Submitted:
01 November 2023
Posted:
02 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Bacterium Strains
2.1.1. Bacterium strains: their origin
2.1.2. AMP-producing Xenorhabdus strains
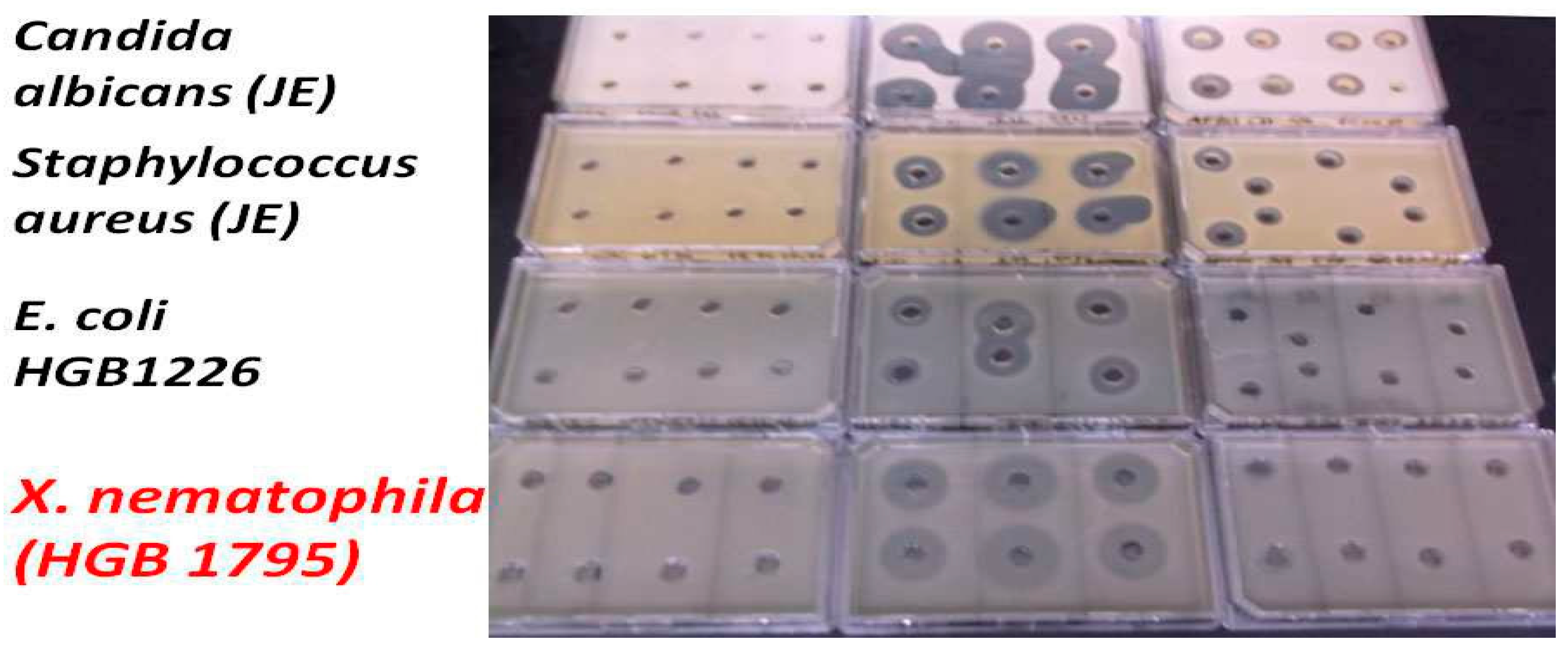
2.1.3. Test organisms
Control organisms
Agrobacterium strains
2.2. Antibiotics-active peptide fractions isolated from EMA CFCM.
2.2.1. Isolation of Antimicrobial Active Peptide Fractions EMA_PF1, and EMA_PF2, were carried out by Amberlite adsorption followed by washing (purification)and eluted by acetone with methanol gradient and eluted by methanol [56] (Furgani,2008), [57] (Böszörményi et al., 2009). 2.2.2. Isolation of Antimicrobial Active Peptide Fraction (EMA30)
