |
Google |
| Mixtral 8x22b |
141B** |
No |
Yes |
Mistral AI |
| Llama3-Med42 |
70B |
Yes |
Yes |
M42 |
4.2.1. Gemini Pro
Gemini Pro is a highly compute-efficient multimodal model capable of recalling and reasoning over fine-grained information from millions of tokens of context [
20]. It achieves near-perfect recall on long-context retrieval tasks across modalities and improves the state-of-the-art in long-document QA, long-video QA, and long-context ASR. Gemini Pro has been shown to match or surpass the performance of other state-of-the-art models on a broad set of benchmarks.
Key features of Gemini Pro include multimodal capabilities, which allows Gemini Pro to process and generate responses across different modalities, such as text and images making it versatile for a wide range of applications. Morever, the model’s ability to handle long contexts makes it ideal for processing detailed clinical reports, which often contain extensive and complex information.
By leveraging Gemini Pro’s advanced capabilities, we aim to achieve high accuracy and efficiency in converting unstructured clinical data into structured FHIR formats.
4.2.2. Mixtral 8x22b
In this study, we have selected Mixtral 8x22b, a powerful variant of Mistral AI’s previous model [
19], Mixtral 8x7b. Mixtral 8x22b is a sparse Mixture-of-Experts (SMoE) model [
21] with 39B active parameters out of 141B, making it a significantly larger model size with 22 times more parameters than its predecessor. This increased parameter count aims to enhance Mixtral’s capabilities in handling complex tasks and understanding nuanced language patterns.
Mixtral 8x22b offers high performance and efficiency and is fluent in multiple languages, including Spanish. Its strong mathematics and coding capabilities make it well-suited for processing complex medical information. Mixtral 8x22b is known for its innovative architecture and efficient processing. By including Mixtral 8x22b in our study, we aim to gain valuable insights into how its unique design influences its ability to grasp information based on clinical cases and convert it to FHIR.
4.2.3. Llama3-Med42
Built on the Llama3 architecture [
36], M42 presents a collection of clinical LLMs designed to overcome the shortcomings of generic models in healthcare contexts [
23]. These models, called Med42 have been fine-tuned using specialized clinical data and undergone multi-stage preference alignment to effectively respond to natural language prompts. Unlike generic models that tend to be overly cautious and avoid answering clinical queries, Med42 is uniquely designed to overcome this limitation and address these clinical concerns. This changes makes it an ideal fit for clinical environments.
The Med42 models outperform across various medical benchmarks, GPT-4 as well as the original Llama3 models in both 8B and 70B parameter configurations. These LLMs are developed to understand clinical queries, perform complex reasoning tasks, and provide valuable assistance in clinical environments.
4.3. Prompting Strategies and Temperature Levels
Prompt Techniques. In this study, we employed various prompting strategies to instruct the LLMs to convert information from clinical reports into the FHIR format. The specific prompts used are provided in
Appendix B. In total, we evaluated the LLMs in six different setups, combining each of the three prompt strategies (basic, two-step reasoning, and chain-of-thought) with both zero-shot and few-shot examples.
The
basic strategy directly asks the LLM to convert the clinical report to FHIR. For the
two-step reasoning strategy, following the approach from Tam et al. [
35], we first asked the LLM to group the information that might be interesting into a FHIR bundle. In a subsequent step, the LLM was tasked with converting this structured information into FHIR. Based on recent advancements in LLMs’ reasoning capabilities, we implemented the iterative self-correcting
Chain-of-Thought (CoT) strategy [
27,
37]. This involved first asking the LLM to perform the conversion and then, in a second call, comparing and correcting the generated FHIR with the clinical report. Details about the prompts and design choice are provided in
Appendix B.1.2
The zero-shot technique involved presenting the LLMs with the text of an unstructured document and asking them to convert it to FHIR following the desired structure. This approach assesses the LLMs’ ability to perform the task without any prior examples, relying solely on their pre-trained knowledge.
The
few-shot technique involved providing the LLMs with a detailed example of an unstructured document and its corresponding FHIR document before asking them to convert a new clinical report. To ensure a robust and representative few-shot learning approach, we iteratively designed a detailed example that covers every resource information. This example is described in
Appendix B.1.1 and was chosen for its representativeness, complexity, and relevance to the study’s objectives.
Temperature. Temperature is a crucial hyperparameter that regulates the randomness of the LLMs’ generated outputs. A temperature of 0 produces deterministic output, whereas a higher temperature value yields more diverse outputs. In this study, we experimented with low (0) and high (2) temperature levels. This decision was informed by a previous study [
38], which suggested that temperature variations within the range of 0 to 1 do not significantly impact LLM performance in problem-solving tasks. By examining the effects of these two temperature levels, we aim to gain insights into the LLMs’ performance and the influence of temperature on the conversion to FHIR based on clinical cases.
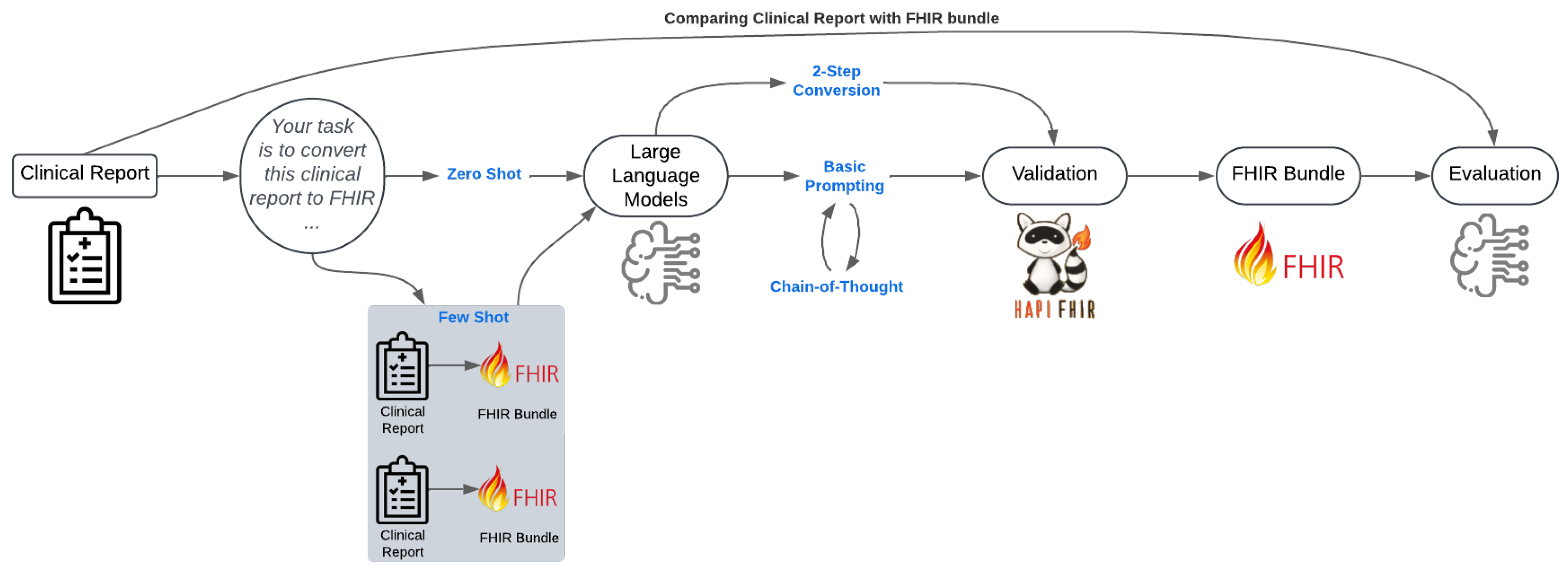
5. Methodology
The workflow for converting unstructured clinical reports into FHIR bundles and evaluating their quality is summarized in
Figure 2. Through this process, we aim to enhance healthcare interoperability by providing insight into the best model, prompt strategies, and parameters to convert unstructured clinical reports to FHIR.
Evaluating the quality of conversions generated by LLMs is a challenging task, especially in the sensitive domain of healthcare [
39]. Evaluation of long-form generations from LLMs remains a nascent area of work [
40]. In this study, we evaluated the quality of the conversion through three methods.
First, we ensured that the resulted data adheres to FHIR standards by implementing data type conversions and value transformations for elements requiring specific data types (e.g., boolean, integer, string, decimal). We call this metric Failure Rate, which corresponds to the proportion of FHIR bundles that respect the FHIR standards using the HAPI FHIR validation tool. A lower failure rate indicates better adherence to FHIR standards.
