Submitted:
05 November 2023
Posted:
07 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Origin of T. cruzi DTUs
3. Intrinsic Characteristics of the DTUs
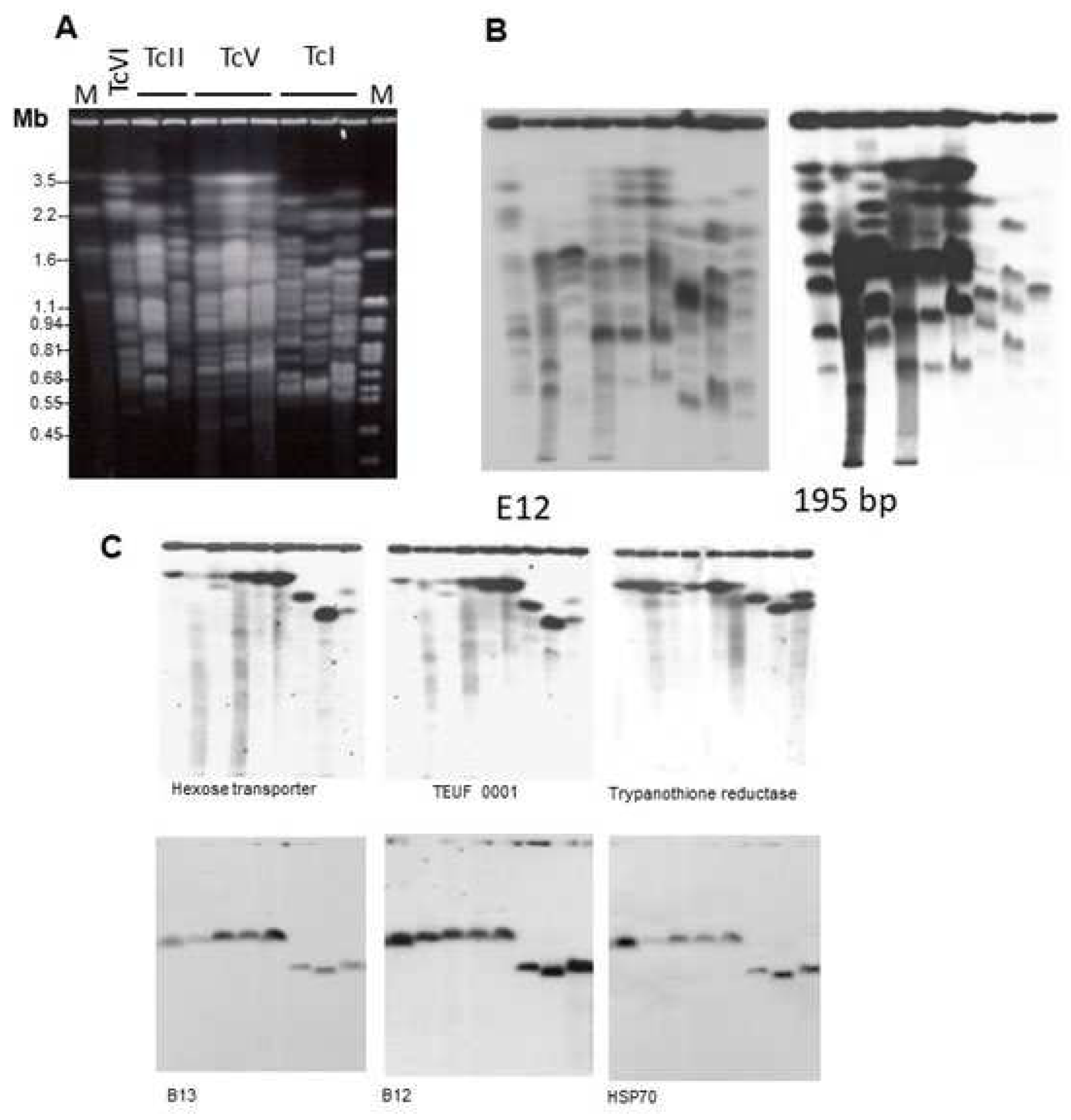
3.1. Genome and Genomics
3.1.1. DTU Nuclear Genomes
3.1.2. DTU Genomics
3.2. In vitro Susceptibility to Drugs
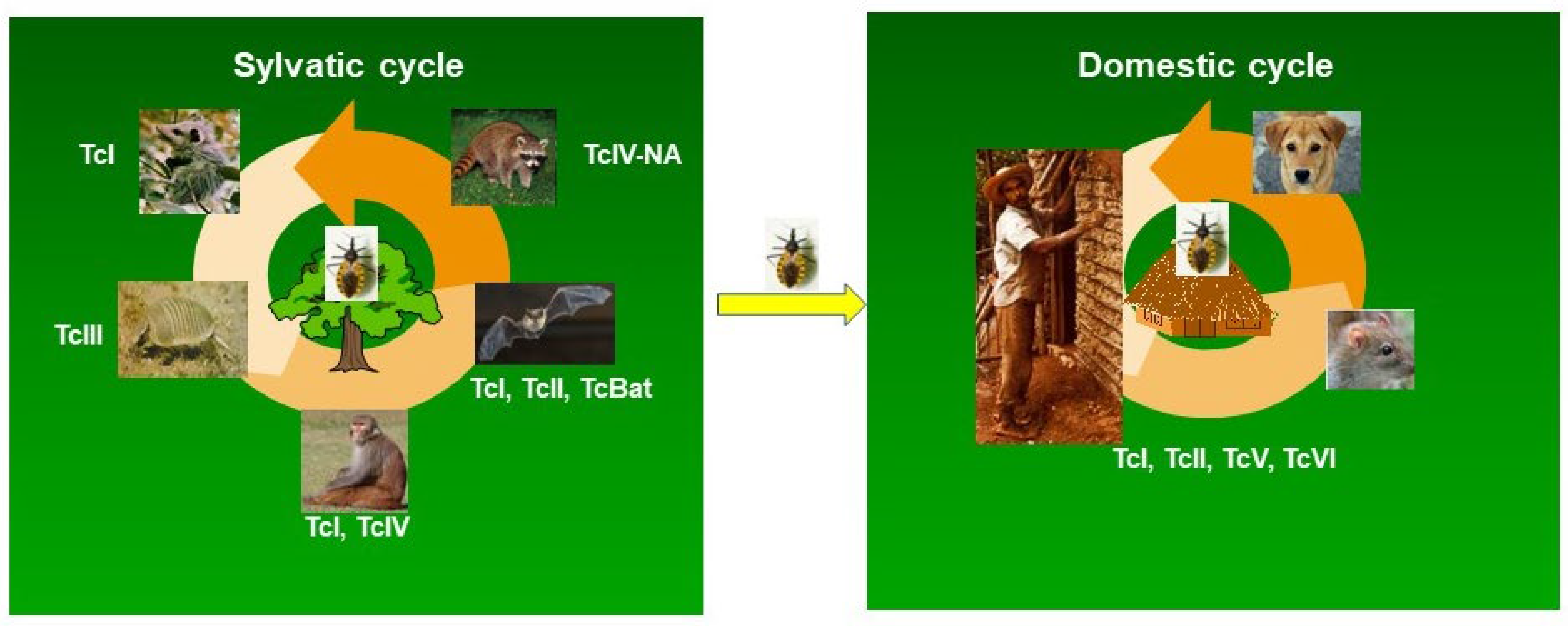
4. Association of the DTUs with Mammalian Reservoirs
5. DTUs and Human Chagas disease
5.1. Geographic Distribution of DTUs in Patients
5.2. Clinical Implications of T. cruzi DTUs
6. DTUs and the Etiological Treatment of Chagas Disease
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Heath Organization Chagas Disease (Also Known as American Trypanosomiasis) 2023, https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis).