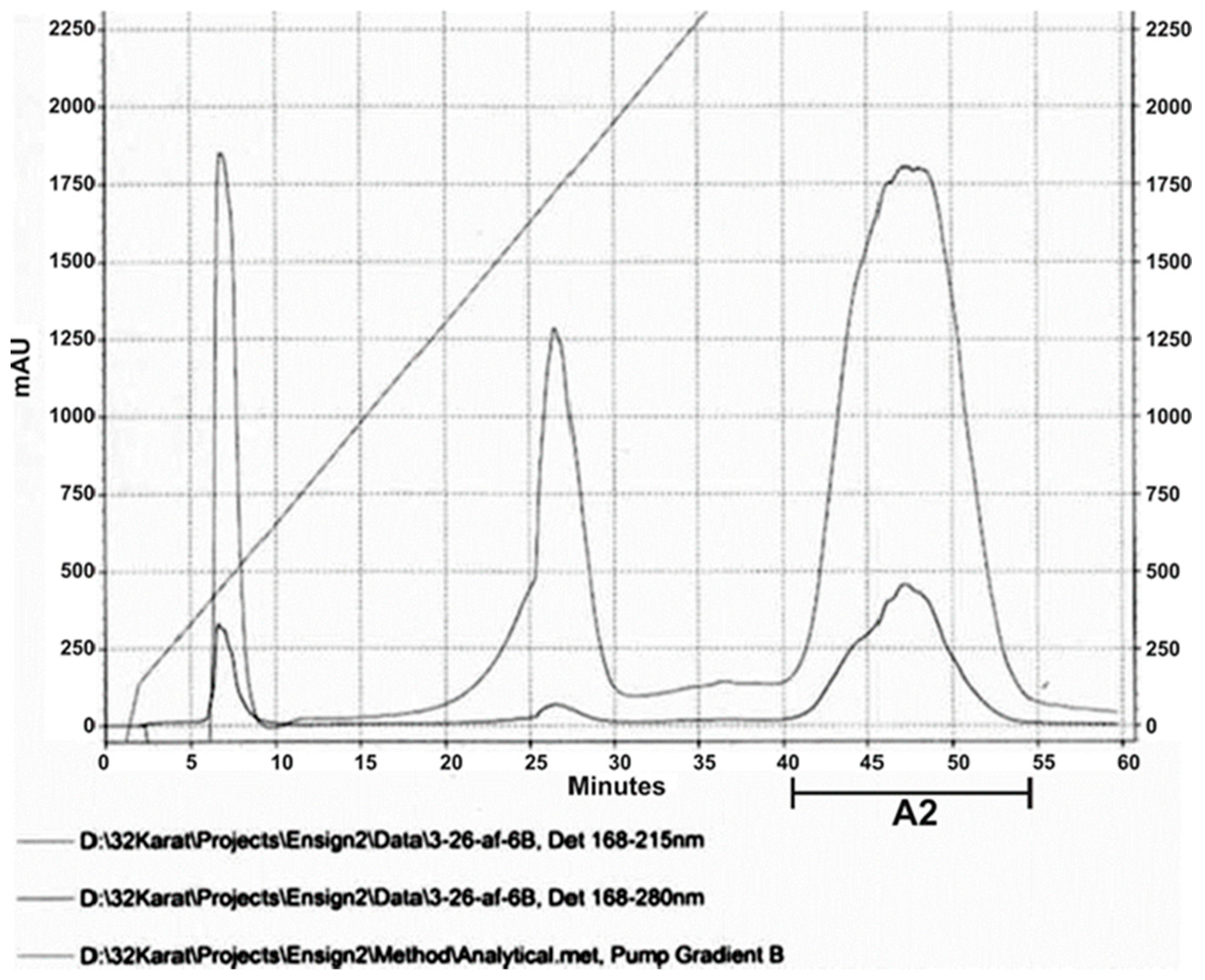
2.2.2. Antimicrobial active HPLC fractions
2.3. Bioassays of Antimicrobial Peptide Fractions from EMA CFCM
2.3.1. Methodology of Liquid Bioassay of EMA AMP-Active Fractions on Agrobacterium strains.
2.3.2. Quantitative evaluations
2.4. Statistics
3. Results
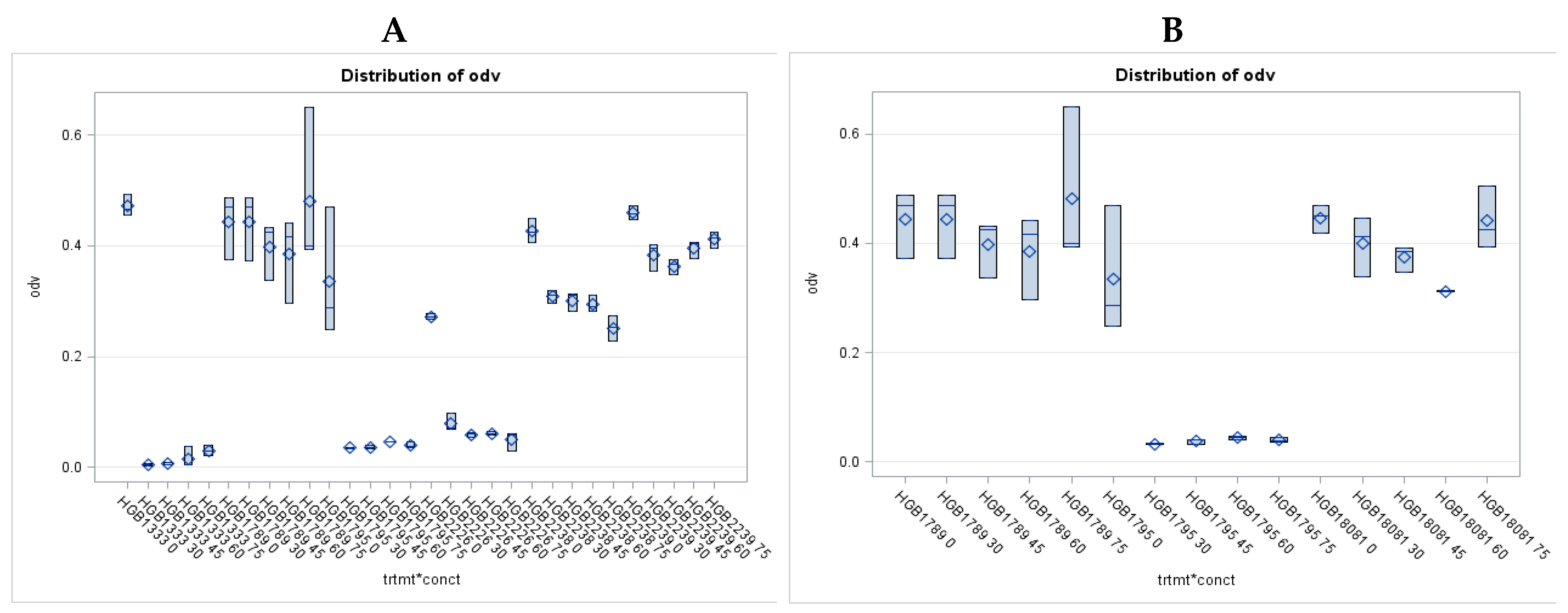
3.1. Antimicrobial Activity of EMA_PF2 on E. coli and Xenorhabdus strains. Results of Liquid Culture Bioassays.
3.2. Antimicrobial Activity Profile EMA_PF2 and EMA 30 peptide fractions
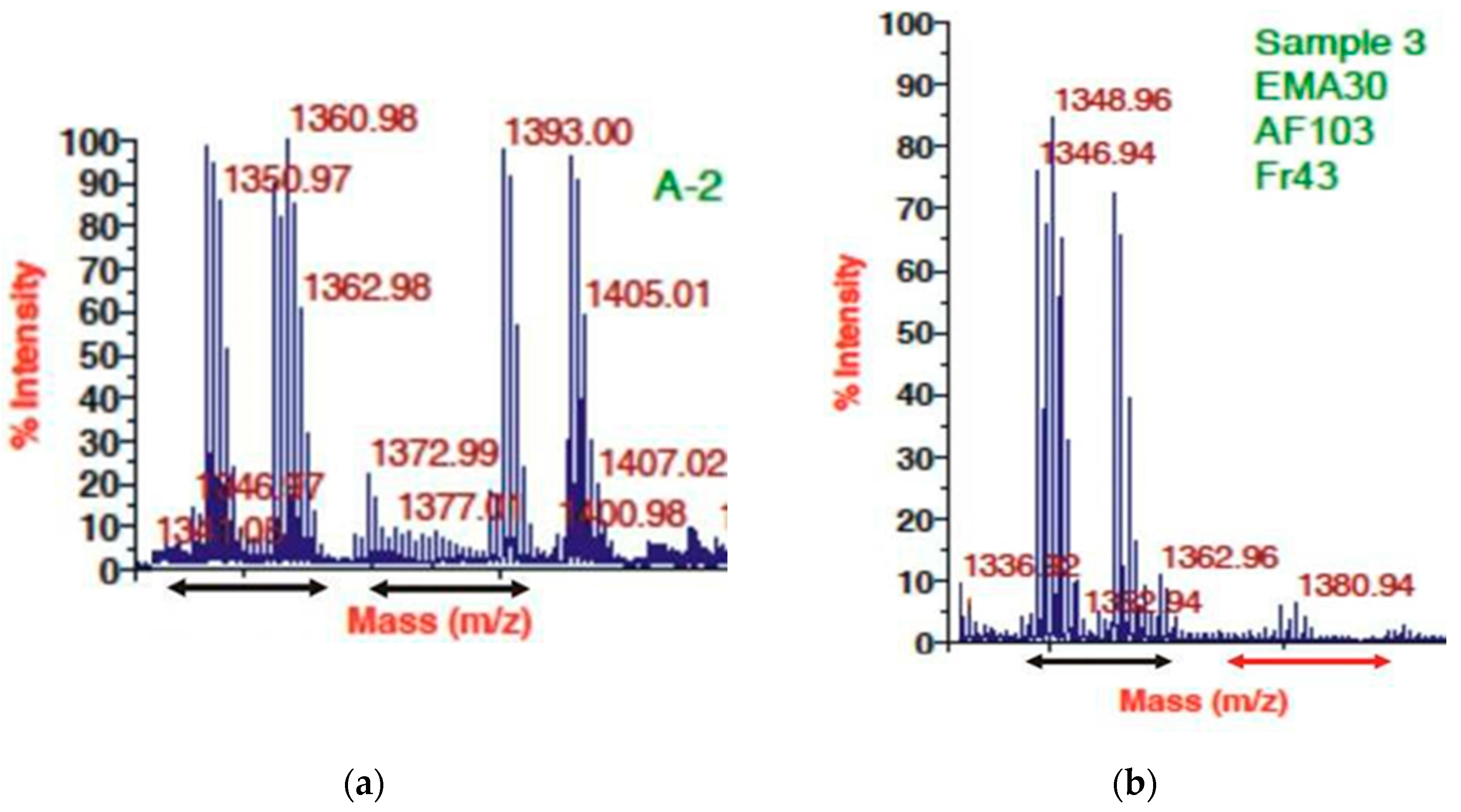
3.3. HPLC and MALDI Profile of EMA_PF2 and EMA 30 peptide fractions
| HPLC run | Staphylococcus aureus J.E. | Escherichia coli HGB2226 J.K |
|---|---|---|
| OD Values at 600 nm for the fractions collected between 40-50 min (Mean ± SD, n=2 at each time) | ||
| 1. | 0.3577±0.0797 | 0.5380± 0.009 |
| 2. | 0.4404±0.0511 | 0.4214±0.0002 |
| 3. | 0.4273±0.0377 | 0.4335±0.0002 |
| 4. | 0.4588±0.0307 | 0.4625±0.001 |
| 5. | 0.4027±0.0285 | 0.48135±0.00063 |
| 6. | 0.3874±0.0510 | 0.4651±0.00198 |
| 7. | 0.4255±0.0571 | 0.4395±0.0004 |
| 8. | 0.0003±0.0247 | 0.00155000±0.0006 |
| 9. | 0.0003±0.0201 | 0.00020000±0.0002 |
| 10. | 0.0081±0.0547 | 0.001±0.0001 |
| 11 | 0.0040±0.0061 | 0.0015±0.0002 |
3.4. Results of Liquid Culture Bioassays of EMA_PF2 on Agrobacterium Strains of Different Genotypes, Opine Types, and Plasmid Genotype
4. Discussion
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fodor A, Abate BA, Deák P, Fodor L, Gyenge E, Klein MG, Koncz Z, Muvevi J, Ötvös L, Székely G, Vozik D, Makrai L. Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides-A Review. Pathogens. 2020 Jun 29;9(7):522. [CrossRef] [PubMed]
- Rice, L. B. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. Journal of Infectious Diseases, 197(8):1079-1081. [CrossRef]
- Williamson R, Calderwood SB, Moellering RC Jr, Tomasz A. 1983. Studies on the mechanism of intrinsic resistance to beta-lactam antibiotics in group D streptococci. Journal of General Microbiology 129:813-822.
- Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Current Opinion in Microbiology 16: 10-16.
- Miller, W. R. , Munita, J. M. and Arias, C. A. 2014. Mechanisms of antibiotic resistance in enterococci Expert Review of Anti-infective Therapy 2014 Oct; 12:1221-1236. [CrossRef]
- Tomasz, A. 1998. Accelerated evolution: the emergence of multidrug-resistant gram-positive bacterial pathogens in the 1990's.Netherlands Journal of Medicine; 52:219-227.
- Tenover FC, Sinner SW, Segal RE, Huang V, Alexandre SS, McGowan JE Jr, Weinstein MP 2009. Characterization of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. International Journal of Antimicrobial Agents, Jun; 33(6):564-568. [CrossRef]
- Shi J, Mao NF, Wang L, Zhang HB, Chen Q, Liu H, Tang X, Jin T, Zhu CT, Li FB, Sun LH, 1109 Xu XM, Xu YQ 2014. Efficacy of combined vancomycin and fosfomycin against methicillin-resistant Staphylococcus aureus in biofilms in vivo. PLoS One 9:e113133. [CrossRef]
- Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, Chmelnitsky I, Schwaber MJ, Carmeli Y, Navon-Venezia S. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. Journal of Clinical Microbiology 47:3261–3265. [CrossRef]
- Schwaber MJ1, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, Shalit I, Carmeli Y; 2011. Israel Carbapenem-Resistant Enterobacteriaceae Working Group Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clinical Infectious Diseases 52:848-855. [CrossRef]
- Vila J, Martí S, Sanchez-Céspedes J. 2007. Porins, efflux pumps, and multidrug resistance in Acinetobacter baumanii. Journal of Antimicrobial Chemotherapy 59:1210–1215. Epub 2007 Feb 26.
- Antunes, L.C.S. , Visca, P., Towner, K.J., 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathogens and Diseases 71:292-301. [CrossRef]
- Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand, Y. Labia R. 1993. Characterization of 1041 a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrobial Agents 1042 and Chemotherapy 37: 962-969. PMID: 8517722 PMCID: PMC187863.
- Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa – a phenomenon of bacterial resistance. Journal of Medical Microbiology 58:1133–1148. [CrossRef]
- Nehme D, Poole K. 2005. Interaction of the MexA and MexB components of the MexAB-Opr Multidrug efflux system of Pseudomonas aeruginosa: identification of MexA extragenic 103suppressors of a T578I mutation in MexB. Antimicrobial Agents and Chemotherapy 49:4375– 4378. [CrossRef]
- Mulcahy, L.R. , Isabella, V.M., and Lewis, K. 2014. Pseudomonas aeruginosa biofilms in disease.Microb Ecol. 68:1. [CrossRef]
- Jeukens J, Kukavica-Ibrulj I, Emond-Rheault JG, Freschi L, Levesque RC. 2017. Comparative genomics of a drug-resistant Pseudomonas aeruginosa panel and the challenges of antimicrobial resistance prediction from genomes. FEMS Microbiology Letters 364, (18), 2 October 2017). [CrossRef] [PubMed]
- Gebreyes, W. A. and Thakur, S. 2005. Multidrug-Resistant Salmonella enterica Serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrobial Agents and Chemotherapy. 49: 503–511. [CrossRef]
- Endimiani A1, Hujer KM, Hujer AM, Bertschy I, Rossano A, Koch C, Gerber V, Francey T, Bonomo RA, Perreten V.. 2011. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data 2011. Journal of Antimicrobial Chemotherapy. 66: 2248-2254. [CrossRef]
- Szmolka A, Nagy B, 2013. Multidrug resistant commensal Escherichia coli in animals and its 1130 impact for public health. Search Results Frontiers in Microbiology. 4: Article 258, 1-13. 1131 Published online 2013 Sep 3. [CrossRef]
- McManus, BA. , Coleman, DC., Deasy, EC., Brennan, GI., O’ Connell, B., Monecke, S., Ehricht, R., Leggett, Leonard, NB., Anna C. Shore, AC. 2015. Comparative Genotypes, Staphylococcal Cassette Chromosome mec (SCCmec) Genes and Antimicrobial Resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus isolates from infections in humans and companion animals. PLoS One 10: e0138079. [CrossRef]
- Rzewuska M, Stefańska I, Kizerwetter-Świda MI, Chrobak M, Chimel D, Szczygielska P, Leśniak M, Binek M. 2015. Characterization of Extended-Spectrum-β-Lactamases Produced by Escherichia coli Strains Isolated from Dogs in Poland. Polish Journal of Microbiology 64: 285–288.
- Marques, C. , Gama, L. T., Belas, A., Bergström, K., Beurlet, S., Briend-Marchal, A., Broens, E.M., Costa, M., Criel, D., Damborg, P., van Dijk, M. A. M., van Dongen, A. M.,Dorsch, R., Espada, C.M., Gerber, B., Kritsepi-Konstantinou, M., Loncaric, I., Mion, D. Misic, D., Movilla, R., Overesch, G., Perreten,V., Roura, X., Steenbergen, J., Timofte, D.,Wolf, G., Zanoni, R. G. Schmitt, S., Guardabassi, L., Pomba, C. 2016. European multicenter study on antimicrobial resistance in bacteria isolated from companion animal urinary tract infections. BMC Veterinary Research 2016; 12:213. [CrossRef]
- Fodor A, Varga I, Hevesi M, Máthé-Fodor A, Racsko J, Hogan JA. 2012. Novel anti-microbial peptides of Xenorhabdus origin against multidrug-resistant plant pathogens, In Bobbarala, V. (Ed. Biochemistry, Genetics and Molecular Biology - A Search for Antibacterial Agents, 872 9.
