Submitted:
16 November 2023
Posted:
17 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
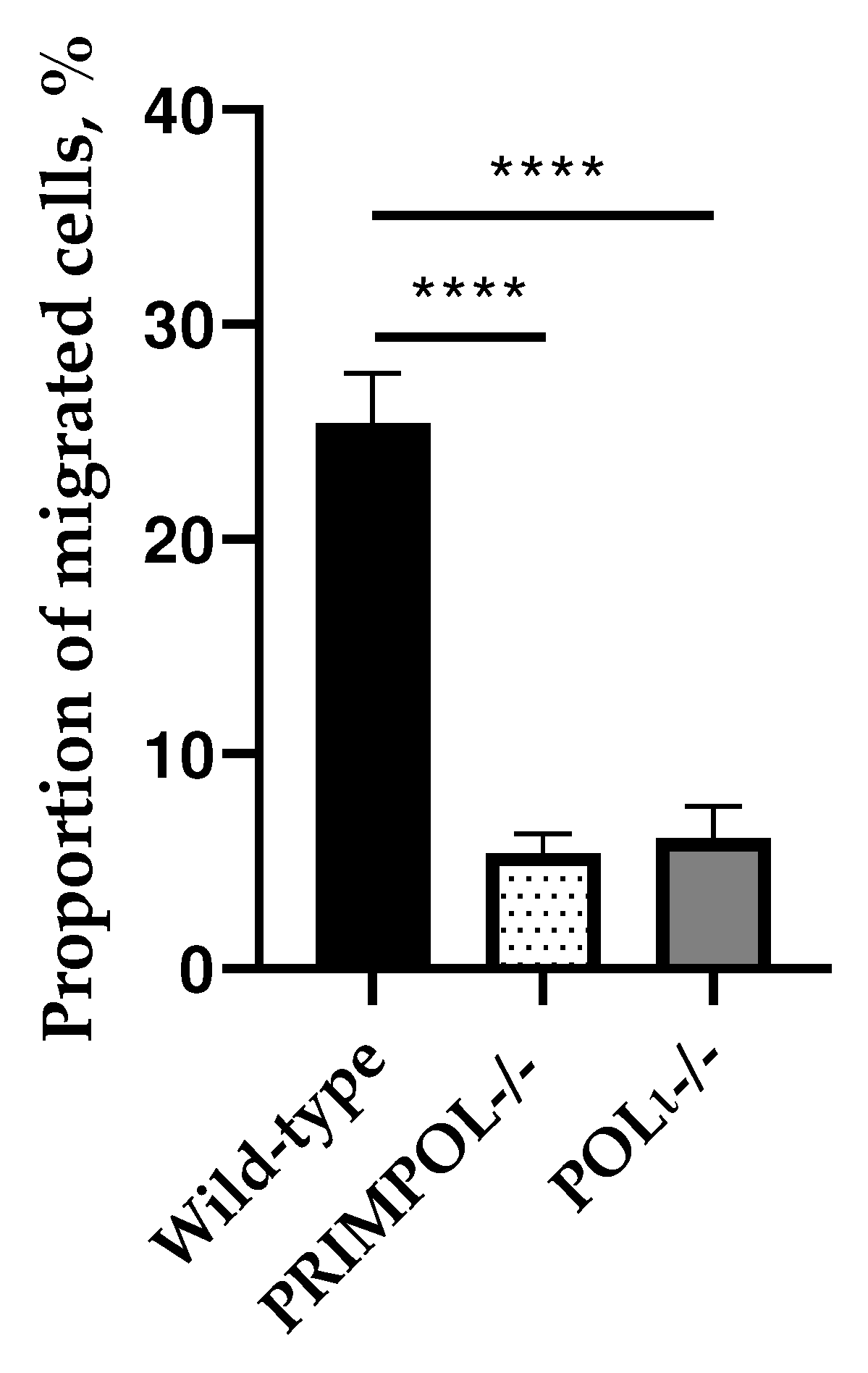
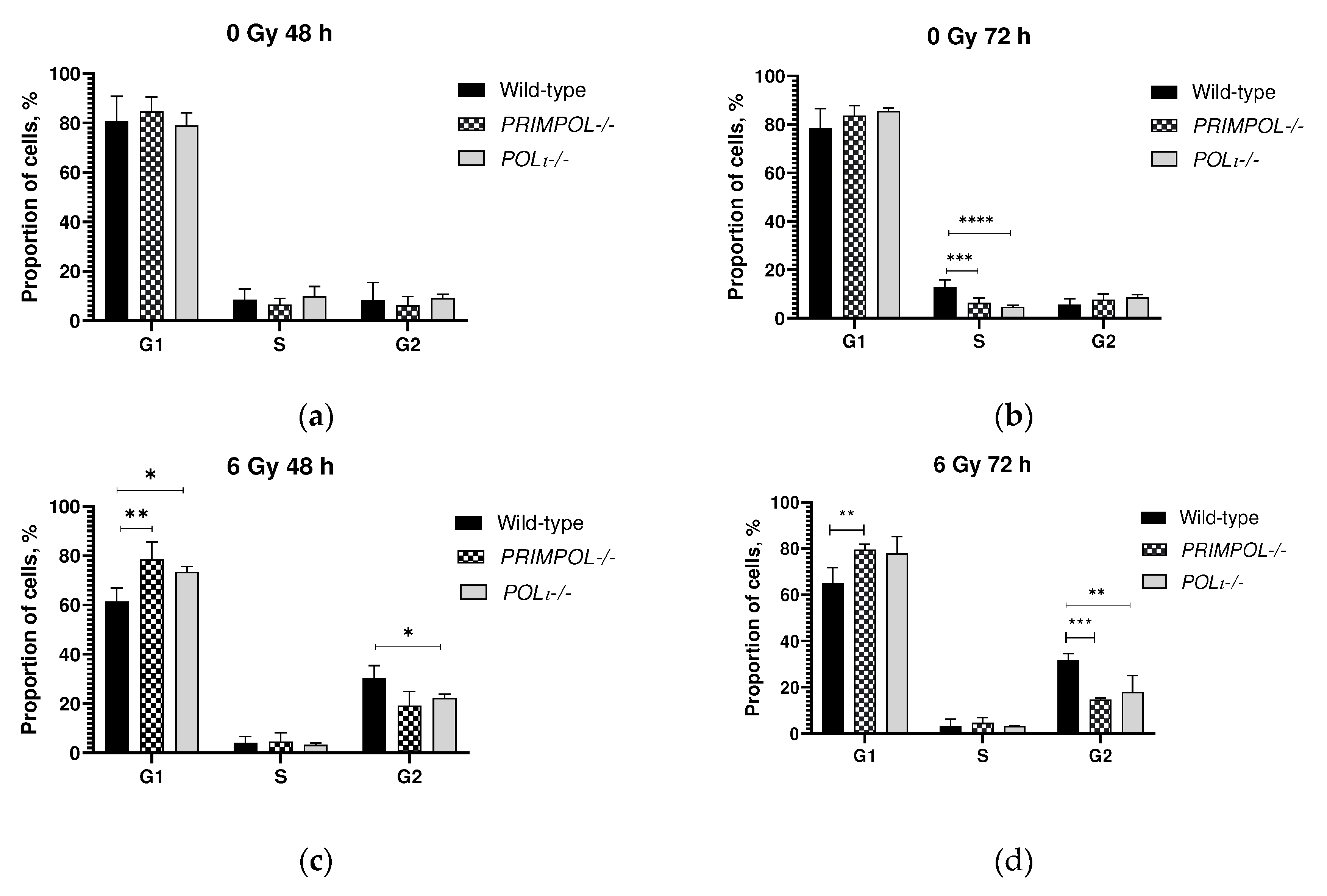
