Submitted:
23 October 2023
Posted:
24 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Screening of nutrients that facilitate the metabolism of strain NCD-2
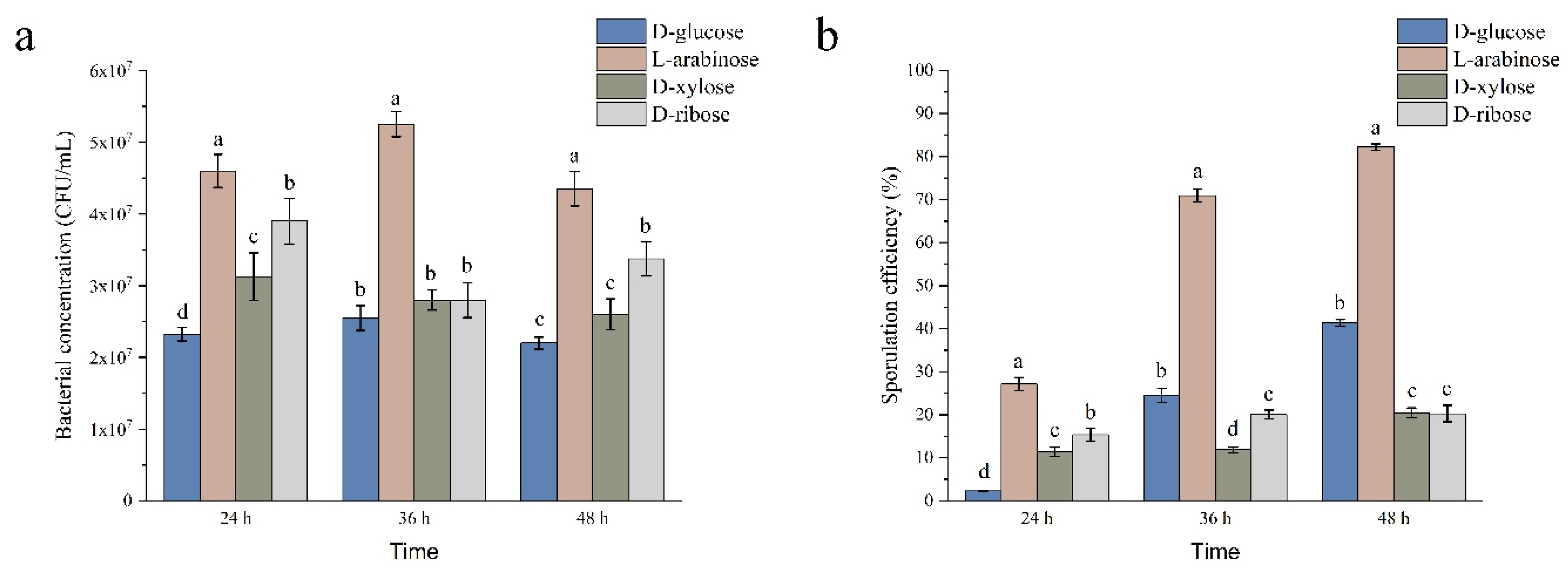
2.2. Effects of L-arabinose, D-ribose, and D-xylose on growth and sporulation efficiency of strain NCD-2
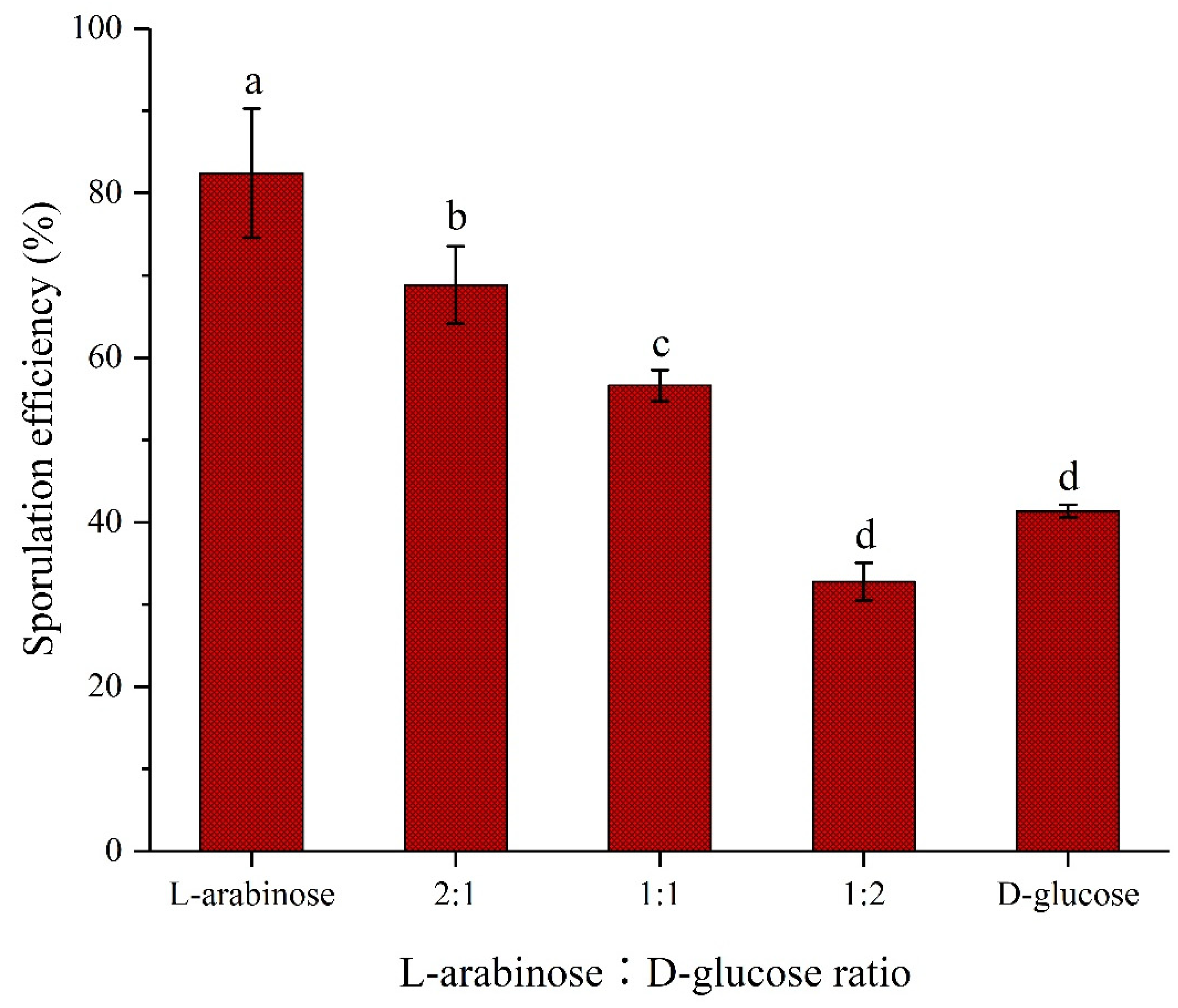
2.3. Different proportions of L-arabinose and D-glucose on sporulation of strain NCD-2
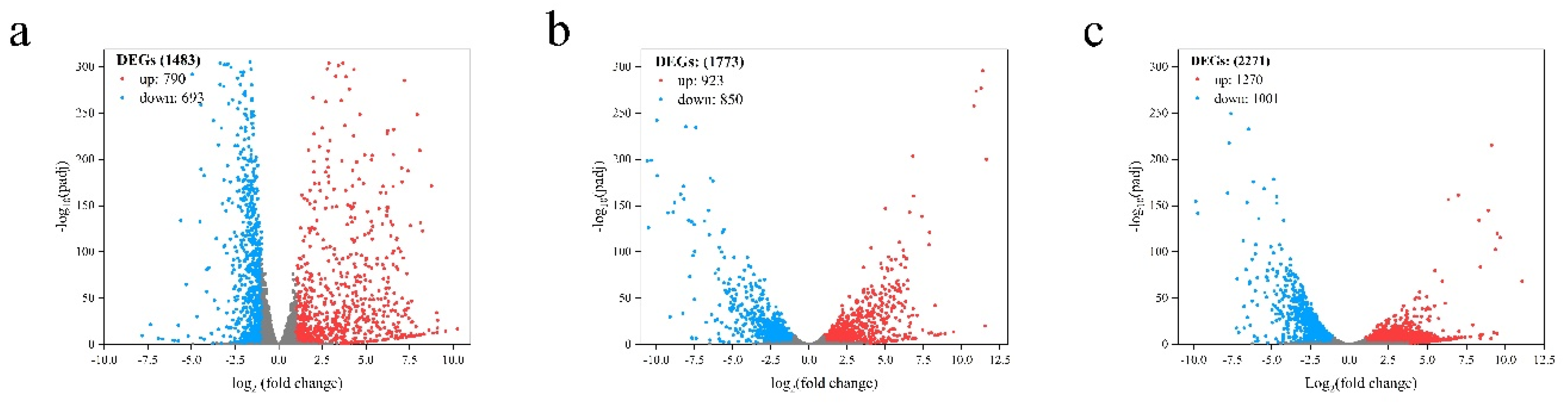
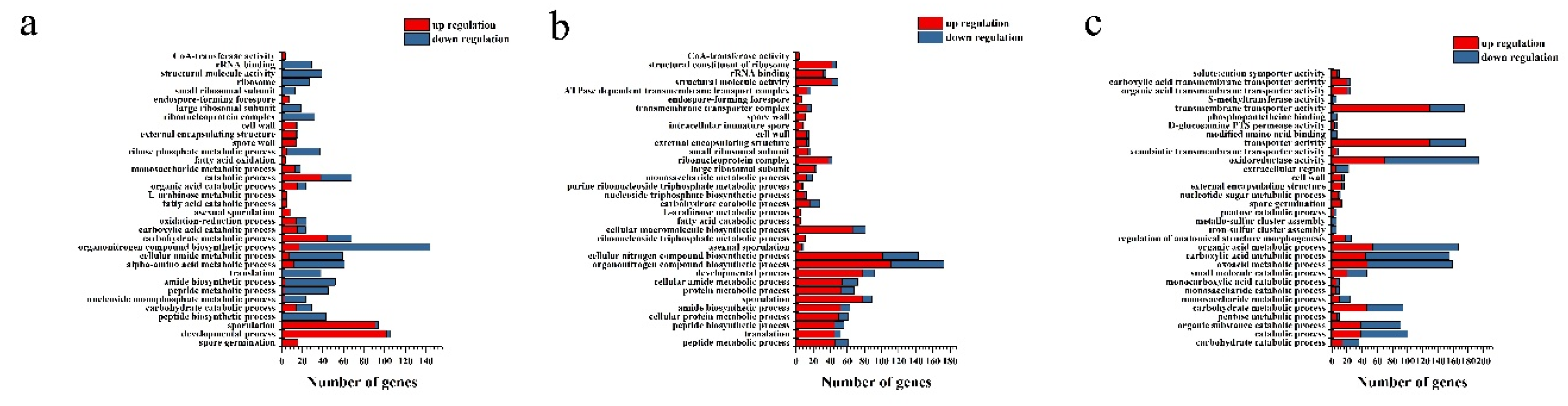
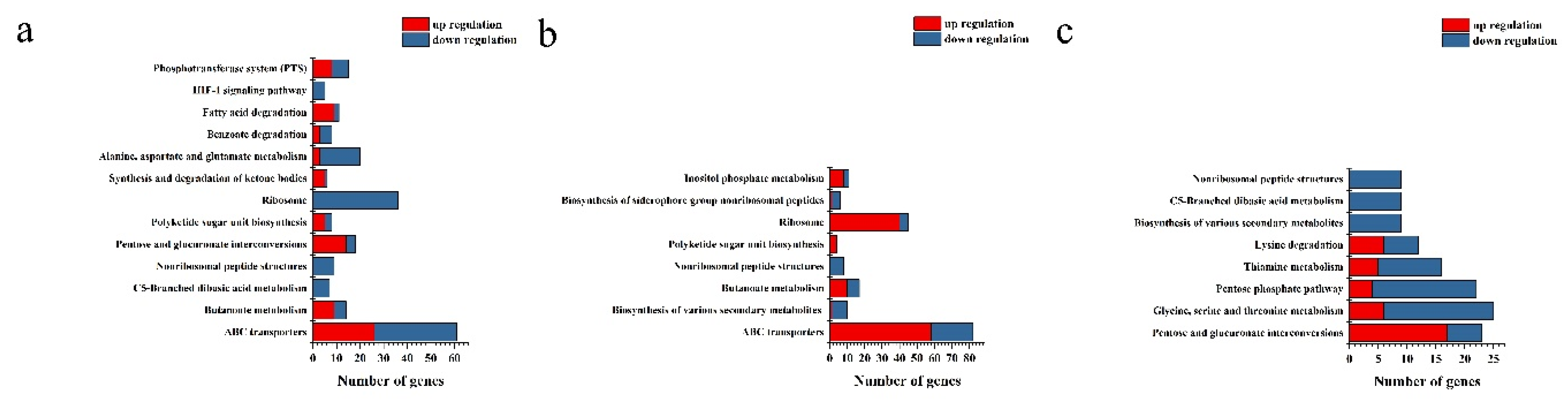
2.4. Transcriptome analysis
2.5. Confirmation of transcriptional results by qRT-PCR
2.6. Analysis of genes associated with sporulation in strain NCD-2
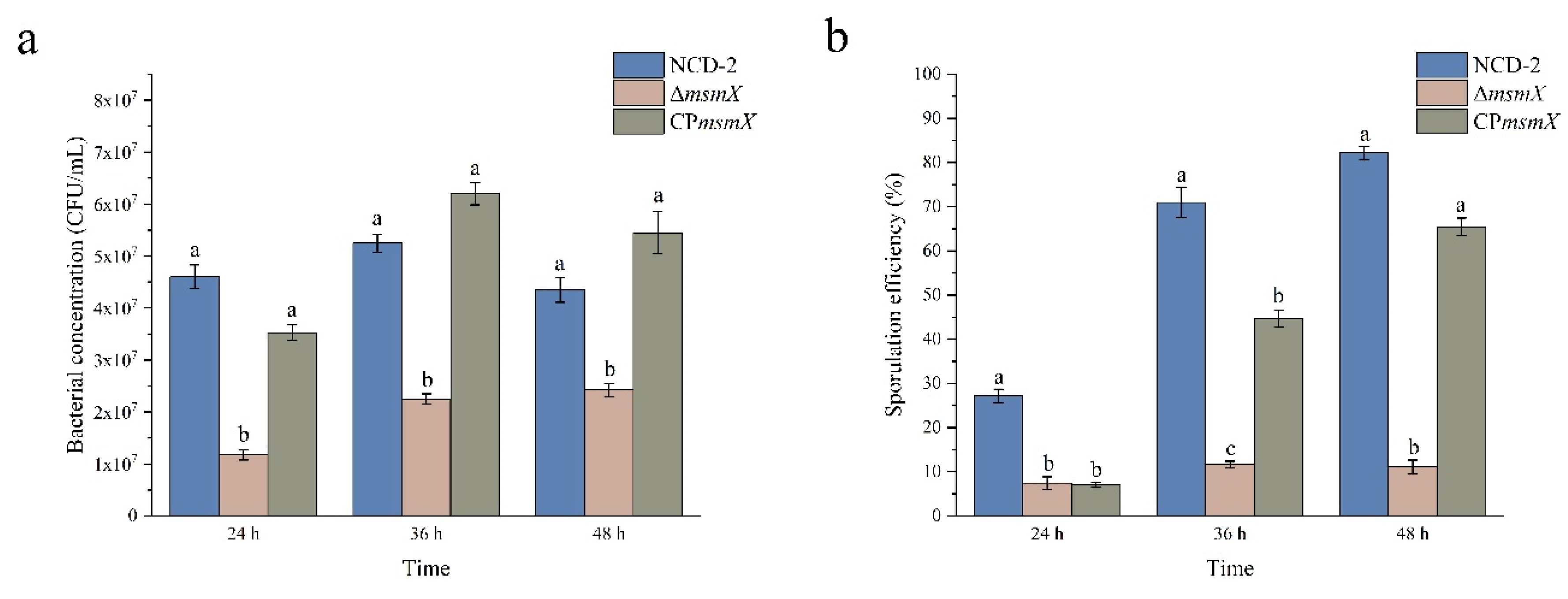
2.7. Deletion of the msmX gene decreased the sporulation efficiency in strain NCD-2
3. Discussion
4. Materials and Methods
4.1. Bacterial strains and growth conditions
4.2. Phenotype microarrays analysis
4.3. Determination of cell concentration and sporulation efficiency
4.4. RNA extraction and RNA sequencing