Second, we developed an approach to automatic evaluation using a multifaceted, iterative design methodology. We leveraged the capabilities of the LLMs themselves to assess the quality of the conversion. Specifically, we used GPT-4o
2 [
41], an advanced external commercial LLM, as
Evaluator. We chose GPT-4o as the evaluator for several reasons. First, GPT-4o is known for its advanced capabilities in understanding and generating coherent, contextually relevant text, making it well-suited for evaluating the quality of the converted outputs. Second, its extensive training on a diverse range of tasks and data ensures that it can provide robust and reliable evaluations across various metrics. Third, using an external evaluator helps mitigate potential biases that might arise from using the same model for both conversion and evaluation. This model evaluates the conversion done by other LLMs, which we refer to as
Convertor LLMs.
We designed the evaluation by instructing the Evaluator to compare the model answers with the original clinical document to determine if the Convertor LLMs show high capacities across our three metrics: (a) precision, (b) hallucination, and (c) resource mapping accuracy:
Precision: Does the converted document correctly identified the information described in the clinical case? High precision indicates accurate identification and representation of clinical information.
Hallucination: Does the converted document included information not described in the clinical case? A low hallucination rate indicates that the conversion accurately reflects the original clinical report without introducing extraneous information.
Resource Mapping Accuracy: Are data elements correctly mapped to their appropriate FHIR resources? High resource mapping accuracy ensures that all relevant data elements are correctly represented in the FHIR bundle.
To perform the automatic evaluation, we used a set of pre-defined prompts to query the Evaluator and elicit their assessments of the converted document (refer to
Appendix B.2 for detailed prompts). In line with Hada et al. [
42], we adjusted the temperature of the Evaluator to 0. For each clinical case, we asked the Evaluator to evaluate the conversion generated by each Convertor LLMs on a scale from 1 to 10 described in
Appendix B.2. We then aggregated the results across clinical cases to obtain a comprehensive evaluation of the performance of each LLM.
Finally, we manually reviewed the FHIR bundle to perform a qualitative analysis and identify trends for each LLM, prompt technique, and temperature level. This comprehensive evaluation approach ensures that the converted FHIR bundles are accurate, reliable, and compliant with FHIR standards, thereby enhancing data interoperability and improving healthcare outcomes.
5.1. Hypotheses
To guide our investigation into the effectiveness of different LLMs and prompting techniques in converting unstructured clinical data to structured FHIR formats, we formulated several hypotheses that we present in this section.
5.1.1. Prompting Technique Effectiveness
Prompting techniques that break down the task into manageable steps (e.g., two-step conversion) or provide a structured reasoning path (e.g., CoT) are expected to improve the model’s ability to accurately map information to FHIR resources. Basic prompting, which lacks such guidance, may result in less structured and less accurate outputs. Thus, we formulated the following hypotheses:
-
H1.1:
Prompting techniques such as Chain-of-Thought (CoT) and two-step conversion will yield more accurate and reliable FHIR bundles compared to basic prompting.
-
H1.2:
Two-step conversion will outperform CoT in terms of resource mapping accuracy and overall completeness of the FHIR bundles.
5.1.2. Impact of Few-Shot vs Zero-Shot Prompting
Providing examples in the prompt (few-shot prompting) can guide the model by showing it the expected format and content. This guidance can help the model identify relevant information and generate more complete FHIR bundles. However, over-reliance on examples may lead to the reproduction of irrelevant or incorrect information.
-
H2.1:
Few-shot prompting will result in more complete and accurate FHIR bundles compared to zero-shot prompting.
-
H2.2:
Few-shot prompting may introduce biases or irrelevant information.
5.1.3. Influence of Temperature
Temperature settings control the randomness of the model’s output. Higher temperatures allow the model more flexibility, potentially generating more diverse and numerous resources. Lower temperatures, on the other hand, restrict the model’s variability, leading to more consistent and precise outputs that better adhere to FHIR standards.
-
H3.1:
Higher temperature will increase the number of resources that the model is generating compared to low temperature.
-
H3.2:
Lower temperature settings will yield more consistent and precise FHIR bundles.
5.1.4. Model Performance
Specialized models that have been fine-tuned on healthcare data are expected to have a better understanding of the domain-specific nuances and terminology, leading to more accurate and precise FHIR bundles. General-purpose models, while powerful, may struggle with the specific requirements of healthcare data conversion. Additionally, models like Mistral, which retain more of the original text, may introduce challenges in standardizing the information for FHIR conversion.
-
H4:
Specialized models fine-tuned on healthcare data (e.g., Llama3-Med42) will perform better than general-purpose models (e.g., Mistral, Gemini) in terms of precision and resource mapping accuracy. However, Mistral and Gemini will exhibit similar performance characteristics, with Mistral retaining more original text, potentially affecting standardization.
6. Results
In our initial experiments, we converted 20 clinical reports (10 from each clinic) to FHIR format using three LLMs: Mistral-8x22b, Gemini 1.5 Pro, and Llama3-Med42, under various prompt settings and temperature levels. We explored combinations of prompting techniques, including straightforward conversion, two-step conversion, and chain-of-thought. Each of these prompting techniques was tested with and without examples. This resulted in 720 conversions. We present the proportion of empty bundles after verifying that the output results adhere to the FHIR standards in
Section 6.1. In
Section 6.2, we analyze the distribution of resource types per bundle, categorized by model and clinic, to provide nuanced insights into the performance of different LLMs. In
Section 6.3 to
Section 6.5, we detail the results obtained for precision, hallucination rate, and resource mapping accuracy of the converted documents, with scores ranging from 1 (low precision, hallucination, and resource mapping accuracy) to 10 (high precision, hallucination, and resource mapping accuracy). Finally, we present our findings on the qualitative analysis in
Section 6.6.
6.1. Failure Rate
We report in
Table 2 the failure rate of each LLM, prompt technique, presence of examples in the prompt, and temperature levels. A green tick (
✓) indicates a full conversion rate, while a red cross (
×) indicates cases where none of the 10 clinical reports were successfully converted to FHIR according to the FHIR standards.
From the table, we observe that the two-step conversion method increases the most the proportion of successful conversions. For instance, Gemini successfully converted every document to FHIR, and Mistral achieved an average failure rate of 10%. Conversely, straightforward or basic conversion techniques resulted in the highest average failure rates, especially with zero-shot prompting.
Additionally, we noted that higher temperatures with few-shot examples reduced the failure rate compared to lower temperatures. However, with zero-shot prompting, there was no clear difference in failure rates across temperature levels.
Finally, these results suggest that the fine-tuned LLM on healthcare data, Llama3-Med42, is not well-suited for the task of converting unstructured data to well-defined and structured FHIR. The model often attempted to interpret the clinical report and provide diagnostic suggestions rather than converting it to FHIR. This behavior might be due to the specific fine-tuning of the LLM for diagnostic tasks. Consequently, we removed Llama3-Med42 from the subsequent analyses since it did not produce any resulting valid FHIR bundles.
