Submitted:
01 November 2023
Posted:
02 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Which process steps need to be performed when harmonizing clinical data or claims data in OMOP CDM?
- What sequence of identified process steps should be followed?
2. Materials and Methods
2.1. Literature Review
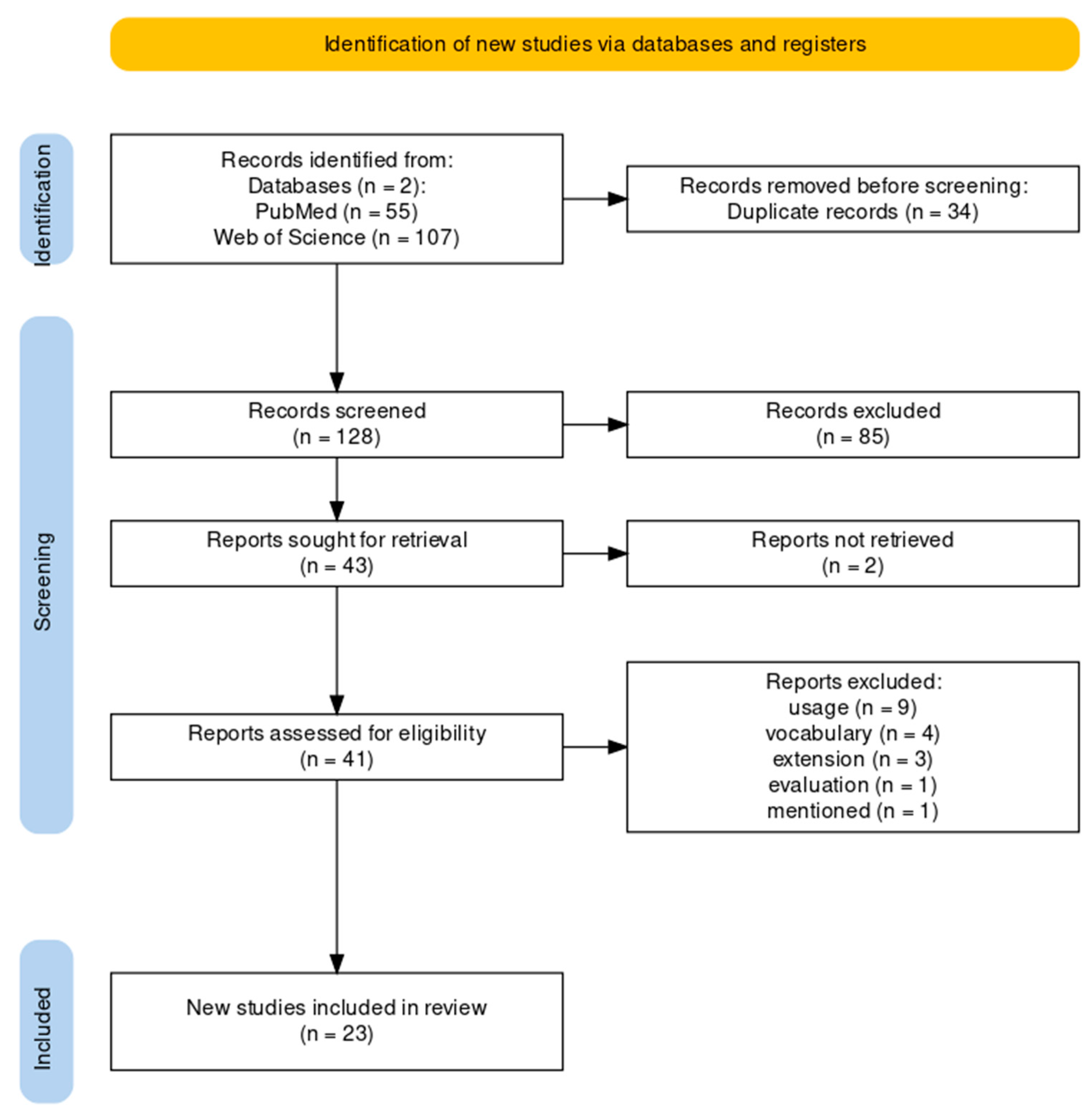
2.1.1. Paper Identification
2.1.2. Paper Exclusion
- Option 1: For a kappa value greater than 0.6 (substantial to almost perfect agreement (interpretation according to Fleiss [12])), the Title-Abstract-Screening for the remaining 80% of the publications found and afterwards, the Full-Text-Screening for the included publications should be divided as follows: 1) reviewer 1 (EH) should screen all publications; 2) reviewer 2 (FB) all included publications and 3) reviewer 3 (MZ) all excluded publications.
- Option 2: If the kappa value is less than or equal to 0.6 (poor to moderate agreement (interpretation according to Fleiss [12])), the remaining 80% of the publications and the full texts had to be screened by all three reviewers.