- Załuga J, Stragier P, Baeyen S, Haegeman A, Van Vaerenbergh J, Maes M, De Vos P. 2014. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics 15. [CrossRef]
- Li, X.-Z. , Plésiat, P., Hiroshi Nikaido, H. 2015. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clinical Microbiology Reviews 28: 337-418. [CrossRef] [PubMed]
- Pitout, JD. 2008. Multiresistant Enterobacteriaceae: new threat of an old problem. Expert Review of Antimicrobial Infection Therapy 6:657–669. [CrossRef]
- Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present,and future. Antimicrobial Agents and Chemotherapy 55:4943-4960. [CrossRef]
- Temkin, E. , Adler, A., Lerner, A. Carmeli, Y. 2014. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. New York Academy of Sciences 1323:22-42. [CrossRef]
- Gupta N, Limbago BM, Patel JB, Kallen AJ.2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clinical Infectious Diseases, 53: 60-67.
- Kádár B, Kocsis B, Nagy, K, Szabó, D. 2013. The renaissance of polymyxins. Current Medicinal Chemistry Journal 20: 3759-3773. [PubMed]
- Otter, J. A. , Doumith, M., Davies, F., Mookerjee, S., Dyakova, E., Gilchrist, M., Brannigan, E. 1055 T., Bamford, K., Galletly, T., Donaldson, H., Aanensen, D. M., Ellington, M. J., Hill, R., 1056 Turton, J. F., Hopkins, K. L., Woodford, N. Holmes, A. 2017. Emergence and clonal spread 1057 of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Scientific Reports 7:12711.
- McManus PS, Stockwell VO, Sundin GW, Jones AL. 2002. Antibiotic use in plant agriculture. Annual Review of Phytopathology 40:443–465. [CrossRef]
- Stockwell VO, Sundin GW. Jones AL. 2002. Antibiotic use in plant agriculture. Annual Review of Phytopathology 40:443–465.
- Aćimović SG, Zeng Q, McGhee GC, Sundin GW, Wise JC. 2015. Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics, and 764 assessment of induction of pathogenesis-related protein genes. Frontiers of Plant Science 6:16. [CrossRef]
- Förster H, McGhee GC, Sundin GW, Adaskaveg JE. 2015. Characterization of streptomycin resistance in isolates of Erwinia amylovora in California. Phytopathology 105:1302–1310. [CrossRef]
- Gusberti M, Klemm U, Meier MS, Maurhofer M, Hunger-Glaser I. 2015. Fire blight control: The struggle goes on. A comparison of different fire blight control methods in Switzerland with respect to biosafety, efficacy and durability. International Journal of Environmental Research and Public Health 12:11422–11447. [CrossRef]
- Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clinical Microbiology Review 19:491–511. [CrossRef]
- Ötvös, L. Jr. D. Wade, J. D. 2014. Current challenges in peptide-based drug discovery. Specialty Grand Challenge Article Frontiers in Chemistry 2, 62. [CrossRef]
- Mojsoska B, Jenssen H. 2015. Peptides and Peptidomimetics for Antimicrobial Drug Design. Pharmaceuticals (Basel). 2015 Jul 13; 8:366-415. [CrossRef]
- Kosikowska P, Lesner A.2016. Antimicrobial peptides (AMPs) as drug candidates: a patent review(2003–2015) Expert Opinion on Therapeutic Patents, 26:689-702. [CrossRef]
- Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015 Jan;20(1):122-8. [CrossRef] [PubMed]
- Sharma, K.; Sharma, K.K.; Sharma, A.; Jain, R. Peptide-based drug discovery: Current status and recent advances. Drug Dis-cov. Today 2023, 28, 103464. [Google Scholar] [CrossRef]
- Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Médigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Research 41:D636 –D647. [CrossRef]
- Bode E, Brachmann AO, Kegler C, Simsek R, Dauth C, Zhou Q, Kaiser M, Klemmt P, Bode HB. 2015a. Simple “on-demand” production of bioactive natural products. ChemBioChem 16:1115–1119.
- Akhurst, RJ. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with 767 insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J GenMicrobiol.128:3061-3065. [CrossRef] [PubMed]
- Forst S, Nealson K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol Rev. 1996 Mar;60(1):21-43. [CrossRef] [PubMed]
- Vivas EI, Goodrich-Blair H. 2001. Xenorhabdus nematophilus as a model for host–bacterium interactions: rpoS is necessary for mutualism with nematodes. Journal of Bacteriology 183:4687–4693.
- Bode, HB. 2009. Entomopathogenic bacteria as a source of secondary metabolites. Current Opinions in Chemistry & Biology 13:224–230. doi.org/10.1016/j.cbpa.2009.02.037.
- Ogier JC, Pagès S, Frayssinet M, Gaudriault S. Entomopathogenic nematode-associated microbiota: from monoxenic paradigm to pathobiome. Microbiome. 2020 Feb 24;8(1):25. [CrossRef] [PubMed]
- Reimer D, Bode HB. 2014. A natural prodrug activation mechanism in the biosynthesis of non- ribosomal peptides. Natural Products Report 31:154-159. [PubMed]
- Park, D. , Ciezki K., van der Hoeven R., Singh S., Reimer D., Bode H.B., Forst S. 2009. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Molecular Microbiology 73:938–949. [CrossRef]
- Gualtieri M, Aumelas A, Thaler JO. 2009. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. Journal of Antibiotics 62:295–302.
- Houard J, Aumelas A, Noël T, Pages S, Givaudan A, Fitton-Ouhabi V, Villain-Guillot P, Gualtieri M. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J Antibiot (Tokyo). 2013 Oct;66(10):617-20. [CrossRef] [PubMed]
- Lengyel K, Lang E, Fodor A, Szállás E, Schumann P, Stackebrandt E. 2005. Description of four 978 novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., 979 Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Systematics of Applied Microbiology 28:115-122. Erratum in: Systematics of Applied Microbiology 30:83; also in March/April 2014 Volume 2 Issue 2 e00190-14 Genome Announcements genomea.asm.org. Xenorhabdus budapestensis sp. Nov., Xenorhabdus ehlersii sp. Nov., Xenorhabdus innexi sp. Nov., and Xenorhabdus szentirmaii sp. Nov. Systematics of Applied Microbiology 28:115–122. [CrossRef]
- Furgani G, Böszörményi E, Fodor A, Máthé-Fodor A, Forst S, Hogan JS, Katona Z, Klein,MG, Stackebrandt E, Szentirmai A, Sztaricskai F, Wolf SL. 2008. Xenorhabdus antibiotics: a 890 comparative analysis and potential utility for controlling mastitis caused by bacteria. Journal of 891 Applied Microbiology 104:745–758. [CrossRef]
- Böszörményi E, Érsek T, Fodor A, Fodor AM, Földes LS, Hevesi M, Hogan JS, Katona Z, 791 Klein MG, Kormány A, Pekár S, Szentirmai A, Sztaricskai F, Taylor RA. 2009. Isolation 792 and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. Journal of Applied Microbiology 107:746–759. [CrossRef]
- Fodor A, Gualtieri M, Zeller M, Tarasco E, Klein MG, Fodor AM, Haynes L, Lengyel K, Forst SA, Furgani GM, Karaffa L, Vellai T. Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC). Pathogens. 2022 Mar 11;11(3):342. [CrossRef] [PubMed]
- Gualtieri M, Ogier J-C, Pagès S, Givaudan A, Gaudriault S. 2014. Draft genome sequence and annotation of the entomopathogenic\ bacterium Xenorhabdus szentirmaii Strain DSM16338. Genome Announcements 2(2): e00190-14 genomea.asm.org 1).