6.2. Resource Distribution Analysis
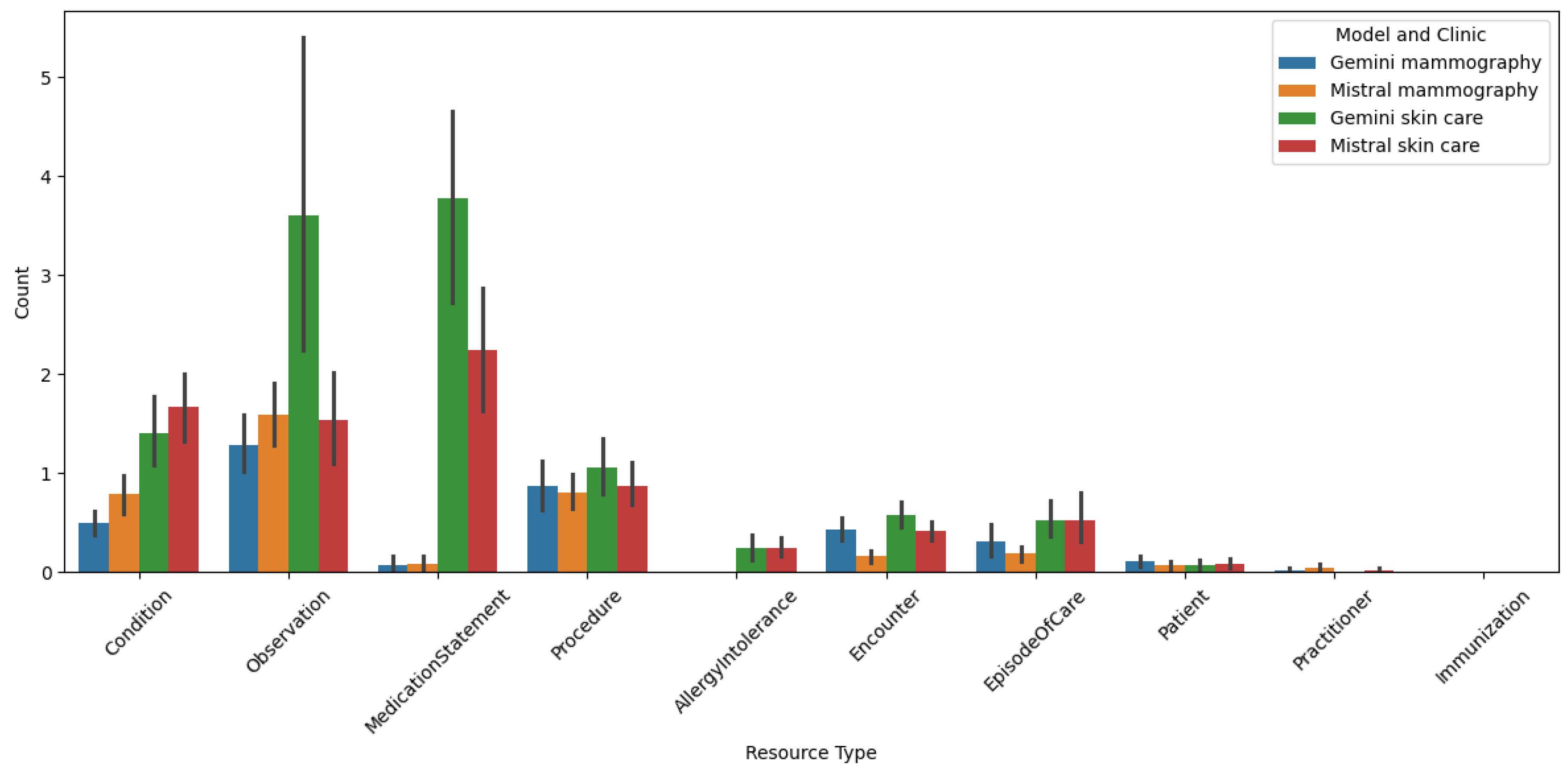
The
Figure 3 presents a bar chart illustrating the distribution of various resource types per bundle, where each bundle corresponds to one clinical report. The data is categorized by different models and clinics, specifically comparing the performance of Gemini and Mistral models in handling mammography and skin care reports from two distinct clinics. The chart highlights the count of different resource types, such as Condition, Observation, MedicationStatement, Procedure, AllergyIntolerance, Encounter, EpisodeOfCare, Patient, Practitioner, and Immunization, across these categories.
This chart provides insights into the types of reports each clinic is producing. Notably, the skin care clinic has a much greater number of MedicationStatement resources, which aim to capture the active or prescribed medication of the patient, compared to the mammography clinic, even when comparing across models. This disparity arises because the documents in the skin clinic consist mainly of diagnoses with proposed treatments, and in the majority of the times, include a summary of the current patient’s situation such as medication, conditions, and allergies. This explains the presence of few AllergyIntolerance resources in the skin clinic’s reports, which are not used in the mammography documents.
In contrast, mammography reports contain more observation-like text, for example findings of nodules and their characteristics, in the majority of them, a BIRADS assessment, and in some cases, procedures that the patient will undergo due to a specific finding. Upon analyzing the chart and comparing the distribution among models, several key observations emerge. The Gemini model, particularly in skin care reports, shows a higher count of resources like Observation and MedicationStatement compared to the Mistral model. This suggests that the Gemini model might be more effective in capturing detailed clinical information in skin care reports. Conversely, the Mistral model demonstrates a more balanced distribution of resource types in mammography reports, indicating its potential versatility in handling diverse clinical data.
The variation in resource counts across different clinics and models underscores the nuanced performance of LLMs in converting unstructured clinical data into structured FHIR formats. These results are particularly interesting, as they reveal the strengths and weaknesses of each model in different clinical contexts, providing valuable insights into optimizing LLM-based data conversion pipelines for enhanced healthcare outcomes.
6.3. Precision
Table 3 indicates variability in the performance of the LLMs depending on the prompt technique used, highlighting the importance of prompt engineering in optimizing the conversion of unstructured clinical data to structured FHIR format.
First, we observe higher precision with few-shot prompting compared to zero-shot techniques. Second, higher temperatures increase this effect compared to lower temperatures. Additionally, tasking the model to structure the information before converting it to FHIR (two-step conversion) yields the best results, even without the presence of examples in the prompt.
Although both models exhibit similar trends, Gemini generally outperforms Mistral across both clinics and various prompt techniques. Both models show the lowest precision with the basic conversion technique.
6.4. Hallucination Rate
We depict in
Table 4 the mean and standard deviation of the hallucination rate for each model’s conversion from unstructured data to FHIR. The table highlights the effectiveness of different prompting techniques and temperature levels in mitigating hallucinations.
Key observations include consistently lowest hallucination rates with the Two-Step conversion across all models and prompting techniques. By first structuring the information and then converting it to FHIR, the model can better understand the context and relevance of the data, leading to more accurate and less hallucinogenic outputs. Then, providing examples in the prompts slightly increases the hallucination rate compared to zero-shot prompting. This suggests that while examples can guide the model, they may also introduce biases or irrelevant information, leading to higher hallucination rates. While higher temperatures generally increase the hallucination rate, especially with zero-shot prompting, we do not observe difference in these experiments.
Finally, Mistral tends to have slightly higher hallucination rates compared to Gemini with zero-shot prompting, while the opposite is observed with few-shot prompting.
6.5. Resource Mapping
The
Table 5 shows that providing examples in the prompt increases the resource mapping accuracy, especially with basic and CoT prompting techniques. We also observe more diversity in the results with few-shot prompting settings, particularly at lower temperatures. Notably, in some situations (e.g., Mistral with few-shot prompting), very high accuracy results are observed with the CoT prompting technique (above 7.5). Additionally, Gemini and Mistral tend to have similar resource mapping accuracy across all prompt techniques and temperature levels.
These observations underscore the importance of careful prompt engineering and temperature control in optimizing the conversion of unstructured clinical data to structured FHIR formats. By understanding these factors, we can better design prompts and adjust parameters to enhance the accuracy and reliability of the converted data.
6.6. Qualitative Analysis
The qualitative analysis involved comparing 480 generated FHIR bundles with their original clinical reports. We examined ten conversions produced by each LLM (Gemini and Mistral) across every combination of prompt techniques (Basic, Two-Step, CoT), temperature settings (low vs. high), presence of examples, and clinics. Our analysis encompassed the raw LLM outputs, the HAPI FHIR-validated bundles, and the original clinical reports. After an initial overview of performance trends for each configuration, we leveraged GPT-4o to provide a qualitative assessment of the conversions, identifying both positive and negative aspects. We then used these evaluations as basis and verified the elements indicated as positive and negative by comparing them with the clinical reports.
6.6.1. Impact of Prompt Techniques
With Basic prompting, the LLM produces structured reports containing important information. However, these reports do not follow the FHIR standard. For instance, they often use non-standard terms (e.g., `Doctor’ instead of `Practitioner’ or `Study’ instead of `Observation’), making automatic conversion to FHIR impossible. The basic approach, while straightforward, lacks the necessary guidance to ensure compliance with FHIR standards, resulting in reports that are informative but not structurally valid.
Using Chain-of-Thought (CoT) prompting, we found that the model accurately identified the relevant information from the source documents. Implementing a second CoT round, intended to allow the model to correct any initial errors, increased the number of valid FHIR bundles generated. However, this approach did not consistently produce fully valid bundles and frequently resulted in a reduction in the number of information elements included.
The two-step conversion approach performs well. This method involves first structuring the information and then converting it to FHIR. It correctly identifies the information and successfully converts it to FHIR, demonstrating a more robust and reliable method for this task. The two-step approach ensures that the information is first organized in a way that the model can understand and then accurately mapped to FHIR resources, resulting in more complete and valid bundles.
6.6.2. Other Factors Influencing Bundle Generation
There are notable differences in the completeness and complexity of the bundles generated for the skin care and mammography clinics. The bundles generated for the skin care clinic are generally more complete and complex, often including detailed patient information, diagnostic notes, and treatment plans. In contrast, the bundles generated for the mammography clinic sometimes include only patient and practitioner names, lacking the comprehensive details required for a full FHIR representation. This disparity may be attributed to the nature of the clinical reports from each clinic as observed in
Section 6.2, with skin care reports containing more structured and detailed information.