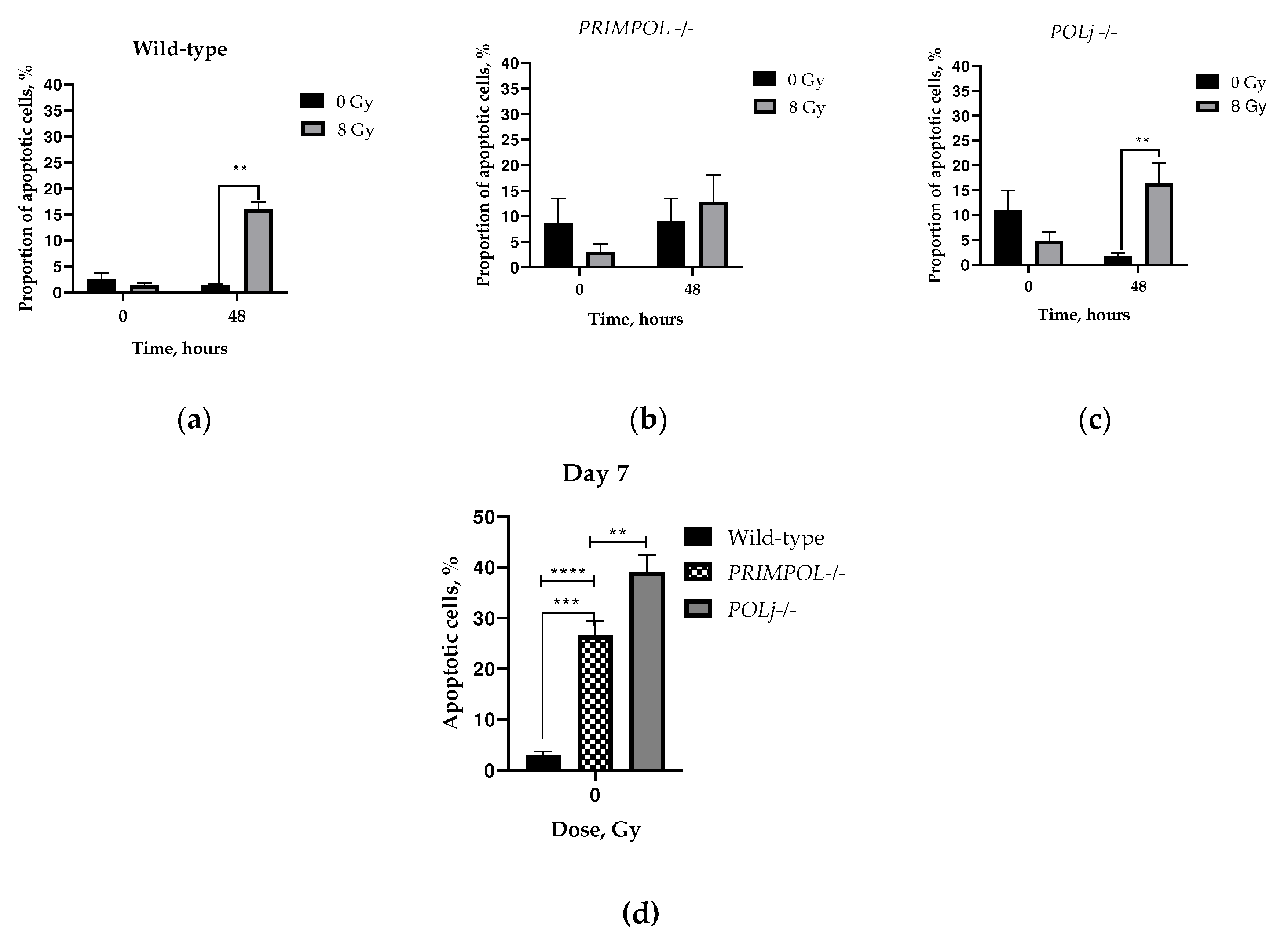
2.1. Evaluation of Invasion, Cell Cycle Progression and Cell Death of PRIMPOL-/- and POLI -/- Cell Lines.
2.2. Radiosensitivity of PRIMPOL-/- and POLI -/- Cell Lines Using Clonogenic Assay