- Fuchs, S.W.; Sachs, C.C.; Kegler, C.; Nollmann, F.I.; Karas, M.; Bode, H.B. Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal. Chem. 2012, 84, 6948–6955 [CrossRef]. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W. Grundmann, F.; Kurz, M.; Kaiser, M.; Bode, H.B. Fabclavines: Bioactive peptide-polyketide-polyamine hybrids from Xenorhabdus. Chembiochem 2014, 15, 512–516 [CrossRef]. [Google Scholar] [CrossRef] [PubMed]
- Wenski, S.L.; Kolbert, D.; Grammbitter, G.L.C.; Bode, H.B. Fabclavine biosynthesis in X. szentirmaii: Shortened derivatives and characterization of the thioester reductase FclG and the condensation domain-like protein FclL. J. Ind. Microbiol. Biotechnol. 2019, 46, 565–572 [CrossRef]. [Google Scholar] [CrossRef] [PubMed]
- Wenski, S.L.; Cimen, H.; Berghaus, N.; Fuchs, S.W.; Hazir, S.; Bode, H.B. Fabclavine diversity in Xenorhabdus bacteria. Fabclavine diversity in Xenorhabdus bacteria. Beilstein J. Org. Chem. 2020, 16, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Fodor A, Fodor AM, Forst S, Hogan J, Hevesi M, Klein MG, Stackebrandt E, Szentirmai A, Sztaricskai F. 2007. New aspects of Xenorhabdus research. In: Abstracts of the 11th Europea Meeting of IOBC.WPRS Working Group “Insect Pathogens and Insect Parasitic Nematodes” in association with COST 862 Bacterial toxins for insect control” Ales (Gard) France, June 03-07 2007.
- Fodor A, Fodor AM, Forst S, Hogan JS, Klein MG, Lehoczky É. 2010. Comparative analysis of antibacterial activities of Xenorhabdus species on related and non-related bacteria in vivo. Journal of Microbiology and Antimicrobials 2:30–35.
- Fodor A, Varga I, Hevesi M, Máthé-Fodor A, Racsko J, Hogan JA. 2012. Novel anti-microbial peptides of Xenorhabdus origin against multidrug resistant plant pathogens, In: Bobbarala, V. (Ed.): Biochemistry, Genetics and Molecular Biology - A Search for Antibacterial Agents, 9:147–196.
- Vozik D, Bélafi-Bakó K, Hevesi M, Böszörményi E, Fodor A. 2015. Effectiveness of a peptide-rich fraction from Xenorhabdus budapestensis culture against fire blight disease on apple blossoms. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 43:547-553. [CrossRef]
- Nester, EW. 2015. Agrobacterium: nature's genetic engineer. Front Plant Sci. 2015 Jan 6;5:730. [CrossRef]
- Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima J, Vagner P, Okura K, Zhou Y, Chen L, Wood GE, Almeida NF Jr., Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Sr., Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li M-J, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, RomeroP, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon- Kamm B, Liao L, Kim S, Hendrick C, Zhao Z-Y, Dola M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciensC58. Science 294:2317–2323. [CrossRef]
- Henkel CV, Dulk-Ras A, Zhang X, Hooykaas PJJ. 2014. Genome sequence of the octopine-type Agrobacterium tumefaciens Strain Ach5. Genome Announcements 2: e00225-14.
- Van Larebeke N, Engler G, Holsters M, Van den Elsacker S, Zaenen I, Schilperoort RA, Schell J. 1974. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252:169–170. (08 November 1974). [CrossRef]
- Currier TC, Nester EW. 1976. Evidence for diverse types of large plasmids in tumor- inducing strains of Agrobacterium. Journal of Bacteriology 126:157–165.
- Schell J, Van Montagu M. The Ti-plasmid of Agrobacterium tumefaciens, a natural vector for the introduction of nif genes in plants? Basic Life Sci. 1977;9:159-79. [CrossRef] [PubMed]
- Hooykaas PJJ. The Ti Plasmid, Driver of Agrobacterium Pathogenesis. Phytopathology. 2023 Apr;113(4):594-604. [CrossRef] [PubMed]
- Chilton M-D, Drummond M, Merlo D, Sciaky D, Montoya A, Gordon M, Nester E. 1977. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11:263–271.
- Koncz Cs, DeGreve H, Andre D, Deboeck F, Van Montagu M, Schell J. 1983. The opine synthase genes carried by Ti plasmids contain all signals necessary for expression in plants. EMBO Journal 2:1597–1603.
- Montoya A, Chilton M-D, Gordon MP, Sciaky D, Nester EW. 1977. Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: role of plasmid genes. Journal of Bacteriology 129:101–107.
- Guyon P, Chilton M-D, Petit A, Tempo J. 1980. Agropine in "null-type" crown gall tumors: Evidence for generality of the opine concept. Proceedings of the National Academy of Science USA 77:2693–2697.
- Tremblay G, Gagliardo R, Chilton WS, Dion P. Diversity among Opine-Utilizing Bacteria: Identification of Coryneform Isolates. Appl Environ Microbiol. 1987 Jul;53(7):1519-24. [CrossRef] [PubMed]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. 1983. A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 926 303:179–180.
- Koekman BP, Ooms G, Klapwijk PM, Schilperoort RA. 1979. Genetic map of an octopine Ti-plasmid. Plasmid 2:346–357.
- Klapwijk PM, Schilperoort RA 1979. Negative control of octopine degradation and transfer genes of octopin Ti plasmids in Agrobacterium tumefaciens. Journal of Bacteriology 132:424-431.
- Ooms G, Klapwijk PM, Poulis JA, Schilperoort RA. 1980. Characterization of Tn904 insertions in octopine Ti-plasmid mutants of Agrobacterium tumefaciens. Journal of Bacteriology 144:82– 1045.
- Ooms G, Hooykaas PJJ, Moolenaar G, Schilperoort RA. 1981. Crown gall tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti-plasmids: Analysis of T-DNA functions. Gene 14:33–50.
- Ooms G, Hooykaas PJJ, Van Veen RJM, Van Beelen P, Regensburg-Tuink AJG, Schilperoort RA. 1982. Octopine Ti plasmid deletion mutants of Agrobacterium tumefaciens with emphasis 1053 on the right side of the T-region. Plasmid 7:15-29.
- Jen GC, Chilton M-D. 1986. Activity of T-DNA borders in plant cell transformation by mini-Ti Plasmids. Journal of Bacteriology 166:491–499.
- Ausubel FM, Brent R, Kingston RE, Moor DD, Seidman JG, Smith JA, Struhl K. (Eds.) 1999. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. pp. 1-1 – 1-6; 5-1 – 6-30. John Wiley & Sons, New York.
- Leclerc MC, Boemare NE. (1991). Plasmids and phase variation in Xenorhabdus spp. Applied Environmental Microbiology 57:2597–3601.
- Wise AA, Liu Z, Binns AN. 2006. Culture and maintenance of Agrobacterium strains. Methods in Molecular Biology 343:3–14. Review.
- Hood EE, Helmer GL, Fraley RT, Chilton M.-D. 1986. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. Journal of Bacteriology 168:1291–1301.
- Lazo GR, Stein PA, Ludwig RA. 1991. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967.
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. 1993. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Research 2:208–218.
- White FF, Nester EW. Hairy root: plasmid encodes virulence traits in Agrobacterium rhizogenes. J Bacteriol. 1980 Mar;141(3):1134-41. [CrossRef] [PubMed]
- Petit A, David C, Dahl GA, Ellis JG, Guyon P, Casse-Delbart F, Tempe J. 1982. Further extension of the opine concept: Plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Molecular and General Genetics 190:204–214.
- Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F. 1986. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes Strain A4. Plasmid 16:124–134.
- Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu, H, Wood DW. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. Journal of Bacteriology 191:2501–2511. [CrossRef]
- Hood EE, Helmer GL, Fraley RT, Chilton MD. 1987. Virulence of Agrobacterium tumefaciens strain A281 on legumes. Plant Physiology 83:529–534.
- Komari T, Halperin W, Nester EW. 1986. Physical and functional map of supervirulent Agrobacterium tumefaciens tumor-inducing plasmid pTiBoS42. Journal of Bacteriology 166:88–94.
- Jin S, Komari T, Milton P, Gordon MP, Nester EW. 1987. Genes Responsible for the supervirulence Phenotype of Agrobacterium tumefaciens A281. Journal of Bacteriology 169:4417–4425.
- Bevan M, 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acid Research 12:8711–8721.
- Taylor CG, Fuchs B, Collier R, Lutke WK. 2006. Generation of composite plants using Agrobacterium rhizogenes. Methods in Molecular Biology 343:155–167. Review.
- Uraji M, Suzuki K, Yoshida K. 2002. A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes and Genetic Systematics 77:1–9.
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of 960 chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics 204:383–396.
- Dessaux Y, Guyon P, Petit A, Tempe J, Demarez M, Legrain C, Tate ME, Farrand SK. 1988. 830 Opine utilization by Agrobacterium spp.: Octopine-type Ti plasmids encode two pathways for mannopinic acid degradation. Journal of Bacteriology 170:2939–2946.
- Jandera, P. Gradient elution in normal-phase high-performance liquid chromatographic systems. J Chromatogr A. 2002 Aug 2;965(1-2):239-61. [CrossRef] [PubMed]
- Josic D, Kovac S. Reversed-phase High Performance Liquid Chromatography of proteins. Curr Protoc Protein Sci. 2010 Aug;Chapter 8:8.7.1-8.7.22. [CrossRef] [PubMed]
- Bowen DJ, Ensign JC. 1998. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Applied Environmental Microbiology 64:3029–3035. [PubMed]
- Bowen DJ, Ensign JC. 2001. Isolation and characterization of intracellular protein inclusions produced by the entomopathogenic bacterium Photorhabdus luminescens. Applied Environmental Microbiology 67:4834–4841.
- Ciche TA, Blackburn M, Carney JR, Ensign JC. Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol. 2003 Aug;69(8):4706-13. [CrossRef] [PubMed]
- Carr D 2002. The Handbook of Analysis and. Purification of Peptides and. Proteins by Reversed- Phase HPLC. Presented by Vydac (The Separations Group). 17434 Mojave Street. Hesperia CA 92345 USA.
- Wiegand, Hilpert & Hancock, 2008; Clinical and Laboratory Standards Institute (CLSI, 2012). M07: Methods for Dilution Antimicrobial Susceptibility Tests... Clinical & Laboratory Standards Institute. https://clsi.org media.
- Incedayi G, Cimen H, Ulug D, Touray M, Bode E, Bode HB, Orenlili Yaylagul E, Hazir S, Cakmak I. Relative potency of a novel acaricidal compound from Xenorhabdus, a bacterial genus mutualistically associated with entomopathogenic nematodes. Sci Rep. 2021 ;11(1):11253. 27 May. [CrossRef] [PubMed]
- Yadav M, Rathore JS. The hipBAXn operon from Xenorhabdus nematophila functions as a bonafide toxin-antitoxin module. Appl Microbiol Biotechnol. 2020 Apr;104(7):3081-3095. [CrossRef] [PubMed]
- Morales-Soto N, Forst SA. The xnp1 P2-like tail synthesis gene cluster encodes xenorhabdicin and is required for interspecies competition. J Bacteriol. 2011 Jul;193(14):3624-32. [CrossRef] [PubMed]