2.1.3. Data Extraction
2.2. Derivation of a Generic Sequence of Process Steps
3. Results
3.1. Flow Diagram of the Literature Review
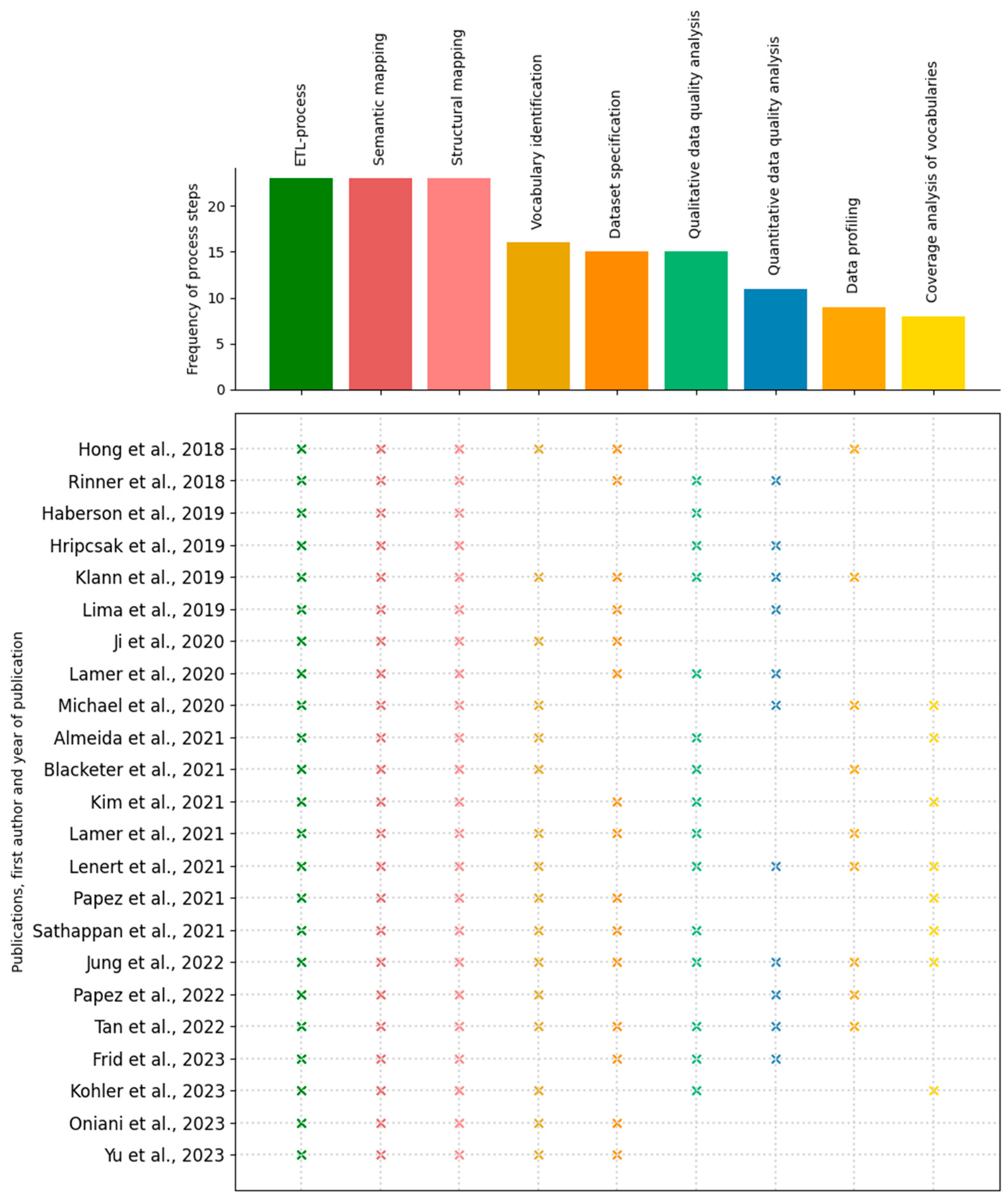
3.2. Process Steps
- Coverage analysis of vocabularies
- Data profiling
- Dataset specification
- Extract-Transform-Load (ETL)-process
- Qualitative data quality analysis
- Quantitative data quality analysis
- Semantic mapping
- Structural mapping