- De Fuentes-Vicente, J.A.; Santos-Hernández, N.G.; Ruiz-Castillejos, C.; Espinoza-Medinilla, E.E.; Flores-Villegas, A.L.; de Alba-Alvarado, M.; Cabrera-Bravo, M.; Moreno-Rodríguez, A.; Vidal-López, D.G. What Do You Need to Know before Studying Chagas Disease? A Beginner’s Guide. Trop Med Infect Dis 2023, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Urbina, J.A. Specific Chemotherapy of Chagas Disease: Relevance, Current Limitations and New Approaches. Acta Trop 2010, 115, 55–68. [Google Scholar] [CrossRef]
- Losada Galván, I.; Alonso-Padilla, J.; Cortés-Serra, N.; Alonso-Vega, C.; Gascón, J.; Pinazo, M.J. Benznidazole for the Treatment of Chagas Disease. Expert Rev Anti Infect Ther 2021, 19, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, L.M.D.; Gollob, K.J.; Zingales, B.; Dutra, W.O. Pathogen Diversity, Immunity, and the Fate of Infections: Lessons Learned from Trypanosoma Cruzi Human–Host Interactions. Lancet Microbe 2022, 3, e711–e722. [Google Scholar] [CrossRef] [PubMed]
- Dutra, W.O.; Menezes, C.A.S.; Villani, F.N.A.; Costa, G.C. da; Silveira, A.B.M. da; Reis, D. d’Ávila; Gollob, K.J. Cellular and Genetic Mechanisms Involved in the Generation of Protective and Pathogenic Immune Responses in Human Chagas Disease. Mem Inst Oswaldo Cruz 2009, 104, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Miles, M.A.; Llewellyn, M.S.; Lewis, M.D.; Yeo, M.; Baleela, R.; FITZPATRICK, S.; GAUNT, M.W.; MAURICIO, I.L. The Molecular Epidemiology and Phylogeography of Trypanosoma Cruzi and Parallel Research on Leishmania: Looking Back and to the Future. Parasitology 2009, 136, 1509–1528. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B. Trypanosoma Cruzi Genetic Diversity: Something New for Something Known about Chagas Disease Manifestations, Serodiagnosis and Drug Sensitivity. Acta Trop 2018, 184, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B.; Bartholomeu, D.C. Trypanosoma Cruzi Genetic Diversity: Impact on Transmission Cycles and Chagas Disease. Mem Inst Oswaldo Cruz 2022, 117. [Google Scholar] [CrossRef]
- Gibson, W.C.; Miles, M.A. The Karyotype and Ploidy of Trypanosoma Cruzi. EMBO J 1986, 5, 1299–1305. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ward, P.; Moya, A.; Ayala, F.J. Natural Populations of Trypanosoma Cruzi, the Agent of Chagas Disease, Have a Complex Multiclonal Structure (Linkage Disequilibrium/Speciation/Evolution/Enzyme Variation/Wagner Network); 1986; Vol. 83.
- Bogliolo, A.R., L. -P.L., G.W.C., 1996. Polymorphisms in Trypanosoma Cruzi: Evidence of Genetic Recombination. Acta Trop 1996, 61, 31–40. [Google Scholar] [CrossRef]
- Carrasco, H.J.; Frame, I.A.; Valente, S.A.; Miles, M.A. Genetic Exchange as a Possible Source of Genomic Diversity in Sylvatic Populations of Trypanosoma Cruzi. Am J Trop Med Hyg 1996, 54, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Souto, R.P.; Fernandes, O.; Macedo, A.M.; Campbell, D.A.; Zingales, B. DNA Markers Define Two Major Phylogenetic Lineages of Trypanosoma Cruzi. Mol Biochem Parasitol 1996, 83, 141–152. [Google Scholar] [CrossRef]
- Brisse, S.; Barnabé, C.; Bañuls, A.L.; Sidibé, I.; Noël, S.; Tibayrenc, M. A Phylogenetic Analysis of the Trypanosoma Cruzi Genome Project CL Brener Reference Strain by Multilocus Enzyme Electrophoresis and Multiprimer Random Amplified Polymorphic DNA Fingerprinting. Mol Biochem Parasitol 1998, 92, 253–263. [Google Scholar] [CrossRef]
- Oliveira, R.P.; Broude, N.E.; Macedo, A.M.; Cantor, C.R.; Smith, C.L.; Pena, S.D.J. Probing the Genetic Population Structure of Trypanosoma Cruzi with Polymorphic Microsatellites. Proceedings of the National Academy of Sciences 1998, 95, 3776–3780. [Google Scholar] [CrossRef] [PubMed]
- Baptista, R. de P.; D’Ávila, D.A.; Segatto, M.; Valle, Í.F. do; Franco, G.R.; Valadares, H.M.S.; Gontijo, E.D.; Galvão, L.M. da C.; Pena, S.D.J.; Chiari, E.; et al. Evidence of Substantial Recombination among Trypanosoma Cruzi II Strains from Minas Gerais. Infection, Genetics and Evolution 2014, 22, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Michael W., Gaunt; Matthew Yeo, I.A.F. J. Russell Stothard; Hernan J. Carrasco; Martin C. Taylor; Susana Solis Mena; Paul Veazey; Graham A. J. Miles; Nidia Acosta; Antonieta Rojas de Arias; et al. Mechanism of Genetic Exchange in American Trypanosomes. Nature 2003, 421, 936–939. Nature 2003, 421, 936–939. [Google Scholar] [CrossRef]
- Alves, C.L.; Repolês, B.M.; da Silva, M.S.; Mendes, I.C.; Marin, P.A.; Aguiar, P.H.N.; Santos, S. da S.; Franco, G.R.; Macedo, A.M.; Pena, S.D.J.; et al. The Recombinase Rad51 Plays a Key Role in Events of Genetic Exchange in Trypanosoma Cruzi. Sci Rep 2018, 8, 13335. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.S.F.; Salazar-Sánchez, R.; Castillo-Neyra, R.; Borrini-Mayorí, K.; Chipana-Ramos, C.; Vargas-Maquera, M.; Ancca-Juarez, J.; Náquira-Velarde, C.; Levy, M.Z.; Brisson, D. Sexual Reproduction in a Natural Trypanosoma Cruzi Population. PLoS Negl Trop Dis 2019, 13, e0007392. [Google Scholar] [CrossRef]
- Schwabl, P.; Imamura, H.; Van den Broeck, F.; Costales, J.A.; Maiguashca-Sánchez, J.; Miles, M.A.; Andersson, B.; Grijalva, M.J.; Llewellyn, M.S. Meiotic Sex in Chagas Disease Parasite Trypanosoma Cruzi. Nat Commun 2019, 10, 3972. [Google Scholar] [CrossRef]
- Zingales, B.; Andrade, S.; Briones, M.; Campbell, D.; Chiari, E.; Fernandes, O.; Guhl, F.; Lages-Silva, E.; Macedo, A.; Machado, C.; et al. A New Consensus for Trypanosoma Cruzi Intraspecific Nomenclature: Second Revision Meeting Recommends TcI to TcVI. Mem Inst Oswaldo Cruz 2009, 104, 1051–1054. [Google Scholar] [CrossRef]
- Marcili, A.; Lima, L.; Cavazzana, M.; Junqueira, A.C.V.; Veludo, H.H.; Maia Da Silva, F.; Campaner, M.; Paiva, F.; Nunes, V.L.B.; Teixeira, M.M.G. A New Genotype of Trypanosoma Cruzi Associated with Bats Evidenced by Phylogenetic Analyses Using SSU RDNA, Cytochrome b and Histone H2B Genes and Genotyping Based on ITS1 RDNA. Parasitology 2009, 136, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.G.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The Revised Trypanosoma Cruzi Subspecific Nomenclature: Rationale, Epidemiological Relevance and Research Applications. Infection, Genetics and Evolution 2012, 12, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Espinosa-Álvarez, O.; Ortiz, P.A.; Trejo-Varón, J.A.; Carranza, J.C.; Pinto, C.M.; Serrano, M.G.; Buck, G.A.; Camargo, E.P.; Teixeira, M.M.G. Genetic Diversity of Trypanosoma Cruzi in Bats, and Multilocus Phylogenetic and Phylogeographical Analyses Supporting Tcbat as an Independent DTU (Discrete Typing Unit). Acta Trop 2015, 151, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M. Genetic Epidemiology of Parasitic Protozoa and Other Infectious Agents: The Need for an Integrated Approach. Int J Parasitol 1998, 28, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Ayala, F.J. Reproductive Clonality in Protozoan Pathogens—Truth or Artifact? A Comment on Ramírez and Llewellyn. Mol Ecol 2015, 24, 5778–5781. [Google Scholar] [CrossRef] [PubMed]