4.5. Transcriptome data and differential gene expression analysis
4.6. Confirmation of transcriptome analysis results
4.7. Function analysis of msmX gene
4.8. Statistical analyses
5. Conclusion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Fiers, M.; Edel-Hermann, V.; Chatot, C.; Hingrat, Y.L.; Steinberg, A.C. Potato soil-borne diseases. A review. Agro. Sustain. Dev. 2012, 32, 93-132. [CrossRef]
- Bisutti, I.L.; Pelz, J.; Büttner, C.; Stephan, D. Field assessment on the influence of RhizoVital® 42 fl. and Trichostar® on strawberries in the presence of soil-borne diseases. Crop Prot. 2017, 96, 195-203. [CrossRef]
- Pérez-García, A.; Romero, D.; de Vicente, A. Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 2011, 22 (2), 187-193. [CrossRef]
- Rao, Y.K.; Tsay, K.J.; Wu, W.S.; Tzeng, Y.M. Medium optimization of carbon and nitrogen sources for the production of spores from Bacillus amyloliquefaciens B128 using response surface methodology. Process Biochem. 2007, 42 (4), 535-541. [CrossRef]
- Posada-Uribe, L.F.; Romero-Tabarez, M.; Villegas-Escobar, V. Effect of medium components and culture conditions in Bacillus subtilis EA-CB0575 spore production. Bioproc. Biosyst. Eng. 2015, 38 (10), 1879-1888. [CrossRef]
- de Vries, Y.; Atmadja, R.; Hornstra, L.; de Vos, W.; Abee, T. Influence of glutamate on growth, sporulation, and spore properties of Bacillus cereus ATCC 14579 in defined medium. App. Environ. Microb. 2005, 71 (6), 3248-3254. [CrossRef]
- Donohue, T.; Bernlohr, R. Effect of cultural conditions on the concentrations of metabolic intermediates during growth and sporulation of Bacillus licheniformis. J. Bacteriol. 1978, 135 (2), 363-372. [CrossRef]
- Singh, R.M. Role of carbon and nitrogen sources in bacterial growth and sporulation. Appl. Microbiol. 1971, 22 (1), 131-132. [CrossRef]
- Lopez, J.M.; Marks, C.L.; Freese, E. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 1979, 587 (2), 238-252. [CrossRef]
- Monteiro, S.M.S.; Clemente, J.J.; Carrondo, M.J.T.; Cunha, A.E. Enhanced Spore Production of Bacillus subtilis Grown in a Chemically Defined Medium. Adv. Microbiol. 2014, 4 (8), 444-454. [CrossRef]