- Vocabulary identification
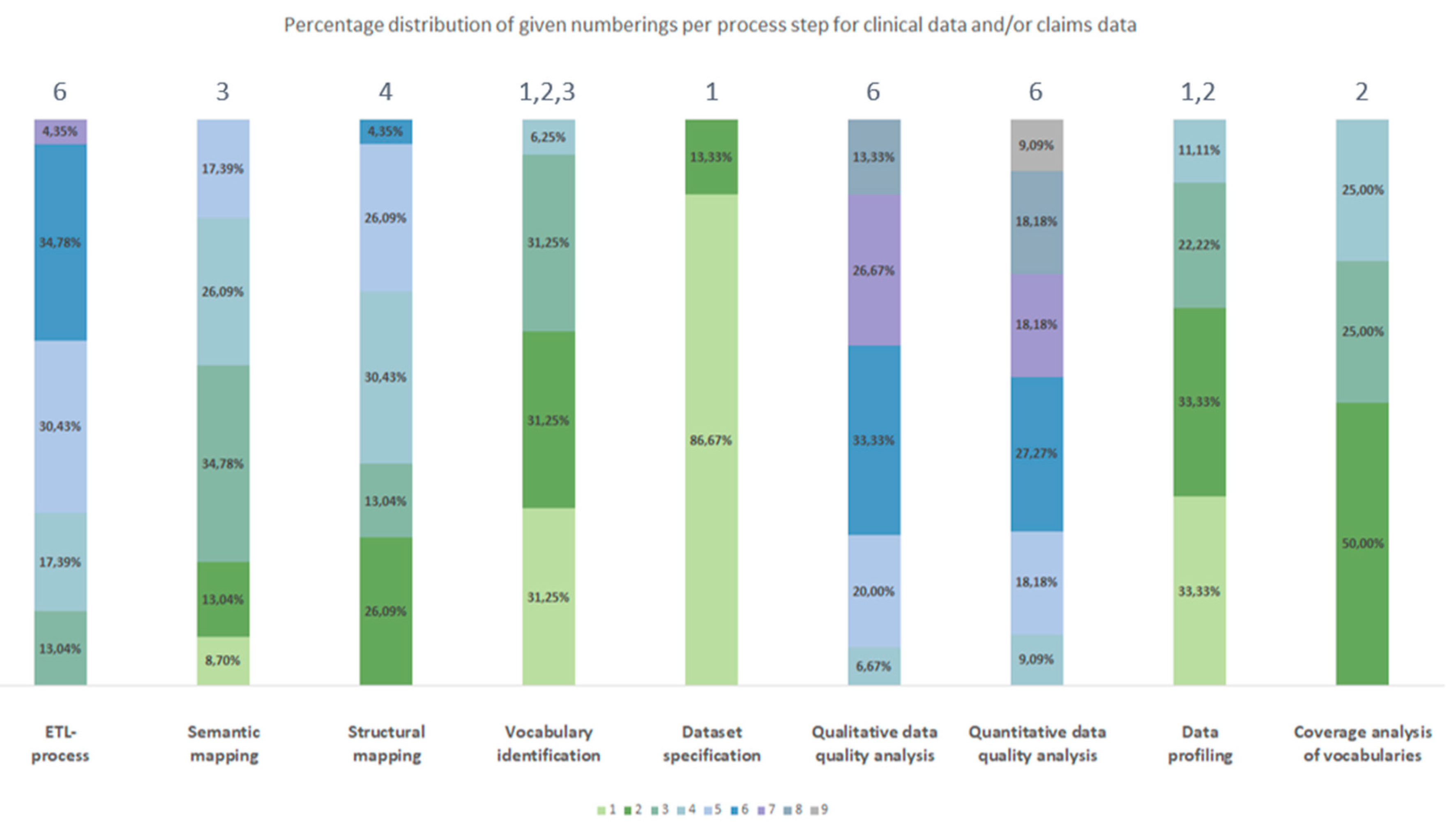
3.2.1. Frequency
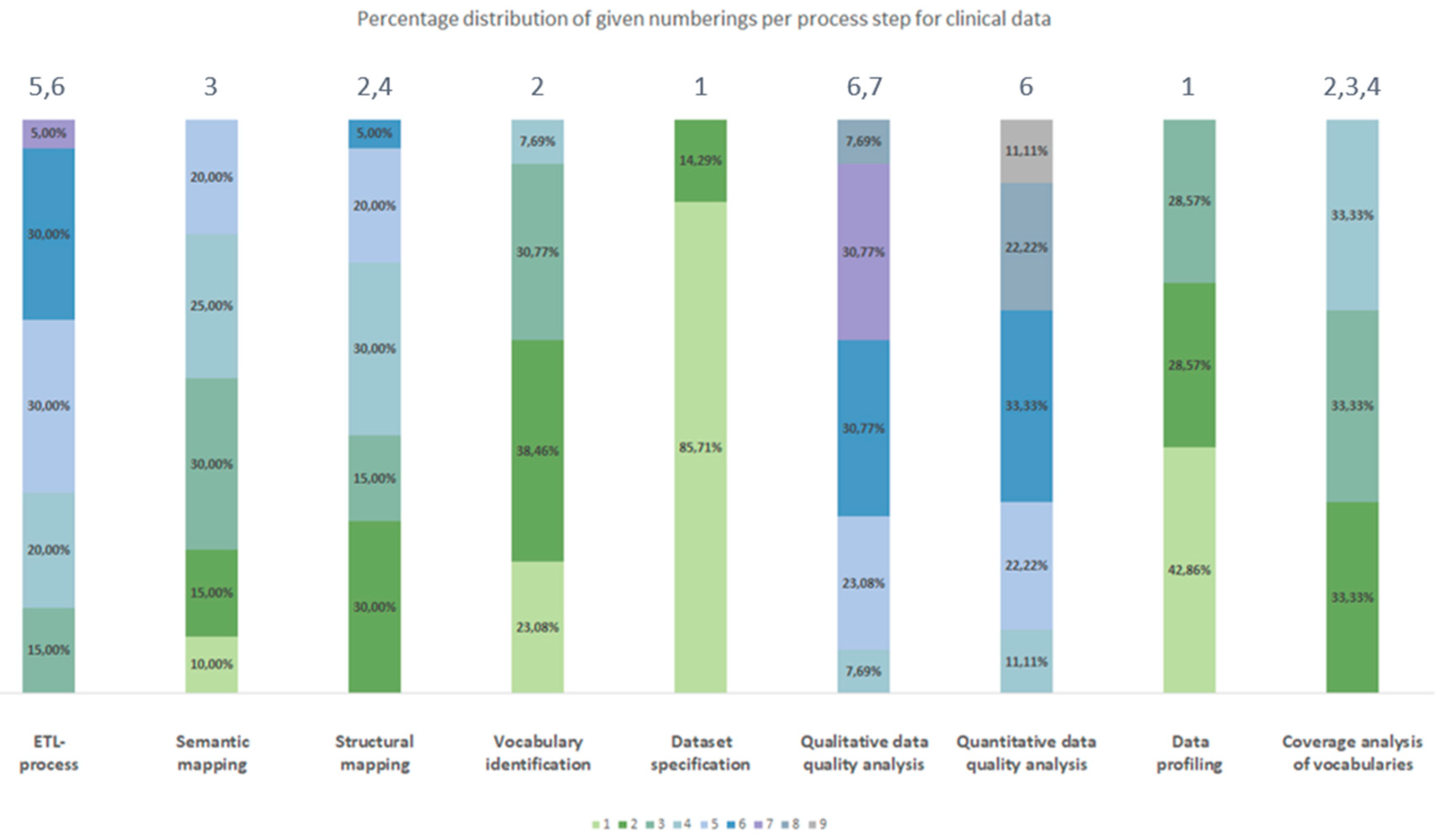
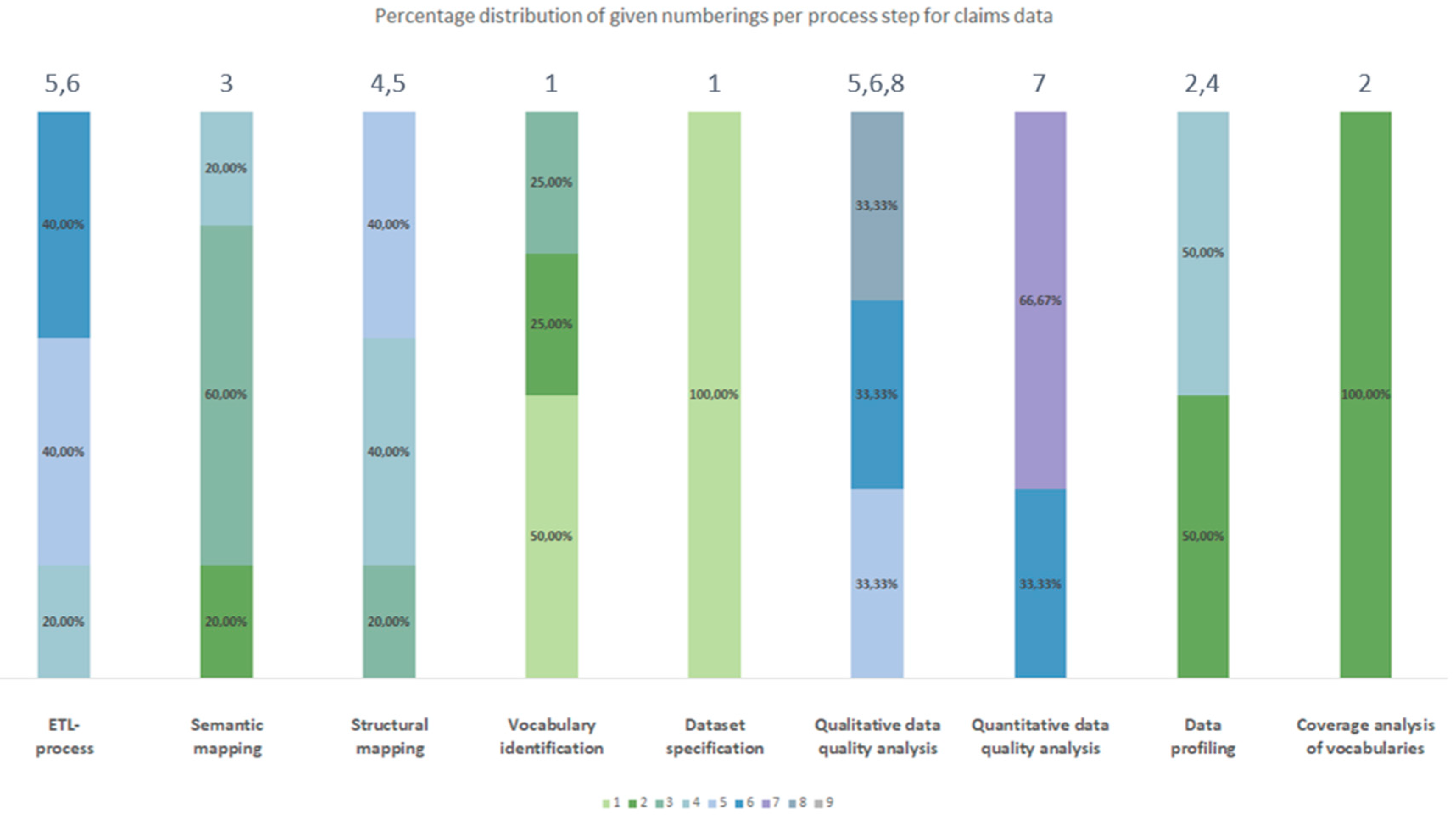
3.2.2. Chronological Order
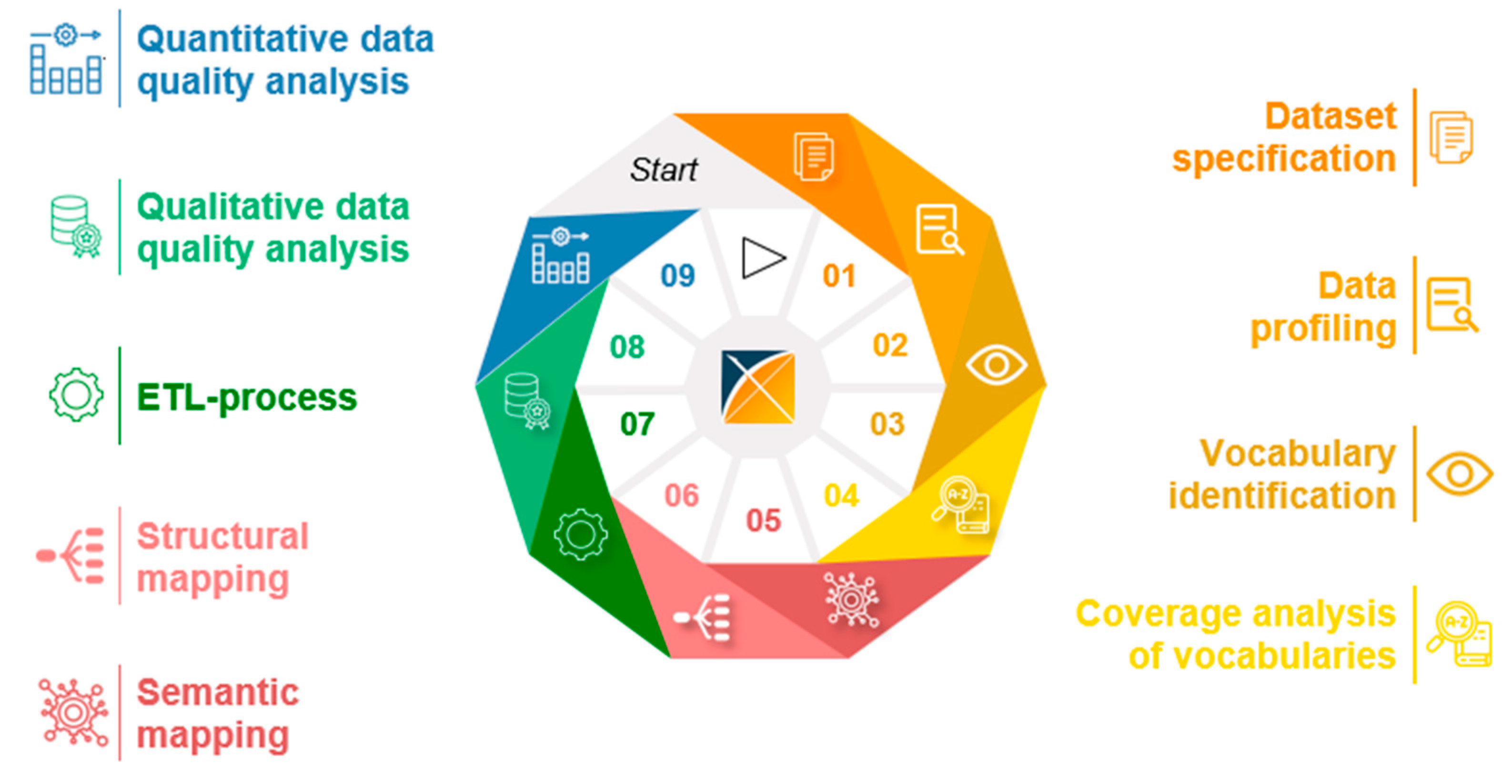
3.3. Generic Data Harmonization Process
- Dataset specification refers to the definition of the scope of the source data for a specific use case. This is usually done by expert teams with clinical expertise. As a result, transformation to OMOP CDM is only performed on source data that is relevant to answering a specific research question.
- To get an overview of the source data including their structure, formats and unique values, a data profiling is performed. For this purpose, OHDSI provides the tool WhiteRabbit [41] to analyzing source data.
- Vocabulary identification focuses on providing a comprehensive compilation of the vocabularies found in the source data, including their scope of application.
- In order to assess the extent to which the vocabularies found in the source data can already be mapped in OMOP CDM, a coverage analysis of the vocabularies of the source data is performed. The analysis helps to identify weaknesses (e.g., missing vocabularies) that would limit a full harmonization of the source data.