Few-shot prompting helps the model identify more information and generate more complete bundles. By providing examples, the model is better guided in understanding the expected format and content of the FHIR bundle. However, a notable issue with few-shot prompting is that the model sometimes follows the example too closely, reproducing the values as they appear in the example without updating them with the actual values from the clinical report. For instance, the model might retain placeholder values like “Date-Time: YYYY-MM-DD’’ instead of inserting the correct date and time from the document. This over-reliance on the example can lead to errors and inaccuracies in the generated bundles.
The impact of temperature on the LLM’s performance appears to be minimal. Temperature settings, which control the randomness of the model’s output, do not seem to significantly affect the resource mapping accuracy or the completeness of the bundles. This suggests that the model’s performance is more influenced by the prompting technique and the presence of examples rather than temperature variations.
Overall, there are few significant differences between Mistral and Gemini, except that Mistral tends to retain more of the original text in Spanish. This characteristic of Mistral may be beneficial in preserving the nuances of the original clinical reports but can also introduce challenges in standardizing the information for FHIR conversion. Gemini, on the other hand, appears to be more adept at translating and structuring the information in a way that aligns with FHIR standards.
7. Discussion
This study investigated the effectiveness of LLMs in converting unstructured clinical reports into standardized FHIR bundles, exploring the impact of prompting strategies, few-shot learning, temperature settings, and LLM selection. Our findings offer valuable insights into optimizing this process for healthcare data interoperability and allow us to validate our initial hypotheses.
7.1. Prompting Strategies and Conversion Success (H1.1 & H1.2)
Our results strongly support H1.1, demonstrating that structured prompting techniques like CoT and two-step conversion significantly outperform basic prompting. The high failure rates observed with basic prompting (
Table 2) highlight its inadequacy for complex FHIR conversion due to a lack of guidance in adhering to FHIR standards. As hypothesized in H1.2, the two-step conversion method proved most effective, achieving near-perfect conversion rates with Gemini and significantly reducing failures with Mistral. This underscores the importance of explicitly structuring the conversion process for LLMs, confirming that organizing information prior to FHIR mapping is crucial for accurate resource mapping and bundle construction. While CoT offered an improvement over basic prompting, it did not match the performance of the two-step approach. This discrepancy suggests that the CoT approach, while effective in breaking down complex tasks, may not be ideally suited for the precise and structured requirements of FHIR conversion. The model’s ability to identify relevant information is not fully translated into accurate FHIR resource mapping, leading to incomplete or incorrect bundles. This finding aligns with the observations of Wu et al. [
43], which demonstrated that LLMs struggle to self-correct reasoning without external feedback and may even experience performance degradation after self-correction attempts. In our context, the lack of explicit structuring in CoT prompts can be seen as a form of limited feedback, hindering the model’s ability to consistently generate valid FHIR.
7.2. The Role of Examples (Few-Shot Learning) (H2.1 & H2.2)
Our findings partially support H2.1, indicating that few-shot prompting generally improves performance, particularly in precision and resource mapping accuracy (
Table 3 and
Table 5). This can be attributed to several factors. Firstly, providing an example helps the LLM understand the desired output format and structure more clearly, reducing ambiguity and guiding the model to generate more accurate FHIR bundles. Secondly, examples serve as a form of contextual learning, allowing the LLM to better interpret the nuances of the clinical data and map them correctly to the corresponding FHIR elements. Lastly, examples may help mitigate hallucinations by anchoring the model’s responses to a concrete reference, thereby reducing the likelihood of generating irrelevant or incorrect information. However, we also confirmed H2.2, identifying the risk of over-reliance on examples, where the model reproduced placeholder values instead of extracting information from the input reports. This highlights the delicate balance between providing sufficient guidance and avoiding overfitting. Interestingly, the failure rate was reduced with few-shot prompting at higher temperature compared to lower temperature. This could be due to the fact that higher temperature allows the model to deviate more from the examples, thus mitigating the over-reliance issue. This finding underscores the importance of incorporating examples in prompts when using LLMs for structured data conversion tasks in healthcare, particularly when dealing with sophisticated and standardized formats like FHIR.
7.3. Influence of Temperature (H3.1 & H3.2)
Our results partially support our hypotheses regarding temperature. While we did observe that higher temperatures sometimes led to the generation of more resources, we did not find a consistent and significant increase as predicted by H3.1. Furthermore, while lower temperatures generally contributed to more consistent outputs, the impact of temperature was less pronounced than that of prompting strategies or the use of examples. This suggests that while temperature plays a role in controlling output variability, its influence on FHIR conversion is secondary to prompt design. However, higher temperatures did increase the risk of hallucinations, especially with zero-shot prompting (
Table 4), partially supporting H3.2, which stated that lower temperature would yield more precise bundles. This aligns with the general understanding that lower temperatures promote more focused and deterministic outputs, which is crucial for tasks requiring adherence to strict standards.
7.4. Model Performance (H4)
Our results strongly refute H4, which hypothesized that the specialized, fine-tuned Llama3-Med42 model would outperform the general-purpose models. Llama3-Med42 proved entirely unsuitable for the FHIR conversion task, consistently prioritizing diagnostic suggestions over accurate data mapping, resulting in no valid FHIR bundles. This highlights the importance of selecting models appropriate for the specific task. Additionally, Gemini generally outperformed Mistral in terms of precision and hallucination rate, particularly with zero-shot prompts. While Mistral did retain more Spanish text, this did not translate to improved FHIR conversion and often introduced standardization challenges.
Standardizing Electronic Health Record (EHR) data using formats like FHIR offers significant potential for enhancing clinical data interoperability, enabling high-throughput computation, and promoting meaningful use. To advance FHIR-based EHR data modeling, this study evaluated the capacity of LLMs to extract, standardize, and integrate information from unstructured clinical narratives into FHIR resources. We believe that LLM-driven modeling of unstructured EHR data can play a crucial role in achieving advanced semantic interoperability across disparate EHR systems. It is important to emphasize that the FHIR format is highly restricted and complex, with clearly defined standards. Our results indicate that LLMs, when used with appropriate prompting strategies, can contribute significantly to this goal. This indicates that the main challenge lies in the conversion of the extracted information into a valid FHIR structure, rather than the initial information identification itself.
7.5. Limits & Future Works
This study has some limitations. The evaluation was conducted using reports in Spanish from only two clinics (skin care and mammography). This limits the generalizability of the findings to other languages, clinical domains, and report types. Future work should include a more diverse dataset of clinical reports to assess the robustness of the proposed methods across different clinical contexts.
While we evaluated several LLMs, including the specialized, healthcare-fine-tuned Llama3-Med42, resource constraints limited our in-depth analysis to this single specialized model. This model proved unsuitable for the FHIR conversion task due to its focus on diagnosis. This highlights the importance of selecting models appropriate for the specific task. Future work should explore LLMs explicitly fine-tuned for FHIR conversion tasks. Furthermore, a key limitation is the lack of control over the training data used for all LLMs. These models are trained on massive text corpora, making it impossible to know precisely what information they have learned and how this might influence their performance on FHIR conversion.
The qualitative analysis, while providing valuable insights, relied on GPT-4o as an evaluator, which exhibited limitations such as occasional hallucinations (identifying non-existent errors) and providing non-substantial feedback, such as “
The hallucination rate was relatively low but could be improved.” or “
The resource mapping could be improved for better accuracy.”. While the overall trends observed by GPT-4o were in line with our own observations, the limitations of LLMs as evaluators should be acknowledged. This aligns with findings from [
42], which suggest evaluating single metrics at a time for more reliable results, although at the cost of increased computational resources.
Our evaluation primarily focused on the structural validity and information content of the generated FHIR bundles. We did not explicitly evaluate the accuracy of coded values mapped to standard terminologies and ontologies (e.g., LOINC, RxNorm, SNOMED CT), or the use of locally maintained dictionaries and look-up tables within FHIR profiles. This is an important aspect of FHIR compliance that should be addressed in future work.
Future research could investigate more advanced prompting techniques, such as prompt chaining or fine-tuning on FHIR-specific data, to further improve conversion accuracy and reduce the risk of over-reliance on examples. Exploring different LLM architectures and sizes could also provide valuable insights. Additionally, developing automated metrics for evaluating FHIR bundle quality would enhance the objectivity and scalability of future studies. Future work should also evaluate the accuracy of mapping clinical concepts to standard terminologies and ontologies within FHIR resources. Finally, future work could investigate the use of FHIR profiles to further constrain and guide the LLM’s output, ensuring adherence to specific implementation requirements.
8. Conclusion
This study explored using LLMs to convert unstructured clinical reports to FHIR bundles, a key step for healthcare data interoperability. We investigated prompting strategies (basic, CoT, two-step), few-shot learning, temperature, and LLM selection (Gemini, Mistral, Llama3-Med42).