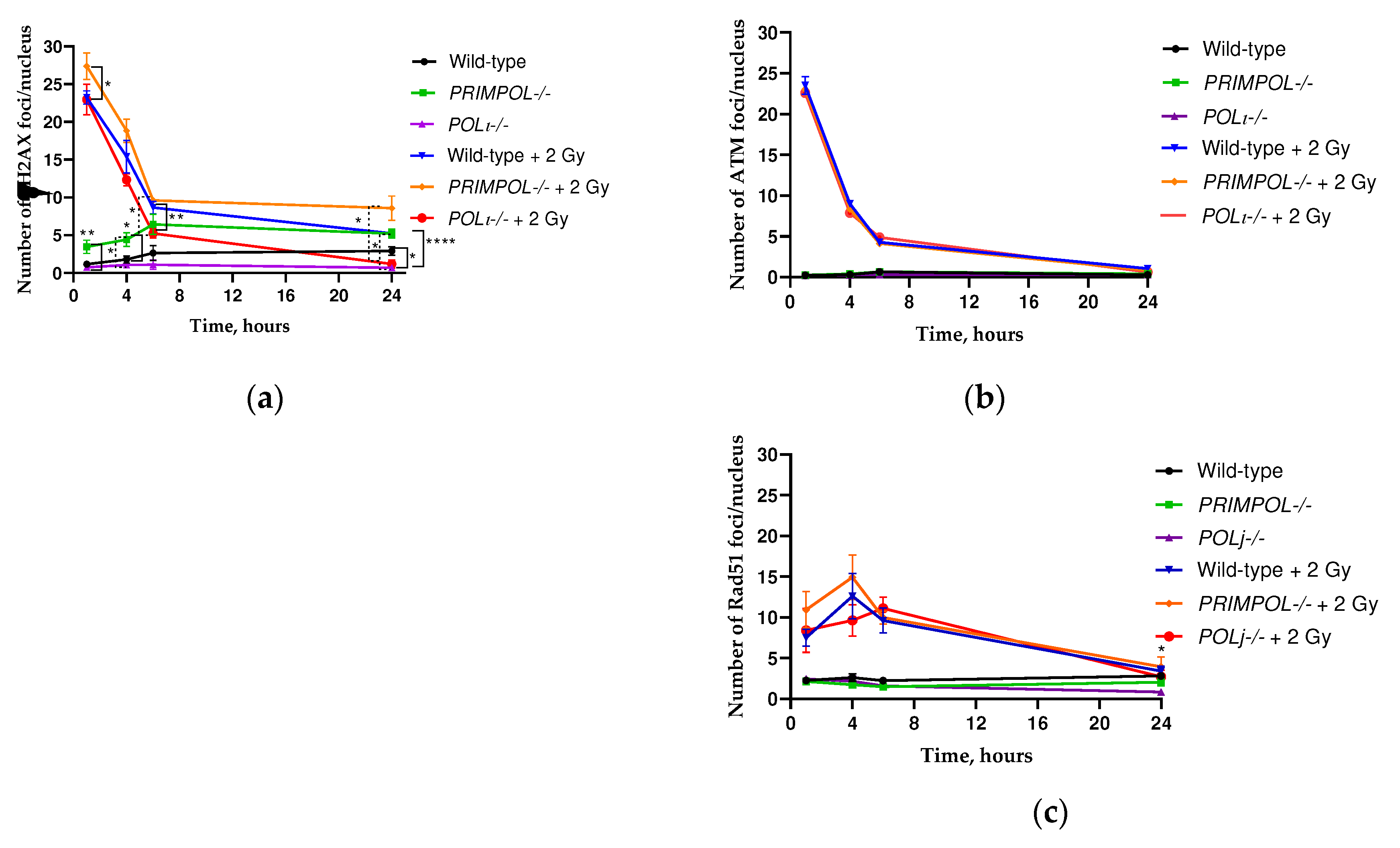
2.3. Comparative Analysis of γH2AX Kinetics, Phosphorylated Ataxia Telangiectasia Mutated (pATM) and Rad51 in wild-type, PRIMPOL-/- and POLI -/- Cell Lines