- Semantic mapping refers to the mapping of local vocabularies to the standardized vocabulary of OMOP CDM [9] (pp. 55-74). This step is necessary to be able to uniquely identify source values by concepts in OMOP CDM and to transfer source values to standard concepts to enable research in an international context. The standardized vocabulary of OMOP CDM is provided by the OHDSI vocabulary repository Athena [42]. Furthermore, the OHDSI tool Usagi [43] supports researchers in semantic mapping of source values to OMOP CDM concepts.
- The technical transformation of the source data into OMOP CDM is realized through the implementation of ETL-processes. ETL-processes enable the reading of source data (Extract), the practical implementation of semantic and structural mapping (Transform), and the final writing of OMOP-compliant source data to the target database (Load).
- After a successful transformation of source data into OMOP CDM, the focus is on data quality analysis. The qualitative data quality analysis examines, in particular, the plausibility, conformity and completeness of the source data in OMOP CDM (according to Kahn et al. [44]). With the Automated Characterization of Health Information at Large-scale Longitudinal Evidence Systems (Achilles) [45] and the Data Quality Dashboard (DQD) [46], two OHDSI tools exist that perform data quality checks on the transformed source data in OMOP CDM.
- The quantitative data quality analysis checks whether the number of data in the source matches the number of records in OMOP CDM.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Semler, S.C.; Wissing, F.; Heyder, R. German Medical Informatics Initiative - A National Approach to Integrating Health Data from Patient Care and Medical Research. Methods Inf Med. 2018, 57 (Suppl 1), e50–e56. [Google Scholar] [CrossRef] [PubMed]
- Green, L.A.; Fryer, G.E.; Yawn, B.P.; Lanier, D.; Dovey, S.M. The Ecology of Medical Care Revisited. N Engl J Med. 2001, 344, 2021–2025. [Google Scholar] [CrossRef]
- Kumar, G.; Basri, S.; Imam, A.A.; Khowaja, S.A.; Capretz, L.F.; Balogun, A.O. Data Harmonization for Heterogeneous Datasets: A Systematic Literature Review. Applied Sciences. 2021, 11, 8275. [Google Scholar] [CrossRef]
- Garza, M.; Del Fiol, G.; Tenenbaum, J.; Walden, A.; Zozus, M.N. Evaluating common data models for use with a longitudinal community registry. J Biomed Inform. 2016, 64, 333–341. [Google Scholar] [CrossRef]
- Reinecke, I.; Zoch, M.; Reich, C.; Sedlmayr, M.; Bathelt, F. The Usage of OHDSI OMOP - A Scoping Review. Stud Health Technol Inform. 2021, 283, 95–103. [Google Scholar] [CrossRef] [PubMed]
- EHDEN European Health Data & Evidence Network. Published 2022. Accessed May 20, 2022. Available online: https://www.ehden.eu/.
- European Medical Agency. Data Analysis and Real World Interrogation Network (DARWIN EU). Published 2023. Accessed May 20, 2022. Available online: https://www.ema.europa.eu/en/about-us/how-we-work/big-data/data-analysis-real-world-interrogation-network-darwin-eu.
- Observational Health Data Sciences and Informatics. HL7 International and OHDSI Announce Collaboration to Provide Single Common Data Model for Sharing Information in Clinical Care and Observational Research. Published 2021. Accessed May 6, 2022. Available online: https://www.ohdsi.org/ohdsi-hl7-collaboration/.
- Observational Health Data Sciences Informatics The Book of, O.H.D.S.I. In: The Book of OHDSI. ; 2021. Accessed April 19, 2022. Available online: https://ohdsi.github.io/TheBookOfOhdsi/.
- Peng, Y.; Henke, E.; Reinecke, I.; Zoch, M.; Sedlmayr, M.; Bathelt, F. An ETL-process design for data harmonization to participate in international research with German real-world data based on FHIR and OMOP CDM. International Journal of Medical Informatics. 2023, 169, 104925. [Google Scholar] [CrossRef] [PubMed]
- Digital Scholar. Zotero. Zotero. Published 2023. Accessed March 31, 2023. Available online: https://www.zotero.org/.
- Fleiss, J.L. Measuring nominal scale agreement among many raters. Psychological Bulletin. 1971, 76, 378–382. [Google Scholar] [CrossRef]
- Signorell, A.; Aho, K.; Alfons, A. DescTools: Tools for Descriptive Statistics. Published online September 6, 2023. Accessed October 25, 2023. Available online: https://cran.r-project.org/web/packages/DescTools/index.html.