- Warriner, K.; Waites, W.M. Enhanced sporulation in Bacillus subtilis grown on medium containing glucose:ribose. Lett. Appl. Microbiol. 1999, 29 (2), 97-102. [CrossRef]
- Nguyen Thi Minh, H.; Durand, A.; Loison, P.; Perrier-Cornet, J.; Gervais, P. Effect of sporulation conditions on the resistance of Bacillus subtilis spores to heat and high pressure. Appl. Microbiol. Biotechnol. 2011, 90 (4), 1409-1417. [CrossRef]
- Bressuire-Isoard, C.; Broussolle, V.; Carlin, F. Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol. Rev. 2018, 42 (5), 614-626. [CrossRef]
- Widderich, N.; Rodrigues, C.D.; Commichau, F.M.; Fischer, K.E.; Ramirez-Guadiana, F.H.; Rudner, D.Z.; Bremer, E. Salt-sensitivity of σ(H) and Spo0A prevents sporulation of Bacillus subtilis at high osmolarity avoiding death during cellular differentiation. Mol. Microbiol. 2016, 100 (1), 108-124. [CrossRef]
- Li, S.Z.; Lu, X.Y.; Ma, P.; Gao, S.G.; Liu, G. Evaluation of biocontrol potential of a bacterial strain NCD-2 against cotton verticillium wilt in field trials. Acta Phytopathol. Sin. 2005, 35, 451-455.