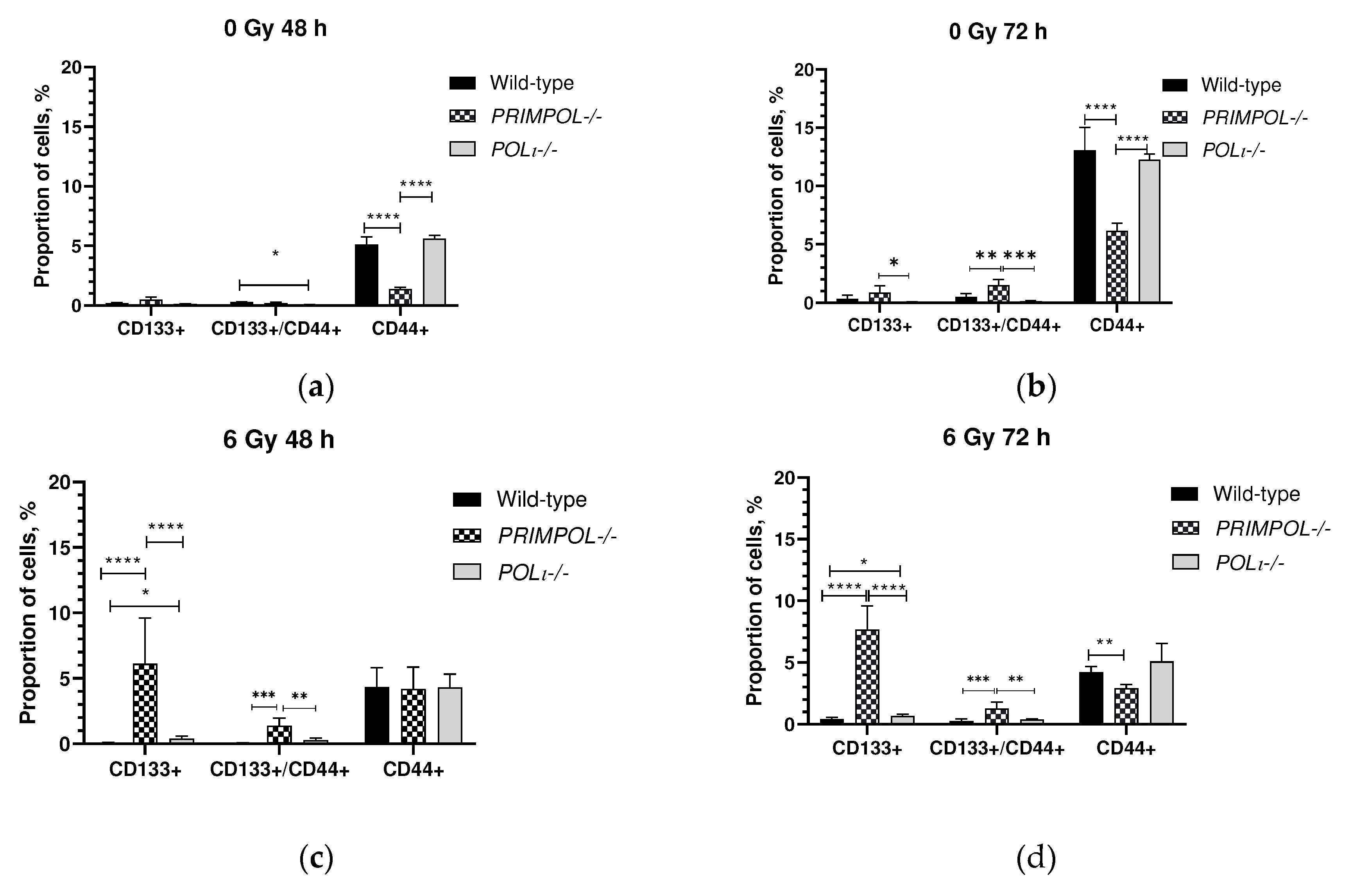
2.4. The Proportion of Cancer Cells with Stem-like Cells in PRIMPOL-/- and POLI -/- Cell Lines with and without IR Exposure.