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: An R package and Shiny app for producing PRISMA 2020-compliant flow diagrams, with interactivity for optimised digital transparency and Open Synthesis. Campbell Systematic Reviews. 2022, 18, e1230. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009, 339, b2700. [Google Scholar] [CrossRef]
- Klann, J.; Joss, M.; Embree, K.; Murphy, S. Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLOS ONE. 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Lamer, A.; Abou-Arab, O.; Bourgeois, A.; et al. Transforming Anesthesia Data Into the Observational Medical Outcomes Partnership Common Data Model: Development and Usability Study. Journal of Medical Internet Research. 2021, 23, e29259. [Google Scholar] [CrossRef]
- Hripcsak, G.; Shang, N.; Peissig, P.L.; et al. Facilitating phenotype transfer using a common data model. JOURNAL OF BIOMEDICAL INFORMATICS 2019, 96. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jiang, G.; Brandt, E.; et al. Integrating real-world data to assess cardiac ablation device outcomes in a multicenter study using the OMOP common data model for regulatory decisions: implementation and evaluation. JAMIA OPEN. 2023, 6. [Google Scholar] [CrossRef] [PubMed]
- Papez, V.; Moinat, M.; Payralbe, S.; et al. Transforming and evaluating electronic health record disease phenotyping algorithms using the OMOP common data model: a case study in heart failure. JAMIA OPEN. 2021, 4. [Google Scholar] [CrossRef]
- Tan, H.; Teo, D.; Lee, D.; et al. Applying the OMOP Common Data Model to Facilitate Benefit-Risk Assessments of Medicinal Products Using Real-World Data from Singapore and South Korea. HEALTHCARE INFORMATICS RESEARCH. 2022, 28, 112–122. [Google Scholar] [CrossRef]
- Papez, V.; Moinat, M.; Voss, E.; et al. Transforming and evaluating the UK Biobank to the OMOP Common Data Model for COVID-19 research and beyond. JOURNAL OF THE AMERICAN MEDICAL INFORMATICS ASSOCIATION. 2022, 30, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Yoo, S.; Kim, S.; et al. Patient-Level Fall Risk Prediction Using the Observational Medical Outcomes Partnership’s Common Data Model: Pilot Feasibility Study. JMIR MEDICAL INFORMATICS. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Silva, J.; Matos, S.; Oliveira, J. A two-stage workflow to extract and harmonize drug mentions from clinical notes into observational databases. JOURNAL OF BIOMEDICAL INFORMATICS. 2021, 120. [Google Scholar] [CrossRef]
- Lima, D.; Rodrigues, J.; Traina, A.; Pires, F.; Gutierrez, M. Transforming Two Decades of ePR Data to OMOP CDM for Clinical Research. In: OhnoMachado L, Seroussi B, eds. Universidade de Sao Paulo. 2019, 264, 233–237. [Google Scholar] [CrossRef]
- Ji, H.; Kim, S.; Yi, S.; Hwang, H.; Kim, J.; Yoo, S. Converting clinical document architecture documents to the common data model for incorporating health information exchange data in observational health studies: CDA to CDM. JOURNAL OF BIOMEDICAL INFORMATICS. 2020, 107. [Google Scholar] [CrossRef]
- Kim, J.; Kim, S.; Ryu, B.; Song, W.; Lee, H.; Yoo, S. Transforming electronic health record polysomnographic data into the Observational Medical Outcome Partnership’s Common Data Model: a pilot feasibility study. SCIENTIFIC REPORTS. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Blacketer, C.; Voss, E.; DeFalco, F. Using the Data Quality Dashboard to Improve the EHDEN Network. APPLIED SCIENCES-BASEL, 2021; 11. [Google Scholar] [CrossRef]
- Rinner, C.; Gezgin, D.; Wendl, C.; Gall, W. A Clinical Data Warehouse Based on OMOP and i2b2 for Austrian Health Claims Data. Stud Health Technol Inform. 2018, 248, 94–99. [Google Scholar] [PubMed]
- Haberson, A.; Rinner, C.; Schöberl, A.; Gall, W. Feasibility of Mapping Austrian Health Claims Data to the OMOP Common Data Model. J Med Syst. 2019, 43, 314. [Google Scholar] [CrossRef] [PubMed]
- Sathappan, S.M.K.; Jeon, Y.S.; Dang, T.K.; et al. Transformation of Electronic Health Records and Questionnaire Data to OMOP CDM: A Feasibility Study Using SG_T2DM Dataset. Appl Clin Inform. 2021, 12, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.L.; Sholle, E.T.; Wulff, R.T.; Roboz, G.J.; Campion, T.R. Mapping Local Biospecimen Records to the OMOP Common Data Model. AMIA Jt Summits Transl Sci Proc. 2020, 2020, 422–429. [Google Scholar] [PubMed]
- Hong, N.; Zhang, N.; Wu, H.; et al. Preliminary exploration of survival analysis using the OHDSI common data model: a case study of intrahepatic cholangiocarcinoma. BMC Med Inform Decis Mak. 2018, 18 (Suppl 5), 116. [Google Scholar] [CrossRef] [PubMed]
- Lamer, A.; Depas, N.; Doutreligne, M.; et al. Transforming French Electronic Health Records into the Observational Medical Outcome Partnership’s Common Data Model: A Feasibility Study. Appl Clin Inform. 2020, 11, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Lenert, L.A.; Ilatovskiy, A.V.; Agnew, J.; et al. Automated production of research data marts from a canonical fast healthcare interoperability resource data repository: applications to COVID-19 research. J Am Med Inform Assoc. 2021, 28, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.; Boscá, D.; Kärcher, F.; et al. Eos and OMOCL: Towards a seamless integration of openEHR records into the OMOP Common Data Model. J Biomed Inform. 2023, 144, 104437. [Google Scholar] [CrossRef]