- Campbell, D.; Westenberger, S.; Sturm, N. The Determinants of Chagas Disease: Connecting Parasite and Host Genetics. Curr Mol Med 2004, 4, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Sturm, N.R.; Campbell, D.A. Alternative Lifestyles: The Population Structure of Trypanosoma Cruzi. Acta Trop 2010, 115, 35–43. [Google Scholar] [CrossRef]
- Brenière, S.F.; Waleckx, E.; Barnabé, C. Over Six Thousand Trypanosoma Cruzi Strains Classified into Discrete Typing Units (DTUs): Attempt at an Inventory. PLoS Negl Trop Dis 2016, 10, e0004792. [Google Scholar] [CrossRef]
- Westenberger, S.J.; Barnabé, C.; Campbell, D.A.; Sturm, N.R. Two Hybridization Events Define the Population Structure of Trypanosoma Cruzi. Genetics 2005, 171, 527–543. [Google Scholar] [CrossRef]
- de Freitas, J.M.; Augusto-Pinto, L.; Pimenta, J.R.; Bastos-Rodrigues, L.; Gonçalves, V.F.; Teixeira, S.M.R.; Chiari, E.; Junqueira, Â.C. V; Fernandes, O.; Macedo, A.M.; et al. Ancestral Genomes, Sex, and the Population Structure of Trypanosoma Cruzi. PLoS Pathog 2006, 2, e24. [Google Scholar] [CrossRef]
- Yeo, M.; Mauricio, I.L.; Messenger, L.A.; Lewis, M.D.; Llewellyn, M.S.; Acosta, N.; Bhattacharyya, T.; Diosque, P.; Carrasco, H.J.; Miles, M.A. Multilocus Sequence Typing (MLST) for Lineage Assignment and High Resolution Diversity Studies in Trypanosoma Cruzi. PLoS Negl Trop Dis 2011, 5, e1049. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, N.; Diosque, P. Evolution of Trypanosoma Cruzi: Clarifying Hybridisations, Mitochondrial Introgressions and Phylogenetic Relationships between Major Lineages. Mem Inst Oswaldo Cruz 2015, 110, 403–413. [Google Scholar] [CrossRef]
- Lewis, M.D.; Llewellyn, M.S.; Yeo, M.; Acosta, N.; Gaunt, M.W.; Miles, M.A. Recent, Independent and Anthropogenic Origins of Trypanosoma Cruzi Hybrids. PLoS Negl Trop Dis 2011, 5, e1363. [Google Scholar] [CrossRef] [PubMed]
- Flores-López, C.A.; Machado, C.A. Analyses of 32 Loci Clarify Phylogenetic Relationships among Trypanosoma Cruzi Lineages and Support a Single Hybridization Prior to Human Contact. PLoS Negl Trop Dis 2011, 5, e1272. [Google Scholar] [CrossRef] [PubMed]
- Barnabé, C.; Mobarec, H.I.; Jurado, M.R.; Cortez, J.A.; Brenière, S.F. Reconsideration of the Seven Discrete Typing Units within the Species Trypanosoma Cruzi, a New Proposal of Three Reliable Mitochondrial Clades. Infection, Genetics and Evolution 2016, 39, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Lima-Cordón, R.A.; Cahan, S.H.; McCann, C.; Dorn, P.L.; Justi, S.A.; Rodas, A.; Monroy, M.C.; Stevens, L. Insights from a Comprehensive Study of Trypanosoma Cruzi: A New Mitochondrial Clade Restricted to North and Central America and Genetic Structure of TcI in the Region. PLoS Negl Trop Dis 2021, 15, e0010043. [Google Scholar] [CrossRef] [PubMed]
- Onofre, T.S.; Loch, L.; Ferreira Rodrigues, J.P.; Macedo, S.; Yoshida, N. Gp35/50 Mucin Molecules of Trypanosoma Cruzi Metacyclic Forms That Mediate Host Cell Invasion Interact with Annexin A2. PLoS Negl Trop Dis 2022, 16, e0010788. [Google Scholar] [CrossRef]
- Martín-Escolano, J.; Marín, C.; Rosales, M.J.; Tsaousis, A.D.; Medina-Carmona, E.; Martín-Escolano, R. An Updated View of the Trypanosoma Cruzi Life Cycle: Intervention Points for an Effective Treatment. ACS Infect Dis 2022, 8, 1107–1115. [Google Scholar] [CrossRef]
- Gonçalves Moreno, C.J.; Freitas Oliveira, J.C.; Castelo Branco, J., Araújo, L.; Queiroz, A.M., et. al., Cell Culture and Maintenance of the Evolutionary Forms of Trypanosoma cruzi for Studies of Parasitic Biology. In Biology of Trypanosoma cruzi. W. de Souza ed. 2019. [CrossRef]
- Callejas-Hernández, F.; Herreros-Cabello, A.; del Moral-Salmoral, J.; Fresno, M.; Gironès, N. The Complete Mitochondrial DNA of Trypanosoma Cruzi: Maxicircles and Minicircles. Front Cell Infect Microbiol 2021, 11. [Google Scholar] [CrossRef]
- Dvorak, J.A.; Hall, T.E.; Crane, M.ST.J.; Engel, J.C.; McDaniel, J.P.; Uriegas, R. Trypanosoma Cruzi: Flow Cytometric Analysis. I. Analysis of Total DNA/Organism by Means of Mithramycin-Induced Fluorescence 1, 2. J Protozool 1982, 29, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.D.; Llewellyn, M.S.; Gaunt, M.W.; Yeo, M.; Carrasco, H.J.; Miles, M.A. Flow Cytometric Analysis and Microsatellite Genotyping Reveal Extensive DNA Content Variation in Trypanosoma Cruzi Populations and Expose Contrasts between Natural and Experimental Hybrids. Int J Parasitol 2009, 39, 1305–1317. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.T.; Lima, F.M.; Barros, R.M.; Cortez, D.R.; Santos, M.F.; Cordero, E.M.; Ruiz, J.C.; Goldenberg, S.; Teixeira, M.M.G.; da Silveira, J.F. Genome Size, Karyotype Polymorphism and Chromosomal Evolution in Trypanosoma Cruzi. PLoS One 2011, 6, e23042. [Google Scholar] [CrossRef] [PubMed]
- Nancy Vargas; Aurélio Pedroso; Bianca Zingales Chromosomal Polymorphism, Gene Synteny and Genome Size in T. Cruzi I and T. Cruzi II Groups. Mol Biochem Parasitol Nov;138(1):131-41. 2004, 138, 131–141. [CrossRef]
- Gonzalez, A.; Prediger, E.; Huecas, M.E.; Nogueira, N.; Lizardi, P.M. Minichromosomal Repetitive DNA in Trypanosoma Cruzi: Its Use in a High-Sensitivity Parasite Detection Assay. Proc Natl Acad Sci U S A 1984, 81, 3356–3360. [Google Scholar] [CrossRef] [PubMed]
- Moser, D.R.; Kirchhoff, L. V; Donelson, J.E. Detection of Trypanosoma Cruzi by DNA Amplification Using the Polymerase Chain Reaction. J Clin Microbiol 1989, 27, 1477–1482. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, J.; Porcel, B.; Rydåker, M.; Ruiz, A.; Sabaj, V.; Galanti, N.; Cazzulo, J.J.; Frasch, A.C.; Pettersson, U. Chromosome Specific Markers Reveal Conserved Linkage Groups in Spite of Extensive Chromosomal Size Variation in Trypanosoma Cruzi. Mol Biochem Parasitol 1995, 73, 63–74. [Google Scholar] [CrossRef]
- El-Sayed, N.M.; Myler, P.J.; Bartholomeu, D.C.; Nilsson, D.; Aggarwal, G.; Tran, A.-N.; Ghedin, E.; Worthey, E.A.; Delcher, A.L.; Blandin, G.; et al. The Genome Sequence of Trypanosoma Cruzi, Etiologic Agent of Chagas Disease. Science 2005, 309, 409–415. [Google Scholar] [CrossRef]
- Berriman, M.; Ghedin, E.; Hertz-Fowler, C.; Blandin, G.; Renauld, H.; Bartholomeu, D.C.; Lennard, N.J.; Caler, E.; Hamlin, N.E.; Haas, B.; et al. The Genome of the African Trypanosome Trypanosoma Brucei. Science 2005, 309, 416–422. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.-A.; Adlem, E.; Aert, R.; et al. The Genome of the Kinetoplastid Parasite, Leishmania Major. Science 2005, 309, 436–442. [Google Scholar] [CrossRef]
- El-Sayed, N.M.; Myler, P.J.; Blandin, G.; Berriman, M.; Crabtree, J.; Aggarwal, G.; Caler, E.; Renauld, H.; Worthey, E.A.; Hertz-Fowler, C.; et al. Comparative Genomics of Trypanosomatid Parasitic Protozoa. Science 2005, 309, 404–409. [Google Scholar] [CrossRef]
- De Pablos, L.M.; Osuna, A. Multigene Families in Trypanosoma Cruzi and Their Role in Infectivity. Infect Immun 2012, 80, 2258–2264. [Google Scholar] [CrossRef]
- Berná, L.; Rodriguez, M.; Chiribao, M.L.; Parodi-Talice, A.; Pita, S.; Rijo, G.; Alvarez-Valin, F.; Robello, C. Expanding an Expanded Genome: Long-Read Sequencing of Trypanosoma Cruzi. Microb Genom 2018, 4. [Google Scholar] [CrossRef]
- Berná, L.; Sebastián Pita; María Laura Chiribao; Adriana Parodi-Talice; Fernando Alvarez-Valin; Carlos Robello Biology of the Trypanosoma Cruzi Genome.; Wanderley De Souza, 2019.