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Cultivation conditions
4.2. Trans-Well Migration Assay Using Boyden Chambers
4.3. Apoptosis Analysis
4.4. Colony Formation and Soft Agar Assay
4.5. γH2AX, pATM and Rad51 Foci Analysis
4.6. Analysis of the Proportion of Cancer Stem Cells by Flow Cytometry
4.7. Cell Cycle Analysis by Flow Cytometry
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, W.D., S.S. Shah, and W.-D. Heyer, Homologous recombination and the repair of DNA double-strand breaks. Journal of Biological Chemistry, 2018. 293(27): p. 10524-10535. [CrossRef]
- Lieber, M.R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem, 2010. 79: p. 181-211. [CrossRef]
- Marteijn, J.A., et al., Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol, 2014. 15(7): p. 465-81. [CrossRef]
- Krokan, H.E. and M. Bjoras, Base excision repair. Cold Spring Harb Perspect Biol, 2013. 5(4): p. a012583.
- Li, G.M., Mechanisms and functions of DNA mismatch repair. Cell Res, 2008. 18(1): p. 85-98. [CrossRef]
- Qiu, S., et al., Replication Fork Reversal and Protection. Front Cell Dev Biol, 2021. 9: p. 670392. [CrossRef]
- Vaisman, A., J.P. McDonald, and R. Woodgate, Translesion DNA Synthesis. EcoSal Plus, 2012. 5(1).
- Sale, J.E., Competition, collaboration and coordination--determining how cells bypass DNA damage. J Cell Sci, 2012. 125(Pt 7): p. 1633-43.
- Vaisman, A. and R. Woodgate, Mysterious and fascinating: DNA polymerase i remains enigmatic 20 years after its discovery. DNA Repair (Amst), 2020. 93: p. 102914. [CrossRef]
- Makarova, A.V. and A.V. Kulbachinskiy, Structure of human DNA polymerase iota and the mechanism of DNA synthesis. Biochemistry (Mosc), 2012. 77(6): p. 547-61. [CrossRef]
- Washington, M.T., et al., Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol, 2004. 24(13): p. 5687-93.
- Vaisman, A. and R. Woodgate, Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J, 2001. 20(22): p. 6520-9. [CrossRef]
- Yang, J., et al., Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res, 2004. 64(16): p. 5597-607.
- Luedeke, M., et al., Predisposition for TMPRSS2-ERG fusion in prostate cancer by variants in DNA repair genes. Cancer Epidemiol Biomarkers Prev, 2009. 18(11): p. 3030-5. [CrossRef]
- Sakiyama, T., et al., Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer, 2005. 114(5): p. 730-7.
- Silvestrov, P., et al., DNArCdb: A database of cancer biomarkers in DNA repair genes that includes variants related to multiple cancer phenotypes. DNA Repair (Amst), 2018. 70: p. 10-17. [CrossRef]
- Yuan, F., et al., Overexpressed DNA polymerase iota regulated by JNK/c-Jun contributes to hypermutagenesis in bladder cancer. PLoS One, 2013. 8(7): p. e69317. [CrossRef]
- Zou, S., et al., DNA polymerase iota (Pol iota) promotes invasion and metastasis of esophageal squamous cell carcinoma. Oncotarget, 2016. 7(22): p. 32274-85. [CrossRef]
- Dumstorf, C.A., et al., Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci U S A, 2006. 103(48): p. 18083-8.
- Ohkumo, T., et al., UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol Cell Biol, 2006. 26(20): p. 7696-706.
- Lee, G.H. and H. Matsushita, Genetic linkage between Pol iota deficiency and increased susceptibility to lung tumors in mice. Cancer Sci, 2005. 96(5): p. 256-9.
- Iguchi, M., et al., The error-prone DNA polymerase iota provides quantitative resistance to lung tumorigenesis and mutagenesis in mice. Oncogene, 2014. 33(27): p. 3612-7.
- Wang, M., et al., Pol iota is a candidate for the mouse pulmonary adenoma resistance 2 locus, a major modifier of chemically induced lung neoplasia. Cancer Res, 2004. 64(6): p. 1924-31.