Prompt engineering emerged as a critical factor. Basic prompting proved inadequate, underscoring the necessity of providing explicit guidance to the LLM. The two-step conversion method consistently outperformed other approaches, demonstrating the value of pre-structuring information before FHIR mapping. While CoT offered some improvement over basic prompting, it did not achieve the same level of effectiveness as the two-step method. Few-shot learning enhanced precision and resource mapping but also introduced the risk of over-reliance on examples, emphasizing the importance of careful example selection. Temperature had a less pronounced effect than prompting, although higher temperatures correlated with an increased risk of hallucinations. The healthcare-fine-tuned Llama3-Med42 proved unsuitable for this task, prioritizing diagnostic suggestions over accurate FHIR mapping, while Gemini generally outperformed Mistral.
Key contributions of this work include: (1) demonstrating the effectiveness of the two-step conversion approach; (2) highlighting the trade-offs inherent in few-shot learning; (3) evaluating the impact of temperature and LLM selection; (4) providing a comprehensive evaluation framework for FHIR conversion using LLMs; and (5) addressing the challenge of converting unstructured EHR data to FHIR.
In conclusion, LLMs demonstrate significant potential for automating FHIR conversion, but careful prompt engineering, appropriate use of few-shot learning, and thoughtful parameter selection are essential for optimal results. The two-step conversion strategy offers a particularly promising path towards generating accurate and valid FHIR bundles, thereby advancing healthcare data interoperability.
Funding
This research received no external funding.
Informed Consent Statement
Not applicable.
Acknowledgments
J.C. work was supported by the Torres Quevedo grant “Asistente Virtual para la mejora del sistema de salud—Virtual Assistant for Better HealthCare -VA4BHC” (ref. PTQ2021-012147).
Conflicts of Interest
Author were employed by the company Top Health Tech.
Abbreviations
The following abbreviations are used in this manuscript:
| LLM |
Large Language Model |
| NLP |
Natural Language Processing |
| ML |
Machine Learning |
| RAG |
Retrieval Augmented Generation |
| CoT |
Chain-of-Thought |
Appendix A. Study Design Choices
Appendix A.1. Resource Selection for FHIR Conversion
We carefully selected a subset of FHIR resource types for the LLMs to use in converting input text. The chosen resource types are Condition, Observation, MedicationStatement, Procedure, AllergyIntolerance, Encounter, Immunization, and EpisodeOfCare.
This decision was based on their collective ability to comprehensively represent the clinical complexities inherent in most possible scenarios. This curated set encompasses a broad spectrum of healthcare activities, from diagnosis and treatment to patient history and follow-up. By focusing on a small set, we have established a robust foundation to ensure that the LLMs have the correct tools not only to accurately model the majority of clinical scenarios in these specialties but also to capture all details due to their intrinsic complexity.
The absence of resources specifically for imaging is justified by the fact that the primary focus of this study is on extracting information from textual reports. Furthermore, the selected set of resources is sufficient for representing the key elements of mammography and dermatology reports. Findings within dermatology and mammography reports can be captured using the Condition resource, while instances related to imaging observations can be recorded using the Observation resource. Procedures, such as biopsies or surgical interventions, can be documented using the Procedure resource. Medications prescribed for treatment can be captured using the MedicationStatement resource, and patient allergies and intolerances can be recorded using the AllergyIntolerance resource. Encounters can be used to document patient visits and the services provided, while immunizations can be recorded using the Immunization resource.
This selection allowed us to ensure that the LLMs follow our guidelines, using the most relevant and comprehensive set of FHIR resources for the given clinical contexts.
Appendix B. Prompt Template
In this section, we detail the diverse prompts we used to ask the Large Language Models (LLMs) to generate a task. We first present in
Appendix B.1, the prompts used to make the models convert a clinical case to FHIR. Then, in
Appendix B.2, we outline the prompts used to evaluate the precision and hallucination rate of the conversion generated by the LLM. Text between `<>’ represents substitutions that are made to the prompt at inference time, for example providing the text of the clinical report.
Appendix B.1. Conversion Prompts
To ensure the accurate conversion of unstructured clinical data into structured FHIR format, we designed a series of prompts to guide the LLM through the process. In this section, we present the prompts used in our study.
For the system prompt, we tested several variations to determine which one would yield the best results. We found that providing the model with a brief explanation of the task helped improve the model’s performance. We also experimented with different levels of detail in the instructions, ranging from very specific to more general. For example, we initially provided detailed instructions on how to structure each FHIR resource, but found that this sometimes led to overly complex outputs. Simplifying the instructions to focus on the key elements of each resource type resulted in more accurate and consistent outputs.
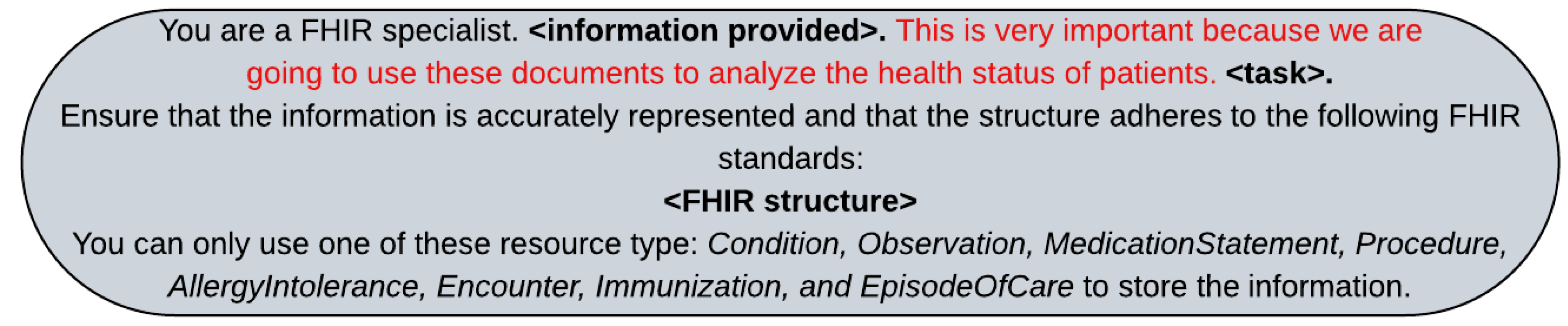
The prompt specifies that the LLM should act as a FHIR specialist and emphasizes the importance of accurately representing the original text. Text in red is the emotional stimulus we have added to the prompt diagnosis, in line with Li et al. [
44]. We replaced the <information provided> and <task> depending on whether we were using zero shot, few shot, two-step reasoning, or Chain-of-Thought (CoT) approaches.
Figure A1.
System prompt used to instruct the LLM on converting unstructured clinical data into structured FHIR format. We replaced <information provided> and <task> by the corresponding text depending on the prompt technique used. Finally, the <FHIR structure> is replaced with detailed instruction of the FHIR standards to be followed.
Figure A1.
System prompt used to instruct the LLM on converting unstructured clinical data into structured FHIR format. We replaced <information provided> and <task> by the corresponding text depending on the prompt technique used. Finally, the <FHIR structure> is replaced with detailed instruction of the FHIR standards to be followed.
Appendix B.1.1. Zero vs Few-Shot Prompting
In case of zero and few shot, we replaced <information provided> with “I will provide you with the plaintext of a file containing unstructured information about a patient and you will return a valid JSON containing the appropriate resources to represent the information contained in that plaintext.” and we replaced <task> by “Display the original text from the clinical case without making any changes or adding any information that does not exist”. For the example-based prompt technique, we filled in the details of the clinical case and the associated FHIR resource. We also tested different variations of the template, such as changing the order of the symptoms or using different phrasing, to see how these changes would affect the model’s performance. We found that providing clear and concise examples helped the model understand the expected format and structure of the FHIR resources. This example was chosen to provide a comprehensive and representative set of FHIR resource types, ensuring the robustness and reliability of our findings.
Appendix B.1.2. Basic vs Two-Step vs Chain-of-Thought Reasoning
For the CoT system prompt, we designed a prompt that instructed the model to correct the generated FHIR bundle based on the original clinical report. Thus, we first replaced <information provided> by “I will provide you with the plaintext of a file containing unstructured information about a patient and a JSON that is a FHIR bundle with these information structured. You will review the provided FHIR bundle and correct any errors or inconsistencies.”. We also replaced <task> with “Ensure that the information is accurately represented and that the structure adheres to FHIR standards.”.
Figure A2.
Example of how the instruction for the LLM is presented in the system prompt for the resource type Condition. This example demonstrates the specific format and structure that the LLM should follow when generating the Condition resource in the FHIR bundle.