- Majeau, A.; Murphy, L.; Herrera, C.; Dumonteil, E. Assessing Trypanosoma Cruzi Parasite Diversity through Comparative Genomics: Implications for Disease Epidemiology and Diagnostics. Pathogens 2021, 10, 212. [Google Scholar] [CrossRef]
- Talavera-López, C.; Messenger, L.A.; Lewis, M.D.; Yeo, M.; Reis-Cunha, J.L.; Matos, G.M.; Bartholomeu, D.C.; Calzada, J.E.; Saldaña, A.; Ramírez, J.D.; et al. Repeat-Driven Generation of Antigenic Diversity in a Major Human Pathogen, Trypanosoma Cruzi. Front Cell Infect Microbiol 2021, 11, 614665. [Google Scholar] [CrossRef]
- Wang, W.; Peng, D.; Baptista, R.P.; Li, Y.; Kissinger, J.C.; Tarleton, R.L. Strain-Specific Genome Evolution in Trypanosoma Cruzi, the Agent of Chagas Disease. PLoS Pathog 2021, 17, e1009254. [Google Scholar] [CrossRef]
- Reis-Cunha, J.L.; Coqueiro-Dos-Santos, A.; Pimenta-Carvalho, S.A.; Marques, L.P.; Rodrigues-Luiz, G.F.; Baptista, R.P.; Almeida, L.V. de; Honorato, N.R.M.; Lobo, F.P.; Fraga, V.G.; et al. Accessing the Variability of Multicopy Genes in Complex Genomes Using Unassembled Next-Generation Sequencing Reads: The Case of Trypanosoma Cruzi Multigene Families. mBio 2022, 13, e0231922. [Google Scholar] [CrossRef]
- Wilkinson, S.R.; Taylor, M.C.; Horn, D.; Kelly, J.M.; Cheeseman, I. A Mechanism for Cross-Resistance to Nifurtimox and Benznidazole in Trypanosomes. Proceedings of the National Academy of Sciences 2008, 105, 5022–5027. [Google Scholar] [CrossRef]
- Müller Kratz, J.; Garcia Bournissen, F.; Forsyth, C.J.; Sosa-Estani, S. Clinical and Pharmacological Profile of Benznidazole for Treatment of Chagas Disease. Expert Rev Clin Pharmacol 2018, 11, 943–957. [Google Scholar] [CrossRef]
- Filardi, L.S.; Brener, Z. Susceptibility and Natural Resistance of Trypanosoma Cruzi Strains to Drugs Used Clinically in Chagas Disease. Trans R Soc Trop Med Hyg 1987, 81, 755–759. [Google Scholar] [CrossRef]
- Andrade, S.G.; Magalhães, J.B.; Pontes, A.L. Terapêutica Da Fase Crônica Da Infecção Experimental Pelo Trypanosoma Cruzi Com o Benzonidazol e o Nifurtimox. Rev Soc Bras Med Trop 1989, 22, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Murta, S.M.; Gazzinelli, R.T.; Brener, Z.; Romanha, A.J. Molecular Characterization of Susceptible and Naturally Resistant Strains of Trypanosoma Cruzi to Benznidazole and Nifurtimox. Mol Biochem Parasitol 1998, 93, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Vela, A.; Coral-Almeida, M.; Sereno, D.; Costales, J.A.; Barnabé, C.; Brenière, S.F. In Vitro Susceptibility of Trypanosoma Cruzi Discrete Typing Units (DTUs) to Benznidazole: A Systematic Review and Meta-Analysis. PLoS Negl Trop Dis 2021, 15, e0009269. [Google Scholar] [CrossRef] [PubMed]
- Toledo, M.J. de O.; Bahia, M.T.; Carneiro, C.M.; Martins-Filho, O.A.; Tibayrenc, M.; Barnabé, C.; Tafuri, W.L.; de Lana, M. Chemotherapy with Benznidazole and Itraconazole for Mice Infected with Different Trypanosoma Cruzi Clonal Genotypes. Antimicrob Agents Chemother 2003, 47, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, D.; Barnabé, C.; Sereno, D.; Tibayrenc, M. Lack of Correlation between in Vitro Susceptibility to Benznidazole and Phylogenetic Diversity of Trypanosoma Cruzi, the Agent of Chagas Disease. Exp Parasitol 2004, 108, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; D’ávila, D.A.; Silva, M.N.; Galvão, L.M.; Macedo, A.M.; Chiari, E.; Gontijo, E.D.; Zingales, B. Trypanosoma Cruzi Benznidazole Susceptibility in Vitro Does Not Predict the Therapeutic Outcome of Human Chagas Disease. Mem Inst Oswaldo Cruz 2010, 105, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.B.; Giardini, M.A.; Kim, H.; Franco, C.H.; Araujo-Junior, A.M.; Schenkman, S.; Chatelain, E.; Freitas-Junior, L.H. Nitroheterocyclic Compounds Are More Efficacious than CYP51 Inhibitors against Trypanosoma Cruzi: Implications for Chagas Disease Drug Discovery and Development. Sci Rep 2014, 4, 4703. [Google Scholar] [CrossRef]
- Revollo, S.; Oury, B.; Vela, A.; Tibayrenc, M.; Sereno, D. In Vitro Benznidazole and Nifurtimox Susceptibility Profile of Trypanosoma Cruzi Strains Belonging to Discrete Typing Units TcI, TcII, and TcV. Pathogens 2019, 8. [Google Scholar] [CrossRef]
- Quebrada Palacio, L.P.; González, M.N.; Hernandez-Vasquez, Y.; Perrone, A.E.; Parodi-Talice, A.; Bua, J.; Postan, M. Phenotypic Diversity and Drug Susceptibility of Trypanosoma Cruzi TcV Clinical Isolates. PLoS One 2018, 13, e0203462. [Google Scholar] [CrossRef]
- Molina, I.; Gómez i Prat, J.; Salvador, F.; Treviño, B.; Sulleiro, E.; Serre, N.; Pou, D.; Roure, S.; Cabezos, J.; Valerio, L.; et al. Randomized Trial of Posaconazole and Benznidazole for Chronic Chagas’ Disease. New England Journal of Medicine 2014, 370, 1899–1908. [Google Scholar] [CrossRef]
- Torrico, F.; Gascon, J.; Ortiz, L.; Alonso-Vega, C.; Pinazo, M.-J.; Schijman, A.; Almeida, I.C.; Alves, F.; Strub-Wourgaft, N.; Ribeiro, I.; et al. Treatment of Adult Chronic Indeterminate Chagas Disease with Benznidazole and Three E1224 Dosing Regimens: A Proof-of-Concept, Randomised, Placebo-Controlled Trial. Lancet Infect Dis 2018, 18, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B.; Miles, M.A.; Moraes, C.B.; Luquetti, A.; Guhl, F.; Schijman, A.G.; Ribeiro, I. Drugs for Neglected Disease Initiative; Chagas Clinical Research Platform Meeting Drug Discovery for Chagas Disease Should Consider Trypanosoma Cruzi Strain Diversity. Mem Inst Oswaldo Cruz 2014, 109, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Duschak, V.G.; Paniz Mondolfi, A.E.; Benaim, G. Editorial: Chagas Disease Novel Drug Targets and Treatments. Front Cell Infect Microbiol 2023, 13. [Google Scholar] [CrossRef]
- Lewis, M.D.; Francisco, A.F.; Taylor, M.C.; Kelly, J.M. A New Experimental Model for Assessing Drug Efficacy against Trypanosoma Cruzi Infection Based on Highly Sensitive In Vivo Imaging. SLAS Discovery 2015, 20, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Nirdé, P.; Larroque, C.; Barnabé, C. Drug-Resistant Epimastigotes of Trypanosoma Cruzi and Persistence of This Phenotype after Differentiation into Amastigotes. C R Acad Sci III 1995, 318, 1239–1244. [Google Scholar] [PubMed]
- Veloso, V.; Carneiro, C.; Toledo, M.; Lana, M.; Chiari, E.; Tafuri, W.; Bahia, M. Variation in Susceptibility to Benznidazole in Isolates Derived from Trypanosoma Cruzi Parental Strains. Mem Inst Oswaldo Cruz 2001, 96, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, D.; Nirdé, P.; Hide, M.; Barnabé, C.; Tibayrenc, M. Differential Gene Expression in Benznidazole-Resistant Trypanosoma Cruzi Parasites. Antimicrob Agents Chemother 2005, 49, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Mejia, A.M.; Hall, B.S.; Taylor, M.C.; Gómez-Palacio, A.; Wilkinson, S.R.; Triana-Chávez, O.; Kelly, J.M. Benznidazole-Resistance in Trypanosoma Cruzi Is a Readily Acquired Trait That Can Arise Independently in a Single Population. J Infect Dis 2012, 206, 220–228. [Google Scholar] [CrossRef]