- Zou, S., et al., DNA polymerase iota (Pol iota) promotes the migration and invasion of breast cancer cell via EGFR-ERK-mediated epithelial to mesenchymal transition. Cancer Biomark, 2019. 24(3): p. 363-370. [CrossRef]
- He, C., et al., Phosphorylation of ETS-1 is a critical event in DNA polymerase iota-induced invasion and metastasis of esophageal squamous cell carcinoma. Cancer Sci, 2017. 108(12): p. 2503-2510. [CrossRef]
- Su, Z., et al., DNA Polymerase Iota Promotes Esophageal Squamous Cell Carcinoma Proliferation Through Erk-OGT-Induced G6PD Overactivation. Front Oncol, 2021. 11: p. 706337. [CrossRef]
- Guilliam, T.A., et al., Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res, 2015. 43(14): p. 6651-64. [CrossRef]
- Gonzalez-Acosta, D., et al., PrimPol-mediated repriming facilitates replication traverse of DNA interstrand crosslinks. EMBO J, 2021. 40(14): p. e106355.
- Piberger, A.L., et al., PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts. Nat Commun, 2020. 11(1): p. 5863.
- Butler, T.J., et al., Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro. Hum Mol Genet, 2020. 29(8): p. 1292-1309. [CrossRef]
- Svikovic, S., et al., R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J, 2019. 38(3).
- Quinet, A., et al., PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells. Mol Cell, 2020. 77(3): p. 461-474 e9. [CrossRef]
- Torregrosa-Munumer, R., et al., PrimPol is required for replication reinitiation after mtDNA damage. Proc Natl Acad Sci U S A, 2017. 114(43): p. 11398-11403. [CrossRef]
- Kobayashi, K., et al., Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides. Cell Cycle, 2016. 15(15): p. 1997-2008.
- Taglialatela, A., et al., REV1-Polzeta maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol Cell, 2021. 81(19): p. 4008-4025 e7.
- Makarova, A.V., et al., In vitro lesion bypass by human PrimPol. DNA Repair (Amst), 2018. 70: p. 18-24. [CrossRef]
- Guilliam, T.A., et al., Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res, 2015. 43(2): p. 1056-68. [CrossRef]
- Zafar, M.K., et al., Kinetic analysis of human PrimPol DNA polymerase activity reveals a generally error-prone enzyme capable of accurately bypassing 7,8-dihydro-8-oxo-2'-deoxyguanosine. Biochemistry, 2014. 53(41): p. 6584-94. [CrossRef]
- Boldinova, E.O., et al., Translesion activity of PrimPol on DNA with cisplatin and DNA-protein cross-links. Sci Rep, 2021. 11(1): p. 17588. [CrossRef]
- Bailey, L.J., et al., PrimPol-deficient cells exhibit a pronounced G2 checkpoint response following UV damage. Cell Cycle, 2016. 15(7): p. 908-18.
- Mouron, S., et al., Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol, 2013. 20(12): p. 1383-9. [CrossRef]
- Schiavone, D., et al., PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol Cell, 2016. 61(1): p. 161-9. [CrossRef]
- Bailey, L.J., J. Bianchi, and A.J. Doherty, PrimPol is required for the maintenance of efficient nuclear and mitochondrial DNA replication in human cells. Nucleic Acids Res, 2019. 47(8): p. 4026-4038. [CrossRef]
- Diaz-Talavera, A., et al., A cancer-associated point mutation disables the steric gate of human PrimPol. Sci Rep, 2019. 9(1): p. 1121. [CrossRef]
- Rechkoblit, O., et al., Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci Adv, 2016. 2(10): p. e1601317. [CrossRef]
- Boldinova, E.O., C.A.C. Manukyan capital A, and C. Makarova capital A, The DNA ligands Arg47 and Arg76 are crucial for catalysis by human PrimPol. DNA Repair (Amst), 2021. 100: p. 103048.
- Kasho, K., et al., A unique arginine cluster in PolDIP2 enhances nucleotide binding and DNA synthesis by PrimPol. Nucleic Acids Res, 2021. 49(4): p. 2179-2191. [CrossRef]
- Carvalho, G., et al., Human PrimPol Discrimination against Dideoxynucleotides during Primer Synthesis. Genes (Basel), 2021. 12(10). [CrossRef]
- Guilliam, T.A., et al., Molecular basis for PrimPol recruitment to replication forks by RPA. Nat Commun, 2017. 8: p. 15222. [CrossRef]
- Grossman, R.L., et al., Toward a Shared Vision for Cancer Genomic Data. N Engl J Med, 2016. 375(12): p. 1109-12. [CrossRef]
- Tate, J.G., et al., COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res, 2019. 47(D1): p. D941-D947. [CrossRef]
- Tonzi, P. and T.T. Huang, Role of Y-family translesion DNA polymerases in replication stress: Implications for new cancer therapeutic targets. DNA Repair (Amst), 2019. 78: p. 20-26. [CrossRef]
- Bertolin, A.P., S.F. Mansilla, and V. Gottifredi, The identification of translesion DNA synthesis regulators: Inhibitors in the spotlight. DNA Repair (Amst), 2015. 32: p. 158-164. [CrossRef]
- Ihle, M., et al., Impact of the interplay between stemness features, p53 and pol iota on replication pathway choices. Nucleic Acids Res, 2021. 49(13): p. 7457-7475.