Figure A2.
Example of how the instruction for the LLM is presented in the system prompt for the resource type Condition. This example demonstrates the specific format and structure that the LLM should follow when generating the Condition resource in the FHIR bundle.
Finally, the system prompt contains information about the FHIR guidelines structure we want the model to follow for each of the accepted resource types.
Figure A2 provides an example of how the instruction for the LLM is presented in the system prompt for the resource type “
Condition”. This example demonstrates the specific format and structure that the LLM should follow when generating the Condition resource in the FHIR bundle.
In our study, we leverage the capabilities of Langchain and the chat structure of the model to facilitate the conversion of unstructured clinical data into structured FHIR format. Once the system prompt is set up, we provide the model with the plain text of the clinical report. For the iterative correction process, we use the prompt detailed in
Figure A3 and we replace
<FHIR generated> with the bundle generated in the previous round and
<clinical case> with the plain text of the clinical report. This iterative approach allows us to refine the conversion process, ensuring that the generated FHIR bundles are accurate and consistent with the original clinical data.
Figure A3.
Prompt used for the iterative process of converting unstructured data to FHIR. The prompt instructs the LLM to correct the generated FHIR bundle based on the original clinical report. It specifies that the LLM should return only the corrected FHIR bundle without adding any additional information and should use only the specified resource types to store the information.
Figure A3.
Prompt used for the iterative process of converting unstructured data to FHIR. The prompt instructs the LLM to correct the generated FHIR bundle based on the original clinical report. It specifies that the LLM should return only the corrected FHIR bundle without adding any additional information and should use only the specified resource types to store the information.
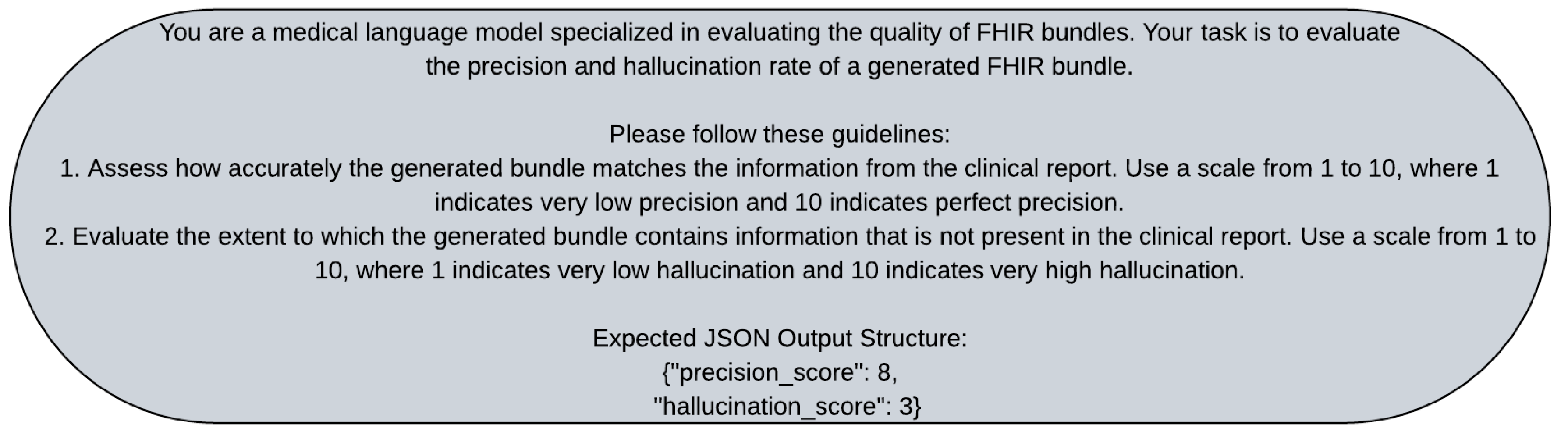
Appendix B.2. Evaluator Prompts
For the evaluation prompt, we used a structured approach to assess the precision and hallucination rate of the generated FHIR bundles. We instructed the evaluator to evaluate the precision and hallucination rate of the generated bundle. We provided in
Figure A4, guidelines for assessing the accuracy and the extent of hallucination in the generated bundle. We tested different variations of the evaluation prompt, such as changing the scale used for scoring or providing additional context for the evaluation. We found that a clear and structured evaluation prompt helped ensure consistent and reliable evaluation results.
Figure A4.
Prompt used by the evaluator to evaluate the precision and hallucination score of the conversion. The prompt instructs the evaluator to evaluate the precision and hallucination rate of the generated bundle. It provides guidelines for assessing the accuracy and the extent of hallucination in the generated bundle.
Figure A4.
Prompt used by the evaluator to evaluate the precision and hallucination score of the conversion. The prompt instructs the evaluator to evaluate the precision and hallucination rate of the generated bundle. It provides guidelines for assessing the accuracy and the extent of hallucination in the generated bundle.
References
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT. Association for Computational Linguistics, 2019, pp. 4171–4186.
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. CoRR 2019. [Google Scholar]
- Sanh, V.; Debut, L.; Chaumond, J.; Wolf, T. DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. ArXiv 2019, abs/1910.01108.
- Vaswani, A.; Shazeer, N.M.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Proceedings of the Proc. in NeurIPS; 2017. [Google Scholar]
- Ayers, J.W.; Poliak, A.; Dredze, M.; Leas, E.C.; Zhu, Z.; Kelley, J.B.; Faix, D.J.; Goodman, A.M.; Longhurst, C.A.; Hogarth, M.; et al. Comparing Physician and Artificial Intelligence Chatbot Responses to Patient Questions Posted to a Public Social Media Forum. JAMA Internal Medicine 2023, 183, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Cusidó, J.; Solé-Vilaró, L.; Marti-Puig, P.; Solé-Casals, J. Assessing the Capability of Advanced AI Models in Cardiovascular Symptom Recognition: A Comparative Study. Applied Sciences 2024, 14. [Google Scholar] [CrossRef]
- Kline, A.; Wang, H.; Li, Y.; Dennis, S.; Hutch, M.; Xu, Z.; Wang, F.; Cheng, F.; Luo, Y. Multimodal machine learning in precision health: A scoping review. npj Digital Medicine 2022, 5, 171. [Google Scholar] [CrossRef] [PubMed]
- Alghamdi, H.; Mostafa, A. Advancing EHR analysis: Predictive medication modeling using LLMs. Information Systems 2025, 102528. [Google Scholar] [CrossRef]
- Hashir, M.; Sawhney, R. Towards unstructured mortality prediction with free-text clinical notes. Journal of Biomedical Informatics 2020, 108, 103489. [Google Scholar] [CrossRef]
- Nan, J.; Xu, L.Q. Designing interoperable health care services based on Fast Healthcare Interoperability Resources: Literature review. JMIR Med. Inform. 2023, 11, e44842. [Google Scholar] [CrossRef]
- Shah, W.F. Exploring the Impact and Evolution of Fast Healthcare Interoperability Resources (FHIR) On Healthcare Systems, Personal Health Records and Data Security. Journal of Health Education Research & Development 2023, 11, 100076. [Google Scholar]
- Hong, N.; Wen, A.; Shen, F.; Sohn, S.; Wang, C.; Liu, H.; Jiang, G. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. JAMIA Open 2019, 2, 570–579. [Google Scholar] [CrossRef]
- Bender, D.; Sartipi, K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In Proceedings of the Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems, 2013, pp. 326–331. [CrossRef]
- Mandel, J.C.; Kreda, D.A.; Mandl, K.D.; Kohane, I.S.; Ramoni, R.B. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. Journal of the American Medical Informatics Association 2016, 23, 899–908. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. In Proceedings of the 1st International Conference on Learning Representations, ICLR 2013, Scottsdale, Arizona, USA, May 2-4, 2013, Workshop Track Proceedings, 2013.
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global Vectors for Word Representation. In Proceedings of the Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014, October 25-29, 2014, Doha, Qatar, A meeting of SIGDAT, a Special Interest Group of the ACL. ACL, 2014, pp. 1532–1543. [CrossRef]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language Models are Unsupervised Multitask Learners. In Proceedings of the Computer Science, Linguistics, 2019.
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. In Proceedings of the Proc. NeurIPS; Larochelle, H.; Ranzato, M.; Hadsell, R.; Balcan, M.; Lin, H., Eds., 2020.