- Campos, M.C.O.; Leon, L.L.; Taylor, M.C.; Kelly, J.M. Benznidazole-Resistance in Trypanosoma Cruzi: Evidence That Distinct Mechanisms Can Act in Concert. Mol Biochem Parasitol 2014, 193, 17–19. [Google Scholar] [CrossRef]
- Campos, M.C.; Phelan, J.; Francisco, A.F.; Taylor, M.C.; Lewis, M.D.; Pain, A.; Clark, T.G.; Kelly, J.M. Genome-Wide Mutagenesis and Multi-Drug Resistance in American Trypanosomes Induced by the Front-Line Drug Benznidazole. Sci Rep 2017, 7, 14407. [Google Scholar] [CrossRef] [PubMed]
- Leprohon, P.; Légaré, D.; Ouellette, M. ABC Transporters Involved in Drug Resistance in Human Parasites. Essays Biochem 2011, 50, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B.; Araujo, R.G.A.; Moreno, M.; Franco, J.; Aguiar, P.H.N.; Nunes, S.L.; Silva, M.N.; Ienne, S.; Machado, C.R.; Brandão, A. A Novel ABCG-like Transporter of Trypanosoma Cruzi Is Involved in Natural Resistance to Benznidazole. Mem Inst Oswaldo Cruz 2015, 110, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Franco, J.; Ferreira, R.C.; Ienne, S.; Zingales, B. ABCG-like Transporter of Trypanosoma Cruzi Involved in Benznidazole Resistance: Gene Polymorphisms Disclose Inter-Strain Intragenic Recombination in Hybrid Isolates. Infection, Genetics and Evolution 2015, 31, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.; Reinhard, K.; Ferreira, L.F.; Pucu, E.; Chieffi, P.P. Paleoparasitology: The Origin of Human Parasites. Arq Neuropsiquiatr 2013, 71, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Barretto, M.P. Estudos Sôbre Reservatórios e Vetores Silvestres Do Trypanosoma Cruzi: XVII. Contribuição Para o Estudo Dos Focos Naturais Da Tripanossomose Americana, Com Especial Referência à Região Nordeste Do Estado de São Paulo, Brasil. Rev Soc Bras Med Trop 1967, 1, 23–36. [Google Scholar] [CrossRef]
- Diotaiuti, L.; Pereira, A.S.; Loiola, C.F.; Fernandes, A.J.; Schofield, J.C.; Dujardin, J.P.; Dias, J.C.P.; Chiari, E. Inter-Relation of Sylvatic and Domestic Transmission of Trypanosoma Cruzi in Areas with and without Domestic Vectorial Transmission in Minas Gerais, Brazil. Mem Inst Oswaldo Cruz 1995, 90, 443–448. [Google Scholar] [CrossRef]
- Deane, M.P.; Lenzi, H.L.; Jansen, A. Trypanosoma Cruzi: Vertebrate and Invertebrate Cycles in the Same Mammal Host, the Opossum Didelphis Marsupialis. Mem Inst Oswaldo Cruz 1984, 79, 513–515. [Google Scholar] [CrossRef]
- Schofield, C. Trypanosoma Cruzi -- the Vector-Parasite Paradox. Mem Inst Oswaldo Cruz 2000, 95, 535–544. [Google Scholar] [CrossRef]
- Jansen, A.M.; Xavier, S.C.C.; Roque, A.L.R. The Multiple and Complex and Changeable Scenarios of the Trypanosoma Cruzi Transmission Cycle in the Sylvatic Environment. Acta Trop 2015, 151, 1–15. [Google Scholar] [CrossRef]
- Jansen, A.M.; Xavier, S.C. das C.; Roque, A.L.R. Landmarks of the Knowledge and Trypanosoma Cruzi Biology in the Wild Environment. Front Cell Infect Microbiol 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.S.H.; Silva, D.J. da; Malheiros, A.F. Mamíferos Como Reservatórios de Trypanosoma Sp. – Uma Abordagem Bibliométrica. Research, Society and Development 2021, 10, e19710716539. [Google Scholar] [CrossRef]
- Izeta-Alberdi, A.; Ibarra-Cerdeña, C.N.; Moo-Llanes, D.A.; Ramsey, J.M. Geographical, Landscape and Host Associations of Trypanosoma Cruzi DTUs and Lineages. Parasit Vectors 2016, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Dumonteil, E.; Ramirez-Sierra, M.-J.; Pérez-Carrillo, S.; Teh-Poot, C.; Herrera, C.; Gourbière, S.; Waleckx, E. Detailed Ecological Associations of Triatomines Revealed by Metabarcoding and Next-Generation Sequencing: Implications for Triatomine Behavior and Trypanosoma Cruzi Transmission Cycles. Sci Rep 2018, 8, 4140. [Google Scholar] [CrossRef] [PubMed]
- Carreira, J.C.A.; Jansen, A.M.; Deane, M.P.; Lenzi, H.L. Histopathological Study of Experimental and Natural Infections by Trypanosoma Cruzi in Didelphis Marsupialis. Mem Inst Oswaldo Cruz 1996, 91, 609–618. [Google Scholar] [CrossRef]
- Yeo, M.; Acosta, N.; Llewellyn, M.; Sánchez, H.; Adamson, S.; Miles, G.A.J.; López, E.; González, N.; Patterson, J.S.; Gaunt, M.W.; et al. Origins of Chagas Disease: Didelphis Species Are Natural Hosts of Trypanosoma Cruzi I and Armadillos Hosts of Trypanosoma Cruzi II, Including Hybrids. Int J Parasitol 2005, 35, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Acosta, N.; López, E.; Lewis, M.D.; Llewellyn, M.S.; Gómez, A.; Román, F.; Miles, M.A.; Yeo, M. Hosts and Vectors of Trypanosoma Cruzi Discrete Typing Units in the Chagas Disease Endemic Region of the Paraguayan Chaco. Parasitology 2017, 144, 884–898. [Google Scholar] [CrossRef]
- Pinto, C.M.; Kalko, E.K.V.; Cottontail, I.; Wellinghausen, N.; Cottontail, V.M. TcBat a Bat-Exclusive Lineage of Trypanosoma Cruzi in the Panama Canal Zone, with Comments on Its Classification and the Use of the 18S RRNA Gene for Lineage Identification. Infection, Genetics and Evolution 2012, 12, 1328–1332. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Hernández, C.; Montilla, M.; Zambrano, P.; Flórez, A.C.; Parra, E.; Cucunubá, Z.M. First Report of Human Trypanosoma Cruzi Infection Attributed to TcBat Genotype. Zoonoses Public Health 2014, 61, 477–479. [Google Scholar] [CrossRef]
- Pinto, C.M.; Ocaña-Mayorga, S.; Tapia, E.E.; Lobos, S.E.; Zurita, A.P.; Aguirre-Villacís, F.; MacDonald, A.; Villacís, A.G.; Lima, L.; Teixeira, M.M.G.; et al. Bats, Trypanosomes, and Triatomines in Ecuador: New Insights into the Diversity, Transmission, and Origins of Trypanosoma Cruzi and Chagas Disease. PLoS One 2015, 10, e0139999. [Google Scholar] [CrossRef]
- Cominetti, M.C.; Csordas, B.G.; Cunha, R.C.; Andreotti, R. Geographical Distribution of Trypanosoma Cruzi in Triatomine Vectors in the State of Mato Grosso Do Sul, Brazil. Rev Soc Bras Med Trop 2014, 47, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Guhl, F.; Auderheide, A.; Ramírez, J.D. From Ancient to Contemporary Molecular Eco-Epidemiology of Chagas Disease in the Americas. Int J Parasitol 2014, 44, 605–612. [Google Scholar] [CrossRef]
- Flores-López, C.A.; Mitchell, E.A.; Reisenman, C.E.; Sarkar, S.; Williamson, P.C.; Machado, C.A. Phylogenetic Diversity of Two Common Trypanosoma Cruzi Lineages in the Southwestern United States. Infection, Genetics and Evolution 2022, 99, 105251. [Google Scholar] [CrossRef] [PubMed]
- Marcili, A.; Lima, L.; Valente, V.C.; Valente, S.A.; Batista, J.S.; Junqueira, A.C.V.; Souza, A.I.; da Rosa, J.A.; Campaner, M.; Lewis, M.D.; et al. Comparative Phylogeography of Trypanosoma Cruzi TCIIc: New Hosts, Association with Terrestrial Ecotopes, and Spatial Clustering. Infection, Genetics and Evolution 2009, 9, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Roellig, D.M.; Brown, E.L.; Barnabé, C.; Tibayrenc, M.; Steurer, F.J.; Yabsley, M.J. Molecular Typing of Trypanosoma Cruzi Isolates, United States. Emerg Infect Dis 2008, 14, 1123–1125. [Google Scholar] [CrossRef] [PubMed]