- Gromova A.S., B.E.O., Kim D.V., Chuprov-Netochin R.N., Leonov S.V., Pustopalova M.V., Zharkov D.O., Makarova A.V. , Response of PRIMPOL-Knockout Human Lung Adenocarcinoma A549 Cells to Genotoxic Stress. Biochemistry, 2023. in press.
- Deng, L., et al., Multi-omics analysis of DNA replication-associated primase polymerase (PRIMPOL) in pan-cancer: a potential target for prognosis and immune response. Eur J Med Res, 2023. 28(1): p. 207. [CrossRef]
- Fujisawa, S., et al., Evaluation of YO-PRO-1 as an early marker of apoptosis following radiofrequency ablation of colon cancer liver metastases. Cytotechnology, 2014. 66(2): p. 259-73. [CrossRef]
- Franken, N.A., et al., Clonogenic assay of cells in vitro. Nat Protoc, 2006. 1(5): p. 2315-9. [CrossRef]
- Mah, L.J., A. El-Osta, and T.C. Karagiannis, gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia, 2010. 24(4): p. 679-86.
- Burma, S., et al., ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem, 2001. 276(45): p. 42462-7. [CrossRef]
- Mason, J.M., et al., Non-enzymatic roles of human RAD51 at stalled replication forks. Nat Commun, 2019. 10(1): p. 4410. [CrossRef]
- Rajendran, V. and M.V. Jain, In Vitro Tumorigenic Assay: Colony Forming Assay for Cancer Stem Cells. Methods Mol Biol, 2018. 1692: p. 89-95.
- Bianchi, J., et al., PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell, 2013. 52(4): p. 566-73. [CrossRef]
- Garcia-Gomez, S., et al., PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell, 2013. 52(4): p. 541-53. [CrossRef]
- Keen, B.A., et al., Molecular dissection of the domain architecture and catalytic activities of human PrimPol. Nucleic Acids Res, 2014. 42(9): p. 5830-45. [CrossRef]
- Mansilla, S.F., et al., Polymerase iota (Pol iota) prevents PrimPol-mediated nascent DNA synthesis and chromosome instability. Sci Adv, 2023. 9(15): p. eade7997.
- Sale, J.E., Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol, 2013. 5(3): p. a012708. [CrossRef]
- Livneh, Z., O. Ziv, and S. Shachar, Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle, 2010. 9(4): p. 729-35. [CrossRef]
- Prakash, S., R.E. Johnson, and L. Prakash, Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem, 2005. 74: p. 317-53. [CrossRef]
- Makridakis, N.M. and J.K. Reichardt, Translesion DNA polymerases and cancer. Front Genet, 2012. 3: p. 174. [CrossRef]
- Pilzecker, B., et al., PrimPol prevents APOBEC/AID family mediated DNA mutagenesis. Nucleic Acids Res, 2016. 44(10): p. 4734-44. [CrossRef]
- Cross, S.E., et al., Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol, 2007. 2(12): p. 780-3.
- Guck, J., et al., Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J, 2005. 88(5): p. 3689-98.
- Swaminathan, V., et al., Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res, 2011. 71(15): p. 5075-80.
- Kraning-Rush, C.M., J.P. Califano, and C.A. Reinhart-King, Cellular traction stresses increase with increasing metastatic potential. PLoS One, 2012. 7(2): p. e32572. [CrossRef]
- Merkher, Y. and D. Weihs, Proximity of Metastatic Cells Enhances Their Mechanobiological Invasiveness. Ann Biomed Eng, 2017. 45(6): p. 1399-1406. [CrossRef]
- Merkher, Y., et al., Rapid Cancer Diagnosis and Early Prognosis of Metastatic Risk Based on Mechanical Invasiveness of Sampled Cells. Ann Biomed Eng, 2020. 48(12): p. 2846-2858. [CrossRef]
- Sun, H., et al., Elevated DNA polymerase iota (Poli) is involved in the acquisition of aggressive phenotypes of human esophageal squamous cell cancer. Int J Clin Exp Pathol, 2015. 8(4): p. 3591-601.
- Li, L., et al., siRNA of DNA polymerase iota inhibits the migration and invasion in the lung cancer cell A549. Acta Biochim Biophys Sin (Shanghai), 2018. 50(9): p. 929-933. [CrossRef]
- Tang, H.M., et al., Molecular signature of anastasis for reversal of apoptosis. F1000Res, 2017. 6: p. 43.
- Chakraborty, S., et al., Integration of EMT and cellular survival instincts in reprogramming of programmed cell death to anastasis. Cancer Metastasis Rev, 2020. 39(2): p. 553-566. [CrossRef]
- Blackford, A.N. and S.P. Jackson, ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell, 2017. 66(6): p. 801-817. [CrossRef]
- Bakkenist, C.J. and M.B. Kastan, DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature, 2003. 421(6922): p. 499-506. [CrossRef]
- Ward, I.M., K. Minn, and J. Chen, UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J Biol Chem, 2004. 279(11): p. 9677-80. [CrossRef]
- Wan, L., et al., hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep, 2013. 14(12): p. 1104-12.