- Völgyi, A. , Fodor A, Forst S. 2000. Inactivation of a novel gene produces a phenotypic variant cell and affects the symbiotic behavior of Xenorhabdus nematophilus. Applied and Environmental Microbiology. 66:1622-1628.
- Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. 2007. The global regulator Lrp contributes to mutualism, pathogenesis, and phenotypic variation in the bacterium Xenorhabdus nematophila. Cellular Microbiology 9:1311-1323. 2007.
- Nuonming P, Khemthong S, Dokpikul T, Sukchawalit R, Mongkolsuk S. Characterization and regulation of AcrABR, a RND-type multidrug efflux system, in Agrobacterium tumefaciens C58. Microbiol Res. 2018 Sep;214:146-155. [CrossRef] [PubMed]
- Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, Posthuma D. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019 Sep;51(9):1339-1348. [CrossRef] [PubMed]
- Ward RD, Tran JS, Banta AB, Bacon EE, Rose WE, Peters JM. Essential Gene Knockdowns Reveal Genetic Vulnerabilities and Antibiotic Sensitivities in Acinetobacter baumannii. bioRxiv [Preprint]. 2023 Aug 2:2023.08.02.551708. [CrossRef] [PubMed]
- Rodrigues SD, Karimi M, Impens L, Van Lerberge E, Coussens G, Aesaert S, Rombaut D, Holtappels D, Ibrahim HMM, Van Montagu M, Wagemans J, Jacobs TB, De Coninck B, Pauwels L. Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc Natl Acad Sci U S A. 2021 Jan 12;118(2):e2013338118. [CrossRef] [PubMed]
- Elston RC, Satagopan J, Sun S. Statistical Genetic Terminology. Methods Mol Biol. 2017;1666:1-9. [CrossRef] [PubMed]
- Kiyokawa K, Yamamoto S, Sakuma K, Tanaka K, Moriguchi K, Suzuki K. Construction of disarmed Ti plasmids transferable between Escherichia coli and Agrobacterium species. Appl Environ Microbiol. 2009 Apr;75(7):1845-51. [CrossRef] [PubMed]
- Stachel SE, Nester EW. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445-54. [CrossRef] [PubMed]
- Palanichelvam K, Oger P, Clough SJ, Cha C, Bent AF, Farrand SK. A second T-region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol Plant Microbe Interact. 2000 Oct;13(10):1081-91. [CrossRef] [PubMed]
- Hattori Y, Iwata K, Suzuki K, Uraji M, Ohta N, Katoh A, Yoshida K. Sequence characterization of the vir region of a nopaline type Ti plasmid, pTi-SAKURA. Genes Genet Syst. 2001 Apr;76(2):121-30. [CrossRef] [PubMed]
- Lacroix B, Citovsky V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016 Jul 12;7(4):e00863-16. [CrossRef] [PubMed]
- Schröder G, Klipp W, Hillebrand A, Ehring R, Koncz C, Schröder J. The conserved part of the T-region in Ti-plasmids expresses four proteins in bacteria. EMBO J. 1983;2(3):403-9. [CrossRef] [PubMed]
- Li YG, Hu B, Christie PJ. Biological and Structural Diversity of Type IV Secretion Systems. Microbiol Spectr. 2019 Mar;7(2):10.1128/microbiolspec.PSIB-0012-2018. [CrossRef] [PubMed]
- Ward JE Jr, Dale EM, Christie PJ, Nester EW, Binns AN. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990 Sep;172(9):5187-99. [CrossRef] [PubMed]
- Bitrián M, Roodbarkelari F, Horváth M, Koncz C. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 2011 Mar;65(5):829-42. [CrossRef] [PubMed]
- Wang X, Teng C, Wei H, Liu S, Xuan H, Peng W, Li Q, Hao H, Lyu Q, Lyu S, Fan Y. Development of a set of novel binary expression vectors for plant gene function analysis and genetic transformation. Front Plant Sci. 2023 Jan 12;13:1104905. [CrossRef] [PubMed]