- Oniani, D.; Parmanto, B.; Saptono, A.; et al. ReDWINE: A clinical datamart with text analytical capabilities to facilitate rehabilitation research. Int J Med Inform. 2023, 177, 105144. [Google Scholar] [CrossRef]
- Frid, S.; Pastor Duran, X.; Bracons Cucó, G.; et al. An Ontology-Based Approach for Consolidating Patient Data Standardized With European Norm/International Organization for Standardization 13606 (EN/ISO 13606) Into Joint Observational Medical Outcomes Partnership (OMOP) Repositories: Description of a Methodology. JMIR Med Inform. 2023, 11, e44547. [Google Scholar] [CrossRef] [PubMed]
- Lamer, A.; Abou-Arab, O.; Bourgeois, A.; et al. Transforming Anesthesia Data Into the Observational Medical Outcomes Partnership Common Data Model: Development and Usability Study. JOURNAL OF MEDICAL INTERNET RESEARCH. 2021, 23. [Google Scholar] [CrossRef] [PubMed]
- Hripcsak, G.; Shang, N.; Peissig, P. Facilitating phenotype transfer using a common data model. JOURNAL OF BIOMEDICAL INFORMATICS 2019, 96. [Google Scholar] [CrossRef] [PubMed]
- Observational Health Data Sciences and Informatics. White Rabbit. Published 2022. Accessed November 11, 2022. Available online: http://ohdsi.github.io/WhiteRabbit/WhiteRabbit.html.
- Observational Health Data Sciences and Informatics. Athena. Athena – OHDSI Vocabularies Repository. Published 2023. Accessed November 25, 2022. Available online: https://athena.ohdsi.org/.
- Schuemie, M. Usagi. Usagi. Published 2021. Available online: http://ohdsi.github.io/Usagi/.
- Kahn, M.G.; Callahan, T.J.; Barnard, J.; et al. A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data. EGEMS (Wash DC). 2016, 4, 1244. [Google Scholar] [CrossRef]
- Observational Health Data Sciences and Informatics. Achilles. Published online August 2, 2022. Accessed April 12, 2023. Available online: https://github.com/OHDSI/Achilles.
- Blacketer, C.; Defalco, F.J.; Ryan, P.B.; Rijnbeek, P.R. Increasing trust in real-world evidence through evaluation of observational data quality. Journal of the American Medical Informatics Association. 2021, 28, 2251–2257. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.; Stöhr, M.R.; Gall, H.; Michel-Backofen, A.; Majeed, R.W. Data Integration into OMOP CDM for Heterogeneous Clinical Data Collections via HL7 FHIR Bundles and XSLT. Digital Personalized Health and Medicine. Published online 2020:138-142. [CrossRef]
- Biedermann, P.; Ong, R.; Davydov, A.; et al. Standardizing registry data to the OMOP Common Data Model: experience from three pulmonary hypertension databases. BMC Medical Research Methodology. 2021, 21, 238. [Google Scholar] [CrossRef]
| Database | Search String |
|---|---|
| PubMed | ((OMOP[Title/Abstract]) OR (OHDSI[Title/Abstract])) AND ((claims data[Title/Abstract]) OR (clinical data[Title/Abstract])) |
| Web of Science | ((TI=(OMOP) OR TI=(OHDSI)) AND (TI=(claims data) OR TI=(clinical data))) OR ((AB=(OMOP) OR AB=(OHDSI)) AND (AB=(claims data) OR AB=(clinical data))) |
| Criterion | Description of criterion |
|---|---|
| no “OMOP” or “OHDSI” | Publication does not mention “OMOP” or “OHDSI” |
| Publication uses “OMOP” or “OHDSI” with other meanings | |
| mentioned | Publication only mentions “OMOP” or “OHDSI” |
| evaluated | Publication focuses on the evaluation of OMOP |
| vocabulary | Publication focuses on vocabularies and their mapping in OMOP or use of OMOP vocabularies |
| extension | Publication focuses on an extension of OMOP or OHDSI tools |
| usage | Publication focuses on the use of OMOP, e.g., for studies, data quality analyses, development of tools or frameworks (e.g., patient level prediction) |
| no full text | Publication is not available as full text |
| foreign language | Publication is written in other languages than English |
| wrong type of source data | Publication focuses on types of source data other than clinical data or claims data |
| Process number Type of source data |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Clinical data | Dataset specification | Data profiling | Vocabulary identification | Coverage analysis of vocabularies | Structural mapping | Semantic mapping | ETL-process | Quantitative data quality analyses | Qualitative data quality analyses |
| Claims data | Dataset specification | Vocabulary identification | Coverage analysis of vocabularies | Data profiling | Semantic mapping | Structural mapping | ETL-process | Qualitative data quality analyses | Quantitative data quality analyses |
| Clinical data and/or claims data | Dataset specification | Data profiling | Vocabulary identification | Coverage analysis of vocabularies | Semantic mapping | Structural mapping | ETL-process | Qualitative data quality analyses | Quantitative data quality analyses |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