- Wold, M.S., Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem, 1997. 66: p. 61-92. [CrossRef]
- Li, X., et al., Polymerase iota (POLI) confers radioresistance of esophageal squamous cell carcinoma by regulating RAD51 stability and facilitating homologous recombination. Cell Death Discov, 2023. 9(1): p. 291. [CrossRef]
- Zellweger, R., et al., Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol, 2015. 208(5): p. 563-79.
- Vallerga, M.B., et al., Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc Natl Acad Sci U S A, 2015. 112(48): p. E6624-33.
- Walcher, L., et al., Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front Immunol, 2020. 11: p. 1280. [CrossRef]
- Yan, X., et al., A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A, 2011. 108(4): p. 1591-6. [CrossRef]
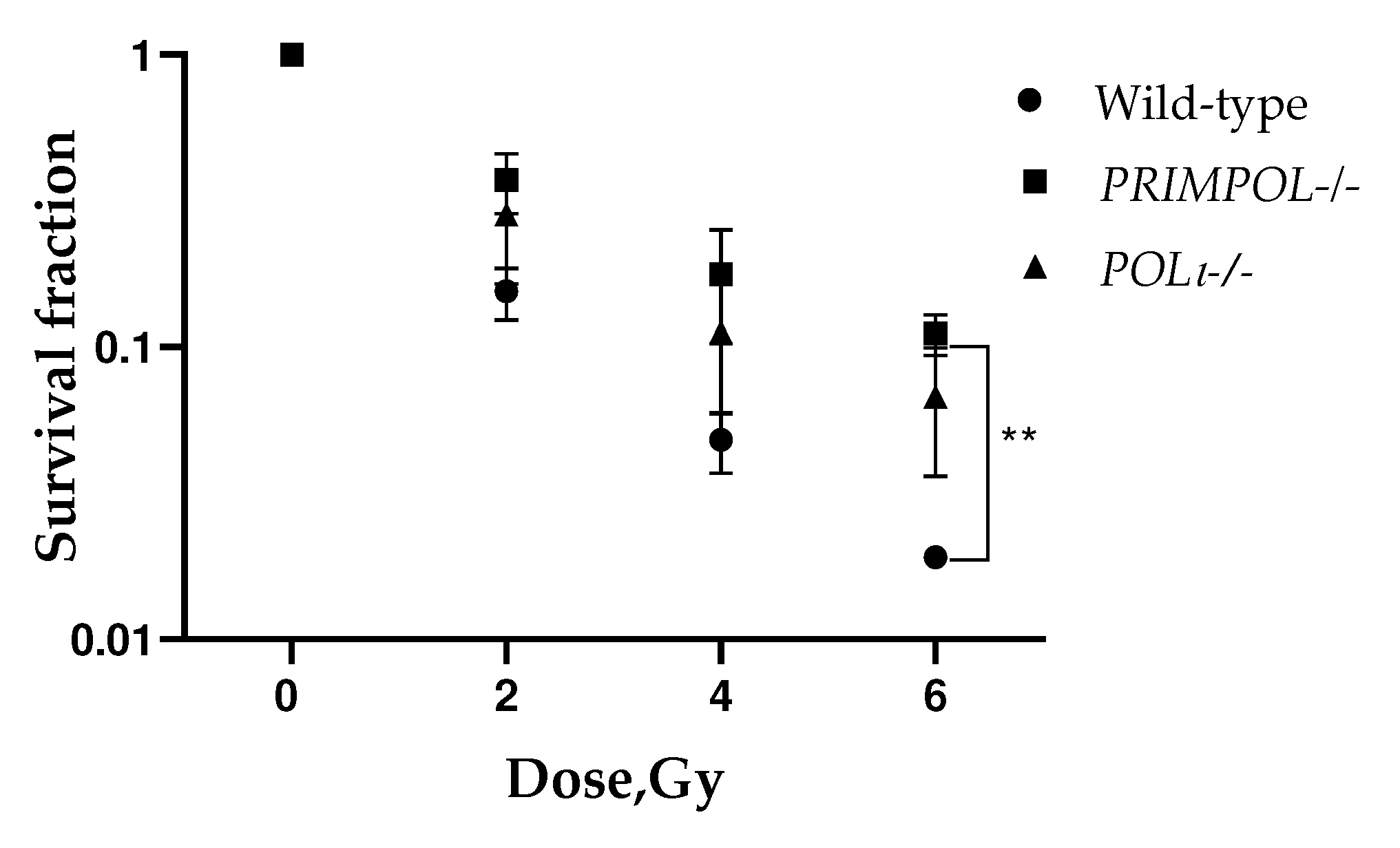
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