- Christie, PJ. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochimica et Biophysica Acta 1694:219–234. [CrossRef]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuïnk TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000 Nov 3;290(5493):979-82. [CrossRef] [PubMed]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Stüve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):832-7. [CrossRef] [PubMed]
- Koncz C, Kreuzaler F, Kalman Z, Schell J. A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J. 1984 May;3(5):1029-37. [CrossRef] [PubMed]
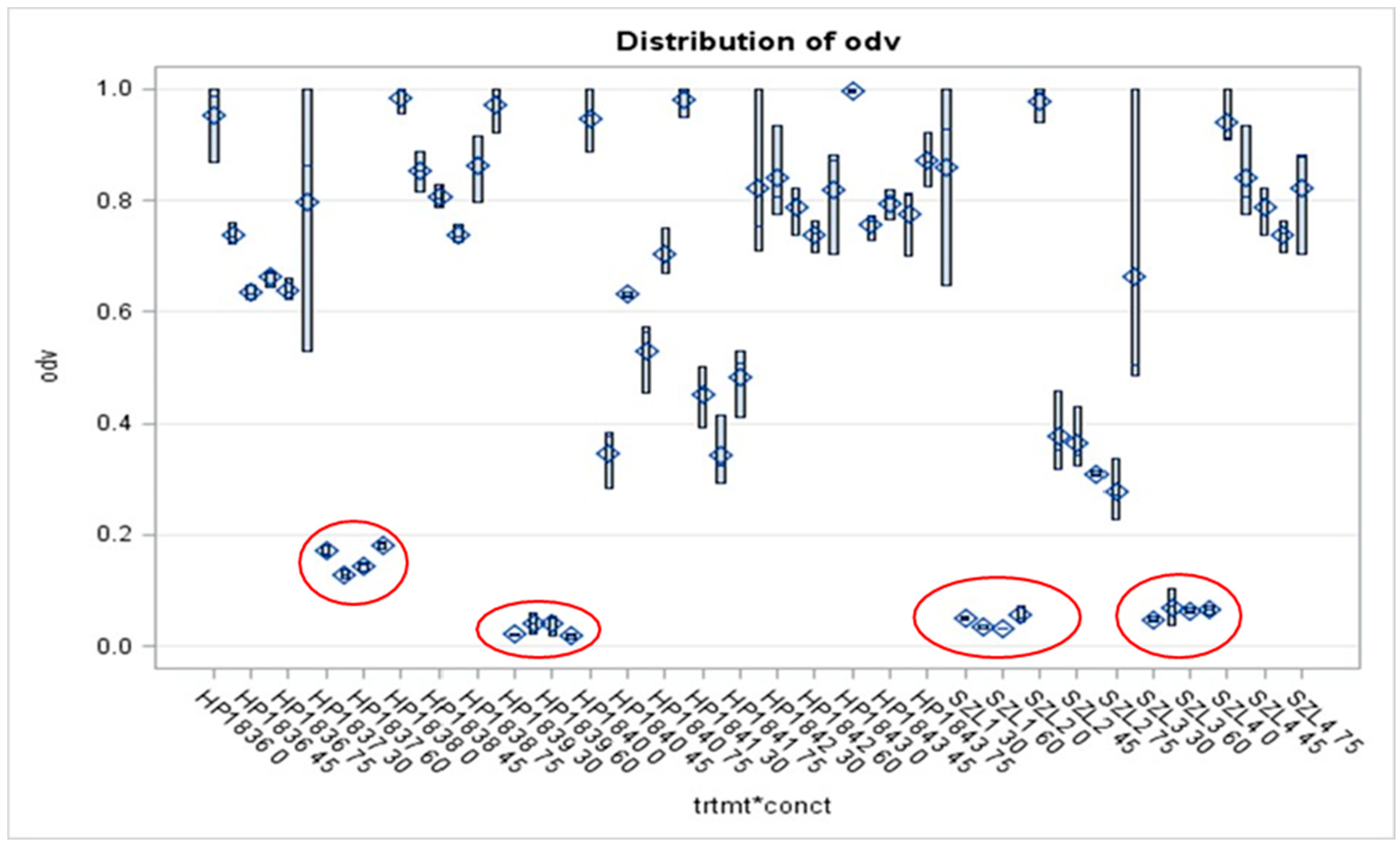
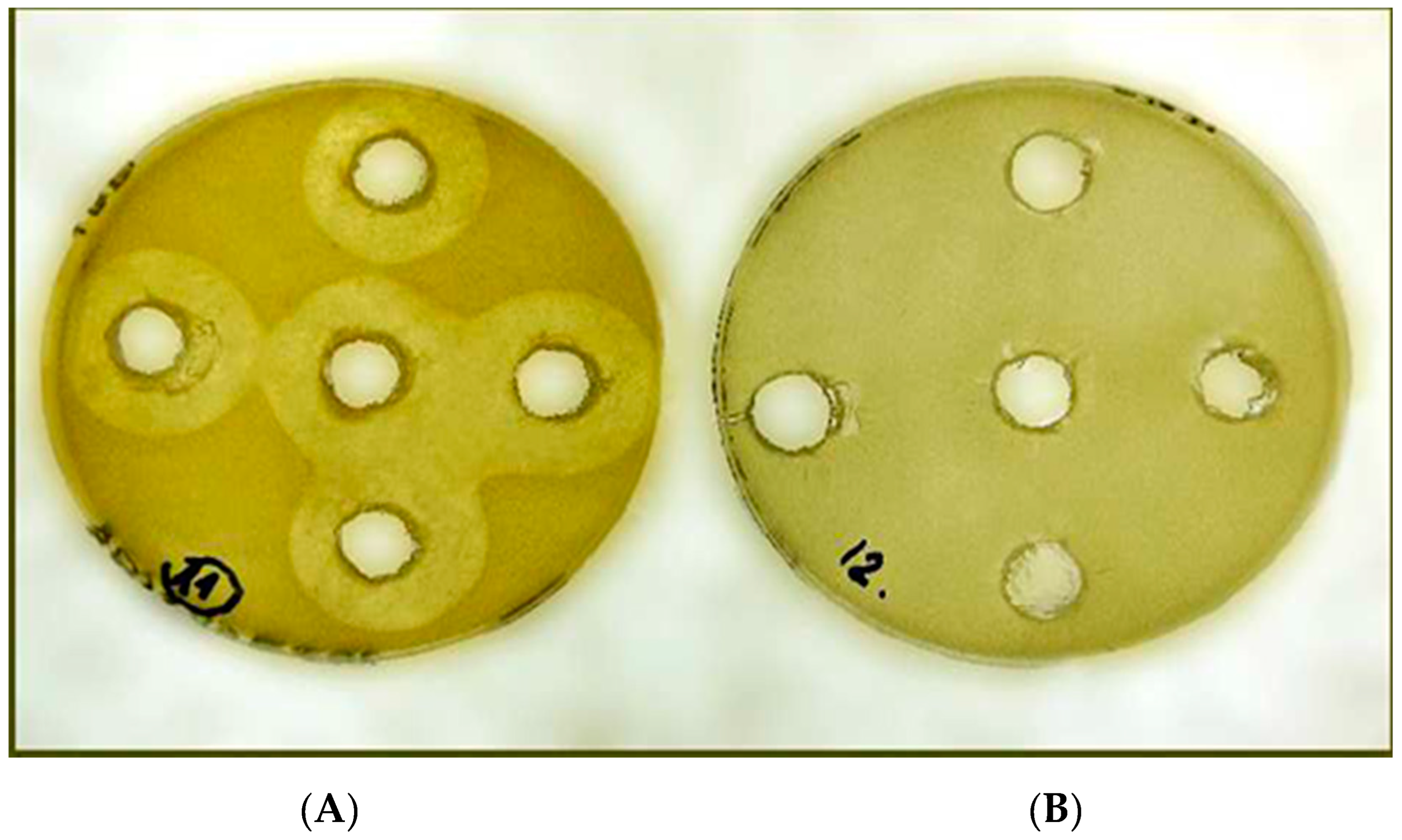
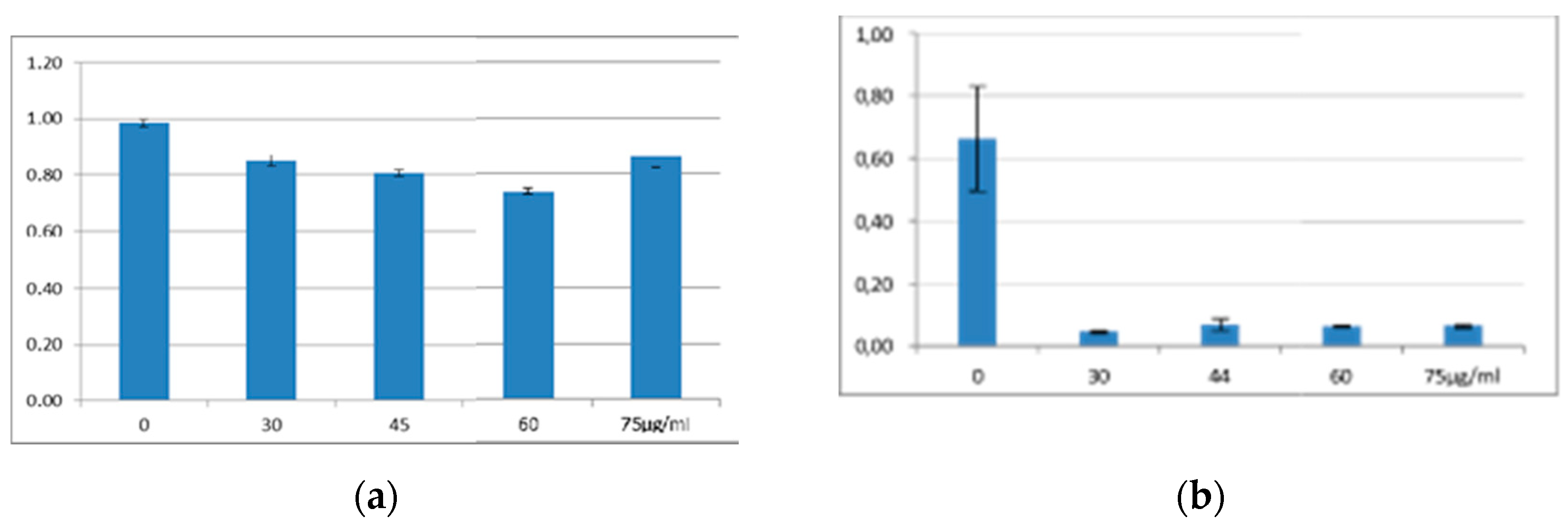
| Name | Genotype | ||||||
|---|---|---|---|---|---|---|---|
|
LAB |
REF |
Genome | Plasmon | T-DNA | Opine | ||
| Chromo some |
Selective marker |
Ti plasmid | BIN | ||||
| HP1836 | C58C*NOP1 | C58C* | NalR | Cured | [T-DNA](-) | NOP | |
| HP1840 | C58C*NOP2 | C58C* | NalR | Cured | [T-DNA](-) | NOP | |
| HP1843 | C58C*NOP3 | C58C* | NalR | Cured | [T-DNA](-) | NOP | |
| HP1841 | C58C1NOP4 | C58C1* | RifR | Cured | [T-DNA](-) | NOP | |
| HP1842 | C58C1NOP5 | C58C1* | RifR | Cured | [T-DNA](-) | NOP | |
| SZL4 | C58C1/ pMP90NOP6 |
C58C1* | RifR | pMP90 GeR | pHP9- Gus101 |
Δ− [T-DNA] | NOP |
| HP1837 | LBA4404/0 OCT1 |
Ach5 | RifR | pAl4404 SmR | Δ− [T-DNA] | OCT | |
| SZL2 | LBA 4404/ pBIN-OCT2 |
Ach5 | RifR | pAl4404 SmR | pBIN | Δ− [T-DNA] | OCT |
| HP1838 | A281 | C58 | RifR | pTiA136 Bo542 |
[T-DNA](+) | AGR | |
| HP1839 | AGL1 | C58C* (AG0) |
CaR RecA(-) |
pEHA101NalR | Δ− [T-DNA] | AGR | |
| SZL1 | EHA 105 | C58C* | RifR | pEHA105NalR | Δ− [T-DNA] | AGR | |
| SZL3 | A4T | RifR | A4T | Δ− [T-DNA] | AGR | ||
| Antimicrobial peptide | . Inactivation zone in mm2 Gram-negative targets | |||
|---|---|---|---|---|
| HGB2226 | HGB1795* | |||
| N | Mean ± SD | N | Mean ± SD | |
| EMA_PF1 | 3 | 3820.00±690.22 | 3 | 4280.00 ±415.69 |
| EMA_PF2 | 4 | 3683.75±799.23 | 2 | 6602.50 456.08 |
| EMA_PF2*20 | 3 | 0 | 3 | 3370.00± 635.83 |
| EMA_PF2*30 | 3 | 0 | 3 | 3119.00 ±842.61 |
| EMA_PF2*40 | 3 | 0 | 3 | 4088.00± 678.70 |
| EMA_PF2*50 | 3 | 0 | 3 | 3821.67 ±214.20 |
| EMA_PF2*70 | 3 | 0 | 3 | 4640.00± 850.97 |
| EMA_PF2*TF | 3 | 0 | 3 | 4172.00±1502.02 |
| EMA_(RPLC)30 | 3 | 1452.50 ±95.45 | 3 | 1761.33± 173.78 |
| AF103_(EMA)_HPLC40 | 3 | 617.00±88.02 | 3 | 1135.33± 119.52 |
| AF103_(EMA)_HPLC43 | 3 | 1614.00± 81.41 | 3 | 2073.33 ±244.32 |
| AF103_(EMA)_HPLC44 | 3 | 1019.33 ±113.52 | 3 | 1385.00± 100.00 |
| Results of agar-diffusion assays - Inactivation zone in mm2 Gram-(+) and fungal targets. | ||||
| Antimicrobial peptide | ||||
| SA | CA | |||
| 3 | 8723.33 ±600.44 | 3 | 11746.67 ±704.37 | |
| EMA_PF1 | 3 | 5931.67± 453.22 | 3 | 6291.6667±627.58134 |
| EMA_PF2 | 3 | 0 | 1 | 530.00 ±0.00 |
| EMA_PF2*20 | 3 | 0 | 3 | 696.33± 279.69 |
| EMA_PF2*30 | 3 | 0 | 3 | 544.67 ±226.68 |
| EMA_PF2*40 | 3 | 0 | 3 | 0.00 |
| EMA_PF2*50 | 3 | 0 | 3 | 623.33 ±175.65 |
| EMA_PF2*70 | 3 | 0 | 558.33± 49.07 | |
| EMA_PF2*TF | 3 | 1656.67± 40.41 | 3 | 1526.00±233.83 |
| EMA_(RPLC)30 | 3 | 1614.00 ±81.41 | 3 | 2289.00±0.000 |
| AF103_(EMA)_HPLC40 | 3 | 1886.33± 66.97 | 3 | 2930.00± 287.51 |
| AF103_(EMA)_HPLC43 | 3 | 1613.67± 81.98 | 3 | 2834.33± 377.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