- Guo, Q.; Dong, W.; Li, S.; Lu, X.; Wang, P.; Zhang, X.; Wang, Y.; Ma, P. Fengycin produced by Bacillus subtilis NCD-2 plays a major role in biocontrol of cotton seedling damping-off disease. Microbio. Res. 2014, 169, 533-540. [CrossRef]
- Zhao, W.S.; Ban, Y.Y..; Su, Z.H.; Li, S.Z.; Liu, X.Y; Guo, Q.G.; Ma, P. Colonization Ability of Bacillus subtilis NCD-2 in Different Crops and Its Effect on Rhizosphere Microorganisms. Microorganisms 2023, 11 (3), 776. [CrossRef]
- Camp, A.H.; Losick, R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 2008, 69 (2), 402-417. [CrossRef]
- Dong, L.H.; Wang, P.; Zhao, W.S.; Su, Z.H.; Zhang, X.Y.; Lu, X.Y.; Guo, Q.G. Surfactin and fengycin contribute differentially to the biological activity of Bacillus subtilis NCD-2 against cotton verticillium wilt. Biol. Control 2022, 174, 104999. [CrossRef]
- Schaeffer, P.; Millet, J.; Aubert, J.P. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 1965, 54 (3), 704-711. [CrossRef]
- Verma, N.; Singh, N.A.; Kumar, N.; Raghu, H.V. Screening of different media for sporulation of Bacillus megaterium. Int. J. Microbiol. Res. Rev 2013, 1, 68-73.
- Monteiro, S.M.; Clemente, J.J.; Henriques, A.O.; Gomes, R.J.; Carrondo, M.J.; Cunha, A.E. A procedure for high-yield spore production by Bacillus subtilis. Biotechnol. Prog. 2005, 21 (4), 1026-1031. [CrossRef]
- Bochner, B.R; Gadzinski, P.; Panomitros, E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001, 11 (7), 1246-1255. [CrossRef]
- Bochner, B.R. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 2003, 4, 309-314. [CrossRef]
- Mackie, A.; Hassan, K.; Paulsen, I.; Tetu, S. Biolog Phenotype Microarrays for phenotypic characterization of microbial cells. Methods Mol. Biol. 2014, 1096, 123-130. [CrossRef]
- Sá-Nogueira, I.; Nogueira, T.V.; Soares, S.; de Lencastre, H. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiol. (Reading) 1997, 143 Pt 3, 957-969. [CrossRef]
- Lepesant, J.A.; Dedonder, R. Metabolism of L-arabinose in Bacillus subtilis Marburg Ind-168. C. R. Acad. Sci. Ser. D. 1967, 264 (23), 2683-2686.