- Jiang, A.Q.; Sablayrolles, A.; Mensch, A.; Bamford, C.; Chaplot, D.S.; de Las Casas, D.; Bressand, F.; Lengyel, G.; Lample, G.; Saulnier, L.; et al. Mistral 7B. CoRR 2023, abs/2310.06825, [2310.06825]. [CrossRef]
- Anil, R.; Borgeaud, S.; Wu, Y.; Alayrac, J.; Yu, J.; Soricut, R.; Schalkwyk, J.; Dai, A.M.; Hauth, A.; Millican, K.; et al. Gemini: A Family of Highly Capable Multimodal Models. CoRR 2023, abs/2312.11805, [2312.11805]. [CrossRef]
- Shazeer, N.; Mirhoseini, A.; Maziarz, K.; Davis, A.; Le, Q.V.; Hinton, G.E.; Dean, J. Outrageously Large Neural Networks: The Sparsely-Gated Mixture-of-Experts Layer. In Proceedings of the In Proc. ICLR, 2017.
- Shazeer, N.; Mirhoseini, A.; Maziarz, K.; Davis, A.; Le, Q.V.; Hinton, G.E.; Dean, J. Outrageously Large Neural Networks: The Sparsely-Gated Mixture-of-Experts Layer. In Proceedings of the 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, Conference Track Proceedings. OpenReview.net, 2017.
- Christophe, C.; Kanithi, P.K.; Raha, T.; Khan, S.; Pimentel, M.A. Med42-v2: A Suite of Clinical LLMs. ArXiv 2024, abs/2408.06142.
- Saab, K.; Tu, T.; Weng, W.H.; Tanno, R.; Stutz, D.; Wulczyn, E.; Zhang, F.; Strother, T.; Park, C.; Vedadi, E.; et al. Capabilities of Gemini Models in Medicine, 2024, [2404.18416].
- McDuff, D.; Schaekermann, M.; Tu, T.; Palepu, A.; Wang, A.; Garrison, J.; Singhal, K.; Sharma, Y.; Azizi, S.; Kulkarni, K.; et al. Towards Accurate Differential Diagnosis with Large Language Models. CoRR 2023. [Google Scholar]
- Van Veen, D.; Van Uden, C.; Blankemeier, L.; Delbrouck, J.B.; Aali, A.; Bluethgen, C.; Pareek, A.; Polacin, M.; Reis, E.P.; Seehofnerová, A.; et al. Adapted large language models can outperform medical experts in clinical text summarization. Nat. Med. 2024, 30, 1134–1142. [Google Scholar] [CrossRef]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Ichter, B.; Xia, F.; Chi, E.H.; Le, Q.V.; Zhou, D. Chain-of-thought prompting elicits reasoning in large language models. In Proceedings of the Proceedings of the 36th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 2024; NIPS ’22.
- Kojima, T.; Gu, S.S.; Reid, M.; Matsuo, Y.; Iwasawa, Y. Large language models are zero-shot reasoners. In Proceedings of the Proceedings of the 36th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 2024; NIPS ’22.
- Denecke, K.; May, R.; LLMHealthGroup.; Rivera Romero, O. Potential of large language models in health care: Delphi study. J. Med. Internet Res. 2024, 26, e52399. [CrossRef]
- Dalmaz, O.; Reinhart, T.; Timmerman, M. Direct Clinician Preference Optimization: Clinical Text Summarization via Expert Feedback-Integrated LLMs. Stanford CS224N Custom Project, 2023. Department of Electrical Engineering, Stanford University; Department of Computer Science, Stanford University; Department of Aeronautics and Astronautics, Stanford University.
- Koo, J.K.; Moyer, L.; Castello, M.A.; Arain, Y. Improving accuracy of handoff by implementing an electronic health record-generated tool: An improvement project in an academic Neonatal Intensive Care Unit. Pediatric Quality & Safety 2020, 5, e329. [Google Scholar]
- Cresswell, K.M.; Bates, D.W.; Sheikh, A. Ten key considerations for the successful implementation and adoption of large-scale health information technology. J. Am. Med. Inform. Assoc. 2013, 20, e9–e13. [Google Scholar] [CrossRef]
- Ayaz, M.; Pasha, M.F.; Alahmadi, T.J.; Abdullah, N.N.B.; Alkahtani, H.K. Transforming healthcare analytics with FHIR: A framework for standardizing and analyzing clinical data. Healthcare (Basel) 2023, 11. [Google Scholar] [CrossRef]
- Dagdelen, J.; Dunn, A.; Lee, S.; Walker, N.; Rosen, A.S.; Ceder, G.; Persson, K.A.; Jain, A. Structured information extraction from scientific text with large language models. Nat. Commun. 2024, 15, 1418. [Google Scholar] [CrossRef]
- Tam, Z.R.; Wu, C.K.; Tsai, Y.L.; Lin, C.Y.; yi Lee, H.; Chen, Y.N. Let Me Speak Freely? A Study on the Impact of Format Restrictions on Performance of Large Language Models, 2024, [arXiv:cs.CL/2408.02442].
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Yang, A.; Fan, A.; et al. The Llama 3 Herd of Models, 2024, [arXiv:cs.AI/2407.21783].
- Liu, G.; Mao, H.; Cao, B.; Xue, Z.; Johnson, K.M.; Tang, J.; Wang, R. On the Intrinsic Self-Correction Capability of LLMs: Uncertainty and Latent Concept. CoRR 2024, abs/2406.02378, [2406.02378]. [CrossRef]
- Renze, M.; Guven, E. The Effect of Sampling Temperature on Problem Solving in Large Language Models. CoRR 2024. [Google Scholar] [CrossRef]
- Rose, S. Machine Learning for Prediction in Electronic Health Data. JAMA Network Open 2018, 1. [Google Scholar] [CrossRef]
- Pfohl, S.R.; Cole-Lewis, H.; Sayres, R.; Neal, D.; Asiedu, M.N.; Dieng, A.; Tomasev, N.; Rashid, Q.M.; Azizi, S.; Rostamzadeh, N.; et al. A Toolbox for Surfacing Health Equity Harms and Biases in Large Language Models. CoRR 2024. [Google Scholar] [CrossRef] [PubMed]
- Kipp, M. From GPT-3.5 to GPT-4.o: A Leap in AI’s Medical Exam Performance. Information 2024, 15. [Google Scholar] [CrossRef]
- Hada, R.; Gumma, V.; de Wynter, A.; Diddee, H.; Ahmed, M.; Choudhury, M.; Bali, K.; Sitaram, S. Are Large Language Model-based Evaluators the Solution to Scaling Up Multilingual Evaluation? In Proceedings of the Findings of EACL; Graham, Y.; Purver, M., Eds. Association for Computational Linguistics, 2024.
- Wu, Z.; Zeng, Q.; Zhang, Z.; Tan, Z.; Shen, C.; Jiang, M. Large Language Models Can Self-Correct with Key Condition Verification. In Proceedings of the Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing; Al-Onaizan, Y.; Bansal, M.; Chen, Y.N., Eds., Miami, Florida, USA, 2024; pp. 12846–12867. [CrossRef]
- Li, C.; Wang, J.; Zhang, Y.; Zhu, K.; Hou, W.; Lian, J.; Luo, F.; Yang, Q.; Xie, X. Large Language Models Understand and Can be Enhanced by Emotional Stimuli, 2023, [2307.11760].
Figure 1.
Artificial example of some information from a clinical report (on the left) converted to FHIR bundle (on the right). Some resources, their ID and reference to the patient have been removed for place.
Figure 1.
Artificial example of some information from a clinical report (on the left) converted to FHIR bundle (on the right). Some resources, their ID and reference to the patient have been removed for place.
Figure 2.
This diagram illustrates the process of converting clinical reports into FHIR bundles using LLMs. The pipeline begins with a clinical report input, which is then tasked for conversion to FHIR. The LLMs employ various prompting techniques, including zero-shot and few-shot, as well as basic, two-step, and chain-of-thought methods. The output is validated using HAPI FHIR to ensure it is a correct FHIR bundle. Finally, an external LLM evaluates the quality of the FHIR bundle by comparing it with the original clinical report.
Figure 2.
This diagram illustrates the process of converting clinical reports into FHIR bundles using LLMs. The pipeline begins with a clinical report input, which is then tasked for conversion to FHIR. The LLMs employ various prompting techniques, including zero-shot and few-shot, as well as basic, two-step, and chain-of-thought methods. The output is validated using HAPI FHIR to ensure it is a correct FHIR bundle. Finally, an external LLM evaluates the quality of the FHIR bundle by comparing it with the original clinical report.
Figure 3.
Comparison of the number of resource types in bundles across clinics and large language models. The different colors indicate the different combinations of model and clinic. The x-axis shows the different resource types, and the y-axis shows the number of resources.
Figure 3.