- Bern, C.; Kjos, S.; Yabsley, M.J.; Montgomery, S.P. Trypanosoma Cruzi and Chagas’ Disease in the United States. Clin Microbiol Rev 2011, 24, 655–681. [Google Scholar] [CrossRef] [PubMed]
- Hodo, C.L.; Hamer, S.A. Toward an Ecological Framework for Assessing Reservoirs of Vector-Borne Pathogens: Wildlife Reservoirs of Trypanosoma Cruzi across the Southern United States. ILAR J 2017, 58, 379–392. [Google Scholar] [CrossRef]
- Roellig, D.M.; Ellis, A.E.; Yabsley, M.J. Genetically Different Isolates of Trypanosoma Cruzi Elicit Different Infection Dynamics in Raccoons (Procyon Lotor) and Virginia Opossums (Didelphis Virginiana). Int J Parasitol 2009, 39, 1603–1610. [Google Scholar] [CrossRef]
- Fernandes, O.; Mangia, R.H.; Lisboa, C. V.; Pinho, A.P.; Morel, C.M.; Zingales, B.; Campbell, D.A.; Jansen, A.M. The Complexity of the Sylvatic Cycle of Trypanosoma Cruzi in Rio de Janeiro State (Brazil) Revealed by the Non-Transcribed Spacer of the Mini-Exon Gene. Parasitology 1999, 118, 161–166. [Google Scholar] [CrossRef]
- Lisboa, C.V.; Monteiro, R.V.; Martins, A.F.; Xavier, S.C. das C.; Lima, V. dos S.; Jansen, A.M. Infection with Trypanosoma Cruzi TcII and TcI in Free-Ranging Population of Lion Tamarins (Leontopithecus Spp): An 11-Year Follow-Up. Mem Inst Oswaldo Cruz 2015, 110, 394–402. [Google Scholar] [CrossRef]
- Ceccarelli, S.; Balsalobre, A.; Medone, P.; Cano, M.E.; Gurgel Gonçalves, R.; Feliciangeli, D.; Vezzani, D.; Wisnivesky-Colli, C.; Gorla, D.E.; Marti, G.A.; et al. DataTri, a Database of American Triatomine Species Occurrence. Sci Data 2018, 5, 180071. [Google Scholar] [CrossRef] [PubMed]
- Gürtler, R.E.; Cardinal, M.V. Reservoir Host Competence and the Role of Domestic and Commensal Hosts in the Transmission of Trypanosoma Cruzi. Acta Trop 2015, 151, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Messenger, L.A.; Miles, M.A.; Bern, C. Between a Bug and a Hard Place: Trypanosoma Cruzi Genetic Diversity and the Clinical Outcomes of Chagas Disease. Expert Rev Anti Infect Ther 2015, 13, 995–1029. [Google Scholar] [CrossRef] [PubMed]
- Messenger, L.A.; Miles, M.A. Evidence and Importance of Genetic Exchange among Field Populations of Trypanosoma Cruzi. Acta Trop 2015, 151, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Velásquez-Ortiz, N.; Herrera, G.; Hernández, C.; Muñoz, M.; Ramírez, J.D. Discrete Typing Units of Trypanosoma Cruzi: Geographical and Biological Distribution in the Americas. Sci Data 2022, 9, 360. [Google Scholar] [CrossRef]
- Abras, A.; Gállego, M.; Muñoz, C.; Juiz, N.A.; Ramírez, J.C.; Cura, C.I.; Tebar, S.; Fernández-Arévalo, A.; Pinazo, M.-J.; de la Torre, L.; et al. Identification of Trypanosoma Cruzi Discrete Typing Units (DTUs) in Latin-American Migrants in Barcelona (Spain). Parasitol Int 2017, 66, 83–88. [Google Scholar] [CrossRef]
- Tavares de Oliveira, M.; Sulleiro, E.; Silgado Gimenez, A.; de Lana, M.; Zingales, B.; Santana da Silva, J.; Marin-Neto, J.A.; Molina, I. Quantification of Parasite Burden of Trypanosoma Cruzi and Identification of Discrete Typing Units (DTUs) in Blood Samples of Latin American Immigrants Residing in Barcelona, Spain. PLoS Negl Trop Dis 2020, 14, e0008311. [Google Scholar] [CrossRef] [PubMed]
- Macedo, A.M.; Segatto, M. Implications of Trypanosoma Cruzi Intraspecific Diversity in the Pathogenesis of Chagas Disease. In American Trypanosomiasis; Elsevier, 2010; pp. 489–522.
- Balouz, V.; Bracco, L.; Ricci, A.D.; Romer, G.; Agüero, F.; Buscaglia, C.A. Serological Approaches for Trypanosoma Cruzi Strain Typing. Trends Parasitol 2021, 37, 214–225. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Guhl, F.; Rendón, L.M.; Rosas, F.; Marin-Neto, J.A.; Morillo, C.A. Chagas Cardiomyopathy Manifestations and Trypanosoma Cruzi Genotypes Circulating in Chronic Chagasic Patients. PLoS Negl Trop Dis 2010, 4, e899. [Google Scholar] [CrossRef]
- Zafra, G.; Mantilla, J.C.; Valadares, H.M.; Macedo, A.M.; González, C.I. Evidence of Trypanosoma Cruzi II Infection in Colombian Chagasic Patients. Parasitol Res 2008, 103, 731–734. [Google Scholar] [CrossRef]
- Mantilla, J.C.; Zafra, G.A.; Macedo, A.M.; González, C.I. Mixed Infection of Trypanosoma Cruzi I and II in a Colombian Cardiomyopathic Patient. Hum Pathol 2010, 41, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Calvopina, M.; Segovia, G.; Cevallos, W.; Vicuña, Y.; Costales, J.A.; Guevara, A. Fatal Acute Chagas Disease by Trypanosoma Cruzi DTU TcI, Ecuador. BMC Infect Dis 2020, 20, 143. [Google Scholar] [CrossRef]
- Burgos, J.M.; Begher, S.; Silva, H.M.V.; Bisio, M.; Duffy, T.; Levin, M.J.; Macedo, A.M.; Schijman, A.G. Molecular Identification of Trypanosoma Cruzi I Tropism for Central Nervous System in Chagas Reactivation Due to AIDS. Am J Trop Med Hyg 2008, 78, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.; Bargues, M.D.; Fajardo, A.; Montilla, M.; Triana, O.; Vallejo, G.A.; Guhl, F. Identifying Four Trypanosoma Cruzi I Isolate Haplotypes from Different Geographic Regions in Colombia. Infect Genet Evol 2007, 7, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Ocaña-Mayorga, S.; Llewellyn, M.S.; Costales, J.A.; Miles, M.A.; Grijalva, M.J. Sex, Subdivision, and Domestic Dispersal of Trypanosoma Cruzi Lineage I in Southern Ecuador. PLoS Negl Trop Dis 2010, 4, e915. [Google Scholar] [CrossRef] [PubMed]
- Cura, C.I.; Mejía-Jaramillo, A.M.; Duffy, T.; Burgos, J.M.; Rodriguero, M.; Cardinal, M. V; Kjos, S.; Gurgel-Gonçalves, R.; Blanchet, D.; De Pablos, L.M.; et al. Trypanosoma Cruzi I Genotypes in Different Geographical Regions and Transmission Cycles Based on a Microsatellite Motif of the Intergenic Spacer of Spliced-Leader Genes. Int J Parasitol 2010, 40, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.S.; Miles, M.A.; Carrasco, H.J.; Lewis, M.D.; Yeo, M.; Vargas, J.; Torrico, F.; Diosque, P.; Valente, V.; Valente, S.A.; et al. Genome-Scale Multilocus Microsatellite Typing of Trypanosoma Cruzi Discrete Typing Unit I Reveals Phylogeographic Structure and Specific Genotypes Linked to Human Infection. PLoS Pathog 2009, 5, e1000410. [Google Scholar] [CrossRef] [PubMed]
- Roman, F.; das Chagas Xavier, S.; Messenger, L.A.; Pavan, M.G.; Miles, M.A.; Jansen, A.M.; Yeo, M. Dissecting the Phyloepidemiology of Trypanosoma Cruzi I (TcI) in Brazil by the Use of High Resolution Genetic Markers. PLoS Negl Trop Dis 2018, 12, e0006466. [Google Scholar] [CrossRef]
- Bosseno, M.-F.; Barnabé, C.; Magallón Gastélum, E.; Lozano Kasten, F.; Ramsey, J.; Espinoza, B.; Brenière, S.F. Predominance of Trypanosoma Cruzi Lineage I in Mexico. J Clin Microbiol 2002, 40, 627–632. [Google Scholar] [CrossRef]
- Guhl, F.; Ramírez, J.D. Trypanosoma Cruzi I Diversity: Towards the Need of Genetic Subdivision? Acta Trop 2011, 119, 1–4. [Google Scholar] [CrossRef]