- Sá-Nogueira, I.; de Lencastre, H. Cloning and characterization of araA, araB, and araD, the structural genes for L-arabinose utilization in Bacillus subtilis. J. Bacteriol. 1989, 171 (7), 4088-4091. [CrossRef]
- Sá-Nogueira, I.; Mota, L.J. Negative regulation of L-arabinose metabolism in Bacillus subtilis: characterization of the araR (araC) gene. J. Bacteriol. 1997, 179 (5), 1598-1608. [CrossRef]
- Sá-Nogueira, I.; Ramos, S.S., Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in L-arabinose utilization. J. Bacteriol. 1997, 179 (24), 7705-7711. [CrossRef]
- Ferreira, M.; Sá-Nogueira, I. A multitask ATPase serving different ABC-type sugar importers in Bacillus subtilis. J. Bacteriol. 2010, 192 (20), 5312-5318. [CrossRef]
- Iyer, J.L.; Shetty, P.; Pai, J. Evaluation of whole cells of Bacillus subtilis as substrate for measurement of autolysin activity. Process Biochem. 2005, 40 (5), 1593-1597. [CrossRef]
- Krogh, S.; Jørgensen, S.T.; Devine, K.M. Lysis genes of the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 1998, 180 (8), 2110-2117. [CrossRef]
- Nandy, S.K.; Venkatesh, K.V. Effect of Carbon and Nitrogen on the cannibalistic behavior of Bacillus subtilis. Appl. Biochem. Biotechnol. 2008, 151, 424-432. [CrossRef]
- Sahoo, S.; Rao, K.K.; Suraishkumar, G.K. Reactive oxygen species induced by shear stress mediate cell death in Bacillus subtilis. Biotechnol.Bioeng. 2006, 94 (1), 118-127. [CrossRef]
- Jolliffe, L.K.; Langemeier, S.O.; Doyle, R.J. Hydrogen ion control of autolysin-dependent functions in Bacillus subtilis. Microbios 1983, 38 (153-154), 187-194.
- González-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by sporulating bacteria. Science (New York, N.Y.) 2003, 301 (5632), 510-513. [CrossRef]
- González-Pastor, J.E. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35 (3), 415-424. [CrossRef]
- Eswaramoorthy, P.; Guo, T.; Fujita, M. In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. J. Bacteriol. 2009, 191 (17), 5358-5368. [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001, 25 (4), 402-408. [CrossRef]
- Liu, P.; Xia, L.Q.; Hu, S.B.; Yan, L.; Ding, X.Z; Zhang, Y.M; Yu, Z. Site-specific integration of heterologous gene into Bacillus thuringiensis chromosome and its expression. Acta Microbiol. Sin. 2008, 48 (5), 661-666.