Comparison of the number of resource types in bundles across clinics and large language models. The different colors indicate the different combinations of model and clinic. The x-axis shows the different resource types, and the y-axis shows the number of resources.
Table 2.
Evaluation of empty bundle proportions post-verification with HAPI FHIR across various clinics, models, prompt techniques, and example presence at different temperatures. Lower proportions indicate better performance. A green tick (✓) signifies a full conversion rate, while a red cross (×) denotes a 100% failure rate.
Table 2.
Evaluation of empty bundle proportions post-verification with HAPI FHIR across various clinics, models, prompt techniques, and example presence at different temperatures. Lower proportions indicate better performance. A green tick (✓) signifies a full conversion rate, while a red cross (×) denotes a 100% failure rate.
| |
Failure Rate |
| Clinic |
Model Name |
Prompt Tech. |
zero-shot |
few-shot |
| Low |
High |
Low |
High |
| Skin Care |
Gemini |
Basic |
73% |
82% |
45% |
18% |
| Two-Step |
✓ |
✓ |
✓ |
✓ |
| CoT |
27% |
27% |
18% |
✓ |
| Mistral |
Basic |
× |
91% |
31% |
✓ |
| Two-Step |
✓ |
10% |
35% |
✓ |
| CoT |
55% |
64% |
✓ |
✓ |
| Llama3-Med42 |
Basic |
× |
× |
× |
× |
| Two-Step |
× |
× |
× |
× |
| CoT |
× |
× |
× |
× |
| Mammography |
Gemini |
Basic |
89% |
78% |
✓ |
✓ |
| Two-Step |
✓ |
✓ |
✓ |
✓ |
| CoT |
56% |
44% |
✓ |
✓ |
| Mistral |
Basic |
× |
× |
29% |
✓ |
| Two-Step |
✓ |
✓ |
29% |
✓ |
| CoT |
44% |
40% |
✓ |
✓ |
| Llama3-Med42 |
Basic |
× |
× |
× |
× |
| Two-Step |
× |
× |
× |
× |
| CoT |
× |
× |
× |
× |
Table 3.
Mean and standard deviation of precision scores for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding indicates value ranges: 1-2.5 (red), 2.5-5 (orange), 5-7.5 (olive green), and 7.5-10 (green). Standard deviations are shown in parentheses. Higher scores are favorable.
Table 3.
Mean and standard deviation of precision scores for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding indicates value ranges: 1-2.5 (red), 2.5-5 (orange), 5-7.5 (olive green), and 7.5-10 (green). Standard deviations are shown in parentheses. Higher scores are favorable.
| |
Precision
|
|
Clinic
|
Model Name
|
Prompt Tech.
|
zero-shot
|
few-shot
|
| Low |
High |
Low |
High |
| Skin Care |
Gemini |
Basic |
2.09 (1.87) |
1.73 (1.62) |
4.45 (3.33) |
6.45 (2.73) |
| Two-Step |
7.64 (0.5) |
7.64 (0.5) |
7.82 (0.4) |
7.45 (0.52) |
| CoT |
5.18 (2.96) |
5.27 (2.94) |
6.27 (2.65) |
7.64 (0.5) |
| Mistral |
Basic |
× |
1.82 (2.71) |
5.56 (3.2) |
7.64 (0.5) |
| Two-Step |
7.18 (0.4) |
6.7 (2.06) |
5.06 (3.11) |
7.44 (0.53) |
| CoT |
3.91 (3.36) |
3.27 (3.17) |
7.55 (0.52) |
7.55 (0.52) |
| Mammography |
Gemini |
Basic |
1.44 (1.33) |
1.56 (1.33) |
7.78 (0.97) |
7.44 (0.53) |
| Two-Step |
7.78 (0.44) |
7.67 (0.5) |
7.67 (0.5) |
7.89 (0.33) |
| CoT |
3.78 (3.42) |
4.33 (3.28) |
8.0 (0.87) |
7.67 (0.5) |
| Mistral |
Basic |
× |
× |
5.86 (3.21) |
7.44 (0.53) |
| Two-Step |
7.89 (0.93) |
7.44 (0.53) |
6.0 (3.35) |
7.56 (0.53) |
| CoT |
4.78 (3.6) |
4.9 (3.38) |
7.62 (0.52) |
7.89 (0.33) |
Table 4.
Average and standard deviation of hallucination rate for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding reflects value ranges: 1-2.5 (green), 2.5-5 (olive green), 5-7.5 (orange), and 7.5-10 (red). Standard deviations are shown in parentheses. Lower values are ideal.
Table 4.
Average and standard deviation of hallucination rate for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding reflects value ranges: 1-2.5 (green), 2.5-5 (olive green), 5-7.5 (orange), and 7.5-10 (red). Standard deviations are shown in parentheses. Lower values are ideal.
| |
Hallucination |
| Clinic |
Model Name |
Prompt Tech. |
zero-shot |
few-shot |
| Low |
High |
Low |
High |
| Skin Care |
Gemini |
Basic |
9.45 (0.93) |
9.82 (0.6) |
7.45 (2.5) |
5.55 (2.34) |
| Two-Step |
3.64 (0.92) |
3.55 (0.82) |
3.36 (1.21) |
4.18 (1.66) |
| CoT |
6.73 (3.2) |
6.55 (3.05) |
5.27 (2.57) |
4.27 (1.68) |
| Mistral |
Basic |
× |
10.0 (0.0) |
6.25 (2.89) |
3.73 (1.27) |
| Two-Step |
4.0 (1.48) |
4.7 (2.16) |
5.88 (3.33) |
4.11 (1.05) |
| CoT |
7.36 (3.38) |
8.27 (2.65) |
3.73 (1.35) |
3.64 (1.36) |
| Mammography |
Gemini |
Basic |
9.78 (0.67) |
9.78 (0.67) |
5.56 (2.24) |
6.44 (2.51) |
| Two-Step |
3.44 (1.24) |
3.67 (1.12) |
3.33 (1.41) |
3.11 (1.27) |
| CoT |
7.44 (3.54) |
7.22 (2.82) |
4.33 (1.41) |
5.11 (2.03) |
| Mistral |
Basic |
× |
× |
5.86 (2.93) |
4.56 (2.3) |
| Two-Step |
3.78 (1.56) |
4.56 (1.74) |
4.86 (3.61) |
4.33 (1.66) |
| CoT |
6.78 (3.15) |
6.1 (3.48) |
4.25 (1.04) |
3.89 (1.45) |
Table 5.
Average and standard deviation of resource mapping accuracy for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding represents value ranges: 1-2.5 (red), 2.5-5 (orange), 5-7.5 (olive green), and 7.5-10 (green). Standard deviations are shown in parentheses. Lower values are desirable.
Table 5.
Average and standard deviation of resource mapping accuracy for different clinics, models, prompt techniques, and example presence at various temperatures. Color coding represents value ranges: 1-2.5 (red), 2.5-5 (orange), 5-7.5 (olive green), and 7.5-10 (green). Standard deviations are shown in parentheses. Lower values are desirable.
| |
Resource Mapping |
| Clinic |
Model Name |
Prompt Tech. |
zero-shot |
few-shot |
| Low |
High |
Low |
High |
| Skin Care |
Gemini |
Basic |
1.91 (1.87) |
1.55 (1.29) |
4.73 (3.58) |
6.09 (2.59) |
| Two-Step |
7.36 (0.5) |
7.27 (0.65) |
7.55 (0.82) |
7.55 (0.52) |
| CoT |
4.91 (2.98) |
5.09 (2.95) |
6.27 (2.69) |
7.18 (1.08) |
| Mistral |
Basic |
× |
1.82 (2.71) |
5.25 (3.0) |
7.27 (0.65) |
| Two-Step |
7.82 (0.4) |
7.0 (2.16) |
5.59 (3.52) |
7.44 (0.73) |
| CoT |
3.73 (3.23) |
3.36 (3.32) |
7.73 (0.9) |
7.64 (0.67) |
| Mammography |
Gemini |
Basic |
1.44 (1.33) |
1.44 (1.33) |
6.78 (1.64) |
6.89 (1.45) |
| Two-Step |
7.67 (0.87) |
7.33 (1.0) |
7.67 (1.0) |
7.44 (1.01) |
| CoT |
3.56 (3.5) |
4.22 (3.49) |
7.44 (1.13) |
7.33 (0.5) |
| Mistral |
Basic |
× |
× |
5.71 (3.17) |
7.44 (1.01) |
| Two-Step |
8.11 (1.05) |
7.89 (0.78) |
6.07 (3.45) |
7.33 (0.71) |
| CoT |
4.44 (3.28) |
4.9 (3.38) |
7.25 (0.71) |
7.89 (1.17) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).