- Falla, A.; Herrera, C.; Fajardo, A.; Montilla, M.; Vallejo, G.A.; Guhl, F. Haplotype Identification within Trypanosoma Cruzi I in Colombian Isolates from Several Reservoirs, Vectors and Humans. Acta Trop 2009, 110, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.M.; Guhl, F.; Zabala, D.; Ramírez, J.D.; Urrea, D.A.; Hernández, D.C.; Cucunubá, Z.; Montilla, M.; Carranza, J.C.; Rueda, K.; et al. The Identification of Two Trypanosoma Cruzi I Genotypes from Domestic and Sylvatic Transmission Cycles in Colombia Based on a Single Polymerase Chain Reaction Amplification of the Spliced-Leader Intergenic Region. Mem Inst Oswaldo Cruz 2013, 108, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Messenger, L.A.; Llewellyn, M.S.; Bhattacharyya, T.; Franzén, O.; Lewis, M.D.; Ramírez, J.D.; Carrasco, H.J.; Andersson, B.; Miles, M.A. Multiple Mitochondrial Introgression Events and Heteroplasmy in Trypanosoma Cruzi Revealed by Maxicircle MLST and Next Generation Sequencing. PLoS Negl Trop Dis 2012, 6, e1584. [Google Scholar] [CrossRef] [PubMed]
- Calzada, J.E.; Samudio, F.; de Juncá, C.; Pineda, V.; Burleigh, B.A.; Saldaña, A. Genetic Diversity of Trypanosoma Cruzi in Panama Inferred by Multi-Locus Sequence Typing of Mitochondrial Genes. Microorganisms 2022, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.N.; Burroughs, H.; Gorchakov, R.; Gunter, S.M.; Dumonteil, E.; Murray, K.O.; Herrera, C.P. Molecular Identification and Genotyping of Trypanosoma Cruzi DNA in Autochthonous Chagas Disease Patients from Texas, USA. Infection, Genetics and Evolution 2017, 49, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Nielebock, M.A.P.; Moreira, O.C.; Xavier, S.C. das C.; Miranda, L. de F.C.; Lima, A.C.B. de; Pereira, T.O. de J.S.; Hasslocher-Moreno, A.M.; Britto, C.; Sangenis, L.H.C.; Saraiva, R.M. Association between Trypanosoma Cruzi DTU TcII and Chronic Chagas Disease Clinical Presentation and Outcome in an Urban Cohort in Brazil. PLoS One 2020, 15, e0243008. [Google Scholar] [CrossRef]
- Lages-Silva, E.; Ramírez, L.E.; Pedrosa, A.L.; Crema, E.; da Cunha Galvão, L.M.; Junho Pena, S.D.; Macedo, A.M.; Chiari, E. Variability of Kinetoplast DNA Gene Signatures of Trypanosoma Cruzi II Strains from Patients with Different Clinical Forms of Chagas’ Disease in Brazil. J Clin Microbiol 2006, 44, 2167–2171. [Google Scholar] [CrossRef] [PubMed]
- Hasslocher-Moreno, A.M.; Salles Xavier, S.; Magalhães Saraiva, R.; Conde Sangenis, L.H.; Teixeira de Holanda, M.; Horta Veloso, H.; Rodrigues da Costa, A.; de Souza Nogueira Sardinha Mendes, F.; Alvarenga Americano do Brasil, P.E.; Sperandio da Silva, G.M.; et al. Progression Rate from the Indeterminate Form to the Cardiac Form in Patients with Chronic Chagas Disease: Twenty-Two-Year Follow-Up in a Brazilian Urban Cohort. Trop Med Infect Dis 2020, 5, 76. [Google Scholar] [CrossRef]
- Gómez-Ochoa, S.A.; Rojas, L.Z.; Echeverría, L.E.; Muka, T.; Franco, O.H. Global, Regional, and National Trends of Chagas Disease from 1990 to 2019: Comprehensive Analysis of the Global Burden of Disease Study. Glob Heart 2022, 17, 59. [Google Scholar] [CrossRef]
- Freitas, J.M.; Lages-Silva, E.; Crema, E.; Pena, S.D.J.; Macedo, A.M. Real Time PCR Strategy for the Identification of Major Lineages of Trypanosoma Cruzi Directly in Chronically Infected Human Tissues. Int J Parasitol 2005, 35, 411–417. [Google Scholar] [CrossRef]
- Tavares de Oliveira, M.; Fuzo, C.A.; da Silva, M.C.; Donadi, E.A.; da Silva, J.S.; Moreira, H.T.; Schmidt, A.; Marin-Neto, J.A. Correlation of TcII Discrete Typing Units with Severe Chronic Chagas Cardiomyopathy in Patients from Various Brazilian Geographic Regions. PLoS Negl Trop Dis 2022, 16, e0010713. [Google Scholar] [CrossRef] [PubMed]
- Virreira, M.; Serrano, G.; Maldonado, L.; Svoboda, M. Trypanosoma Cruzi: Typing of Genotype (Sub)Lineages in Megacolon Samples from Bolivian Patients. Acta Trop 2006, 100, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, M. V.; Lauricella, M.A.; Ceballos, L.A.; Lanati, L.; Marcet, P.L.; Levin, M.J.; Kitron, U.; Gürtler, R.E.; Schijman, A.G. Molecular Epidemiology of Domestic and Sylvatic Trypanosoma Cruzi Infection in Rural Northwestern Argentina. Int J Parasitol 2008, 38, 1533–1543. [Google Scholar] [CrossRef]
- Monje-Rumi, M.M.; Brandán, C.P.; Ragone, P.G.; Tomasini, N.; Lauthier, J.J.; Alberti D’Amato, A.M.; Cimino, R.O.; Orellana, V.; Basombrío, M.A.; Diosque, P. Trypanosoma Cruzi Diversity in the Gran Chaco: Mixed Infections and Differential Host Distribution of TcV and TcVI. Infection, Genetics and Evolution 2015, 29, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Yager, J.E.; Lozano Beltran, D.F.; Torrico, F.; Gilman, R.H.; Bern, C. Prevalence of Chagas Heart Disease in a Region Endemic for <I>Trypanosoma Cruzi</I>: Evidence From a Central Bolivian Community. Glob Heart 2015, 10, 145. [Google Scholar] [CrossRef]
- Forsyth, C.J. “I Cannot Be Worried”: Living with Chagas Disease in Tropical Bolivia. PLoS Negl Trop Dis 2017, 11, e0005251. [Google Scholar] [CrossRef] [PubMed]
- Bizai, M.L.; Romina, P.; Antonela, S.; Olivera, L. V.; Arias, E.E.; Josefina, D.C.; Silvia, M.; Walter, S.; Diana, F.; Cristina, D. Geographic Distribution of Trypanosoma Cruzi Genotypes Detected in Chronic Infected People from Argentina. Association with Climatic Variables and Clinical Manifestations of Chagas Disease. Infection, Genetics and Evolution 2020, 78, 104128. [Google Scholar] [CrossRef]
- Luquetti, A.O.; Tavares, S.B. do N.; Siriano, L. da R.; Oliveira, R.A. de; Campos, D.E.; Morais, C.A. de; Oliveira, E.C. de Congenital Transmission of Trypanosoma Cruzi in Central Brazil. A Study of 1,211 Individuals Born to Infected Mothers. Mem Inst Oswaldo Cruz 2015, 110, 369–376. [Google Scholar] [CrossRef]
- Bustos, P.L.; Milduberger, N.; Volta, B.J.; Perrone, A.E.; Laucella, S.A.; Bua, J. Trypanosoma Cruzi Infection at the Maternal-Fetal Interface: Implications of Parasite Load in the Congenital Transmission and Challenges in the Diagnosis of Infected Newborns. Front Microbiol 2019, 10. [Google Scholar] [CrossRef]
- Herrera, C.; Truyens, C.; Dumonteil, E.; Alger, J.; Sosa-Estani, S.; Cafferata, M.L.; Gibbons, L.; Ciganda, A.; Matute, M.L.; Zuniga, C.; et al. Phylogenetic Analysis of Trypanosoma Cruzi from Pregnant Women and Newborns from Argentina, Honduras, and Mexico Suggests an Association of Parasite Haplotypes with Congenital Transmission of the Parasite. The Journal of Molecular Diagnostics 2019, 21, 1095–1105. [Google Scholar] [CrossRef]