- Arnaud, M.; Chastanet, A.; Débarbouillé, M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70 (11), 6887-6891. [CrossRef]
- Guo, Q.; Li, S.; Lu, X.Y.; Li, B.Q.; Ma, P. PhoR/PhoP two component regulatory system affects biocontrol capability of Bacillus subtilis NCD-2. Genet. Mol. Biol. 2010, 33 (2), 333-340. [CrossRef]
| Accession ID | Gene nane | Log2(Ara/Glc) | Production | ||
|---|---|---|---|---|---|
| 8 h | 12 h | 16 h | |||
| WP_003231833.1 | cotE | 7.65 | 5.81 | 2.56 | outer spore coat protein CotE |
| WP_003243364.1 | cotF | 6.59 | 9.47 | 5.54 | spore coat protein CotF |
| WP_080344234.1 | cotG | 3.55 | 5.59 | 3.82 | spore coat protein CotG |
| ADV92699.1 | cotM | 5.01 | 1.53 | 4.21 | spore coat protein (outer) |
| WP_047183078.1 | cotS | 3.98 | 6.01 | 4.53 | spore coat protein CotS |
| PSM02245.1 | cotT | 5.88 | 5.95 | 8.39 | spore coat protein |
| AGE63031.1 | cotV | 4.37 | 6.32 | 5.46 | spore coat protein (insoluble fraction) |
| WP_069486390.1 | cotW | 4.53 | 6.81 | 5.73 | spore coat protein |
| WP_014476454.1 | cotX | 4.28 | 6.83 | 6.13 | spore coat protein |
| WP_003231888.1 | dpaA | 6.29 | 6.84 | 4.81 | dipicolinic acid synthetase subunit A |
| WP_003231884.1 | dpaB | 5.76 | 6.12 | 4.21 | dipicolinate synthase subunit B |
| WP_015383228.1 | yheD | 5.17 | 3.35 | 2.27 | spore coat associated protein YheD |
| WP_063336053.1 | gerBA | 4.55 | 2.20 | 4.73 | spore germination protein GerKA |
| WP_003184172.1 | gerE | 3.94 | 5.01 | 3.91 | spore germination protein GerE |
| WP_014478336.1 | gerQ | 7.95 | 4.77 | 1.32 | spore coat protein GerQ |
| WP_047182746.1 | gerT | 5.10 | 6.02 | 6.03 | spore germination protein GerT |
| AKE24397.1 | sigK | 6.45 | 4.08 | 3.26 | RNA polymerase sporulation-specific sigma factor |
| WP_047182864.1 | spoIIIAE | 3.84 | 2.72 | 1.41 | stage III sporulation protein AE |
| WP_003221804.1 | spoIIID | 10.25 | 6.81 | 4.40 | sporulation transcriptional regulator SpoIIID |
| WP_004398593.1 | spoIIM | 1.78 | 1.84 | 2.35 | stage II sporulation protein M |
| WP_047183325.1 | spoIIQ | 5.69 | 1.78 | -1.82 | stage II sporulation protein SpoIIQ |
| WP_004398697.1 | spoIVB | 6.19 | 1.77 | 2.12 | SpoIVB peptidase |
| WP_015483522.1 | spoIVFB | 1.25 | 1.39 | 1.84 | stage IV sporulation protein SpoIVFB |
| WP_003230465.1 | spoVAD | 6.29 | 1.70 | 4.08 | stage V sporulation protein AD |
| AGE63365.1 | spoVD | 2.67 | 3.20 | 2.32 | penicillin-binding protein |
| WP_047182441.1 | yjcA | 6.69 | 5.32 | 5.67 | sporulation protein YjcA |
| WP_015383520.1 | ykvU | 3.60 | 3.01 | 4.18 | sporulation protein YkvU |
| WP_003223491.1 | sspA | 7.22 | 3.14 | 3.04 | alpha/beta-type small acid-soluble spore protein |
| WP_003233287.1 | sspB | 7.39 | 3.43 | 2.66 | alpha/beta-type small acid-soluble spore protein |
| WP_003218568.1 | sspD | 7.57 | 3.93 | 4.00 | alpha/beta-type small acid-soluble spore protein |
| BAI84385.2 | sspE | 6.59 | 4.34 | 3.30 | gamma-type small acid-soluble spore protein |
| WP_003244950.1 | sdpC | -1.40 | -5.57 | -6.81 | sporulation delaying protein family toxin |
| WP_003228357.1 | sdpI | -4.53 | -3.43 | -4.90 | immunity protein SdpI |
| WP_003243541.1 | sdpR | -4.51 | -3.34 | -2.64 | sporulation delaying system autorepressor SdpR |
| Strain | Genotype | Source |
|---|---|---|
| WT | Bacillus subtilis NCD-2 wild type | Lab stock |
| ΔmsmX | NCD-2 mutant, msmX deletion mutant | This study |
| CPmsmX | Complementary of ΔmsmX by intact msmX, CmR | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