- Hakim, J.M.C.; Waltmann, A.; Tinajeros, F.; Kharabora, O.; Machaca, E.M.; Calderon, M.; del Carmen Menduiña, M.; Wang, J.; Rueda, D.; Zimic, M.; et al. Amplicon Sequencing Reveals Complex Infection in Infants Congenitally Infected With Trypanosoma Cruzi and Informs the Dynamics of Parasite Transmission. J Infect Dis 2023, 228, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Vicco, M.H.; Bontempi, I.; Ortiz, S.; Solari, A.; Bottasso, O.A.; Marcipar, I. Chronic Chagas Disease with Cardiodigestive Involvement and the TcVI Infective Form of Trypanosoma Cruzi. A Case Report. Parasitol Int 2012, 61, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, W.M.; Magalhães, L.K.; Santana Filho, F.S.; Borborema, M.; Silveira, H.; Barbosa, M. das G. V. Trypanosoma Cruzi TcIII / Z3 Genotype as Agent of an Outbreak of Chagas Disease in the Brazilian Western Amazonia. Tropical Medicine & International Health 2010, no-no. [CrossRef]
- Câmara, A.C.J.; Varela-Freire, A.A.; Valadares, H.M.S.; Macedo, A.M.; D’Ávila, D.A.; Machado, C.R.; Lages-Silva, E.; Chiari, E.; Galvão, L.M.C. Genetic Analyses of Trypanosoma Cruzi Isolates from Naturally Infected Triatomines and Humans in Northeastern Brazil. Acta Trop 2010, 115, 205–211. [Google Scholar] [CrossRef]
- Valente, S.A. da S.; da Costa Valente, V.; das Neves Pinto, A.Y.; de Jesus Barbosa César, M.; dos Santos, M.P.; Miranda, C.O.S.; Cuervo, P.; Fernandes, O. Analysis of an Acute Chagas Disease Outbreak in the Brazilian Amazon: Human Cases, Triatomines, Reservoir Mammals and Parasites. Trans R Soc Trop Med Hyg 2009, 103, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, W.M.; Magalhães, L.K.C.; de Sá, A.R.N.; Gomes, M.L.; Toledo, M.J. de O.; Borges, L.; Pires, I.; de Oliveira Guerra, J.A.; Silveira, H.; Barbosa, M. das G.V. Trypanosoma Cruzi IV Causing Outbreaks of Acute Chagas Disease and Infections by Different Haplotypes in the Western Brazilian Amazonia. PLoS One 2012, 7, e41284. [Google Scholar] [CrossRef] [PubMed]
- Freitas, V.L.T. de; Piotto, M.R.; Esper, H.R.; Nakanishi, E.Y.S.; Fonseca, C. de A.; Assy, J.G.P.L.; Berreta, O.C.P.; França, F.O. de S.; Lopes, M.H. Detection of Trypanosoma Cruzi DTUs TcI and TcIV in Two Outbreaks of Orally-Transmitted Chagas Disease in the Northern Region of Brazil. Rev Inst Med Trop Sao Paulo 2023. [CrossRef]
- Miles, M.A.; Souza, A.; Povoa, M.; Shaw, J.J.; Lainson, R.; Toye, P.J. Isozymic Heterogeneity of Trypanosoma Cruzi in the First Autochthonous Patients with Chagas’ Disease in Amazonian Brazil. Nature 1978, 272, 819–821. [Google Scholar] [CrossRef]
- Iwagami, M.; Higo, H.; Miura, S.; Yanagi, T.; Tada, I.; Kano, S.; Agatsuma, T. Molecular Phylogeny of Trypanosoma Cruzi from Central America (Guatemala) and a Comparison with South American Strains. Parasitol Res 2007, 102, 129–134. [Google Scholar] [CrossRef]
- Carrasco, H.J.; Segovia, M.; Llewellyn, M.S.; Morocoima, A.; Urdaneta-Morales, S.; Martínez, C.; Martínez, C.E.; Garcia, C.; Rodríguez, M.; Espinosa, R.; et al. Geographical Distribution of Trypanosoma Cruzi Genotypes in Venezuela. PLoS Negl Trop Dis 2012, 6, e1707. [Google Scholar] [CrossRef]
- Yun, O.; Lima, M.A.; Ellman, T.; Chambi, W.; Castillo, S.; Flevaud, L.; Roddy, P.; Parreño, F.; Albajar Viñas, P.; Palma, P.P. Feasibility, Drug Safety, and Effectiveness of Etiological Treatment Programs for Chagas Disease in Honduras, Guatemala, and Bolivia: 10-Year Experience of Médecins Sans Frontières. PLoS Negl Trop Dis 2009, 3, e488. [Google Scholar] [CrossRef]
- Morillo, C.A.; Marin-Neto, J.A.; Avezum, A.; Sosa-Estani, S.; Rassi, A.; Rosas, F.; Villena, E.; Quiroz, R.; Bonilla, R.; Britto, C.; et al. Randomized Trial of Benznidazole for Chronic Chagas’ Cardiomyopathy. New England Journal of Medicine 2015, 373, 1295–1306. [Google Scholar] [CrossRef]
- Cancado, J.R. Long Term Evaluation of Etiological Treatment of Chagas Disease with Benznidazole. Rev Inst Med Trop Sao Paulo 2002, 44, 29–37. [Google Scholar] [CrossRef]
- Pinazo, M.-J.; Thomas, M.-C.; Bustamante, J.; Almeida, I.C. de; Lopez, M.-C.; Gascon, J. Biomarkers of Therapeutic Responses in Chronic Chagas Disease: State of the Art and Future Perspectives. Mem Inst Oswaldo Cruz 2015, 110, 422–432. [Google Scholar] [CrossRef]
- Cortes-Serra, N.; Losada-Galvan, I.; Pinazo, M.-J.; Fernandez-Becerra, C.; Gascon, J.; Alonso-Padilla, J. State-of-the-Art in Host-Derived Biomarkers of Chagas Disease Prognosis and Early Evaluation of Anti-<i>Trypanosoma Cruzi<i> Treatment Response. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2020, 1866, 165758. [Google Scholar] [CrossRef]
- Francisco, A.F.; Saade, U.; Jayawardhana, S.; Pottel, H.; Scandale, I.; Chatelain, E.; Liehl, P.; Kelly, J.M.; Zrein, M. Comparing in Vivo Bioluminescence Imaging and the Multi-Cruzi Immunoassay Platform to Develop Improved Chagas Disease Diagnostic Procedures and Biomarkers for Monitoring Parasitological Cure. PLoS Negl Trop Dis 2022, 16, e0010827. [Google Scholar] [CrossRef] [PubMed]
| Mammalian orders | T. cruzi DTU | |||||||
|---|---|---|---|---|---|---|---|---|
| TcI | TcII | TcIII | TcIV | TcV | TcVI | TcBat | Total | |
| Carnivora | 46 | 36 | 82 | |||||
| Chiroptera | 57 | 21 | 59 | 137 | ||||
| Cingulata | 78 | 78 | ||||||
| Didelphimorphia | 262 | 262 | ||||||
| Primata | 43 | 10 | 10 | 63 | ||||
| Rodentia | 91 | 37 | 10 | 24 | 20 | 182 | ||
| Total | 499 | 68 | 88 | 46 | 24 | 20 | 59 | 804 |
| DTU | Transmission Cycle | Geographic Distribution | Major Clinical Aspects |
|---|---|---|---|
| TcI | Predominant in sylvatic and/or domestic cycles depending on the endemic region | North and Central Americas and Amazon region | Associated with cardiac Chagas disease in Venezuela and Colombia. Associated with oral transmission and severe acute cases in Brazil. Associated with neuroencephalitis in immunocompromised patients. |
| TcII | Predominant in domestic cycle; rare in sylvatic cycles | Southern Cone region of South America | Primary cause of severe cardiac Chagas disease in Brazil. Associated with megaesophagus and megacolon in Brazil. |
| TcIII | Predominant in sylvatic cycle; rare in domestic cycles | Amazon region | Rarely causes human Chagas disease, most of them in Brazil associated with oral transmission and acute cases. |
| TcIV | Predominant in sylvatic cycle; rare in domestic cycles | Amazon region | Few strains have been isolated from chronic infected humans, most of them in Venezuela. Associated with oral transmission and acute cases |
| TcV | Predominant in domestic cycle; rare in sylvatic cycles | Bolivia, Chile, northern Argentina and southern Brazil | Probably associated with milder cardiac chronic Chagas disease. Associated with megaesophagus and megacolon in Bolivia. Possibly associated to higher risk for congenital transmission. |
| TcVI | Predominant in domestic cycle; rare in sylvatic cycles | Southern Cone region of South America | Primary cause of severe chronic cardiac Chagas disease in Argentina. |
| TcBat | Predominant in sylvatic cycle | Panama, central and southeast Brazil and Colombia | So far, only one isolated case of human infection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





