Submitted:
19 October 2023
Posted:
19 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Sample collection
2.2. Library preparation and sequencing
2.3. Sequence data and analysis
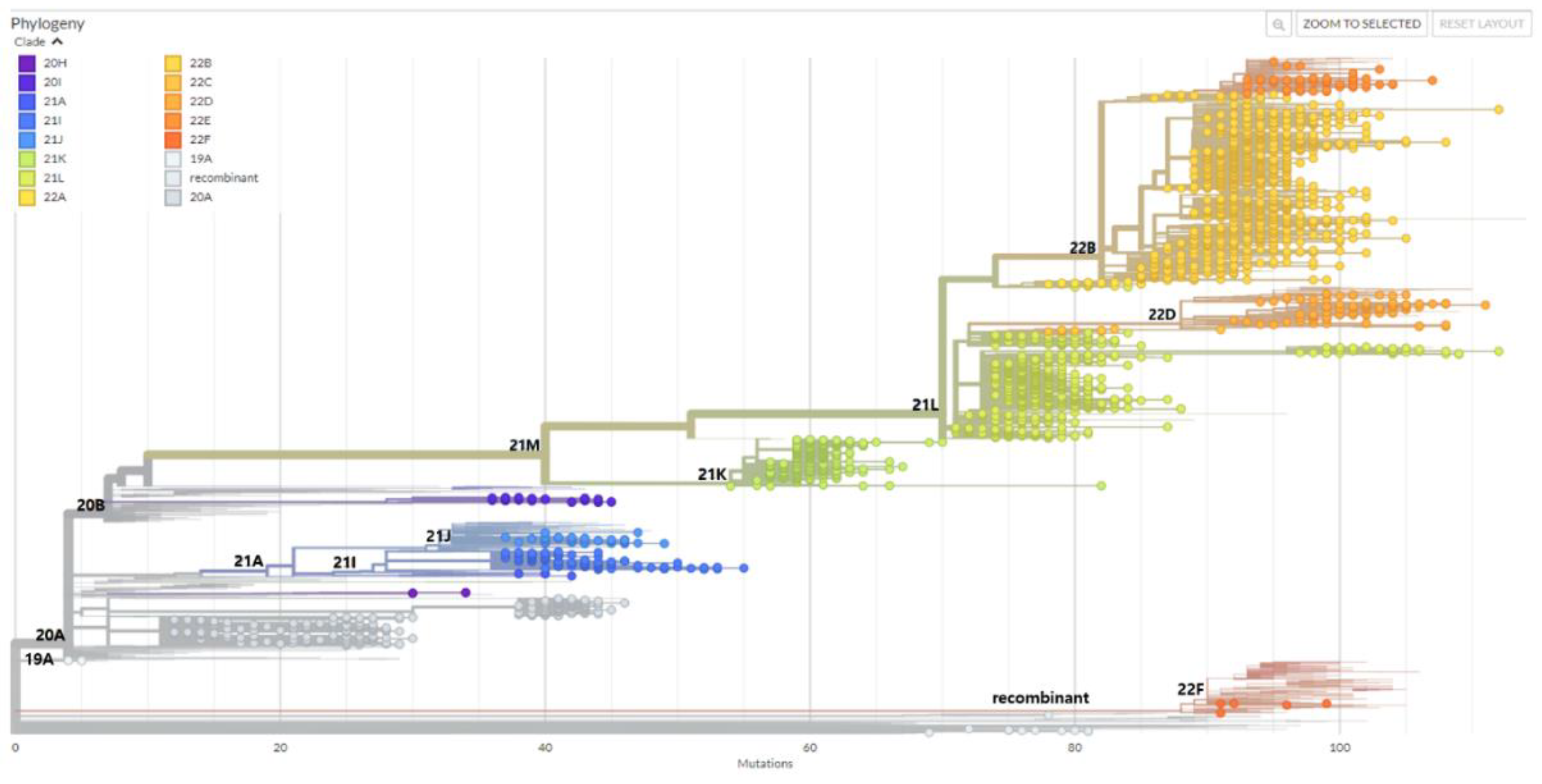
2.4. Phylogenetic analysis
3. Results
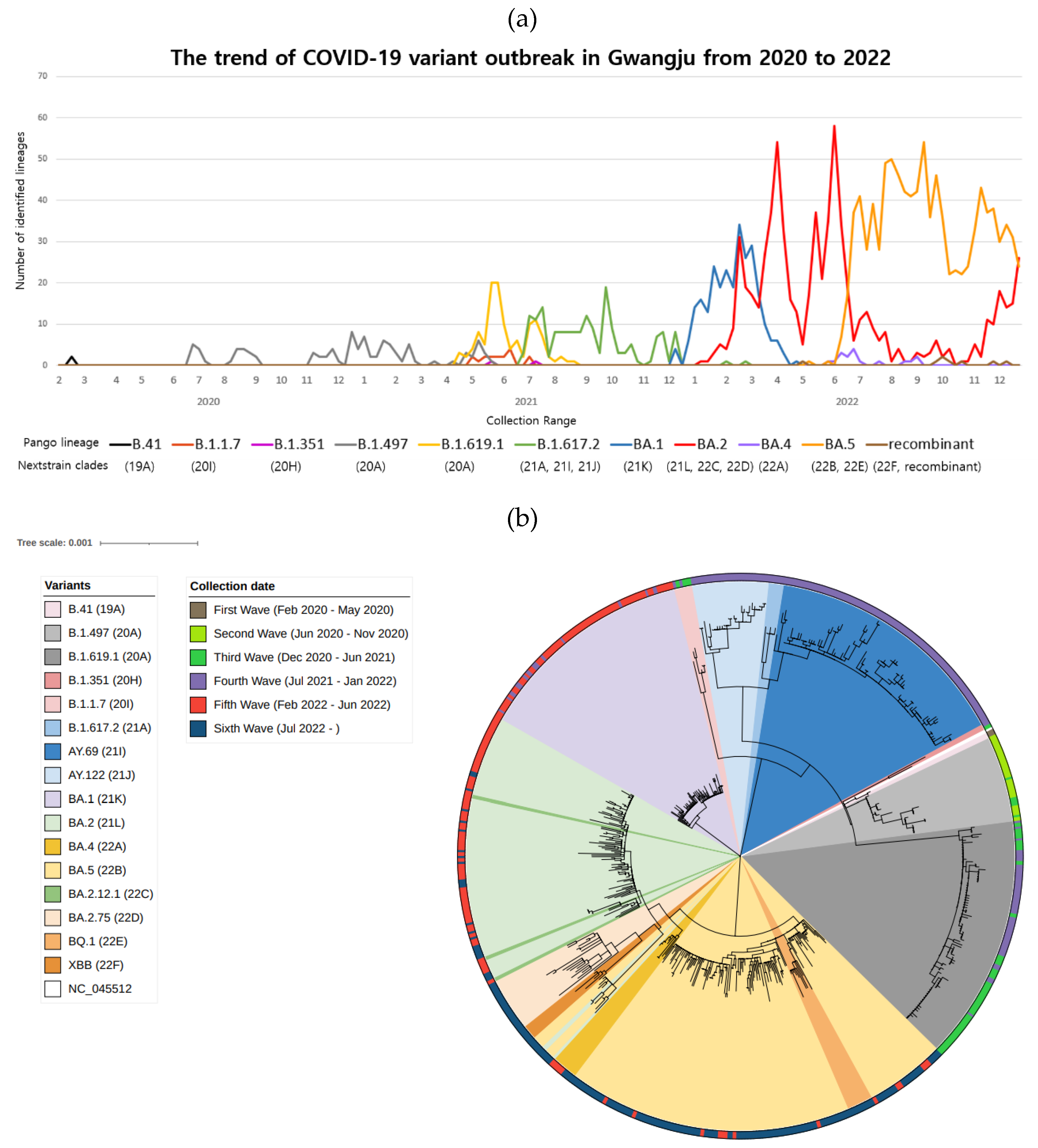
3.1. SARS-CoV-2 cases during pandemic waves in Gwangju, South Korea
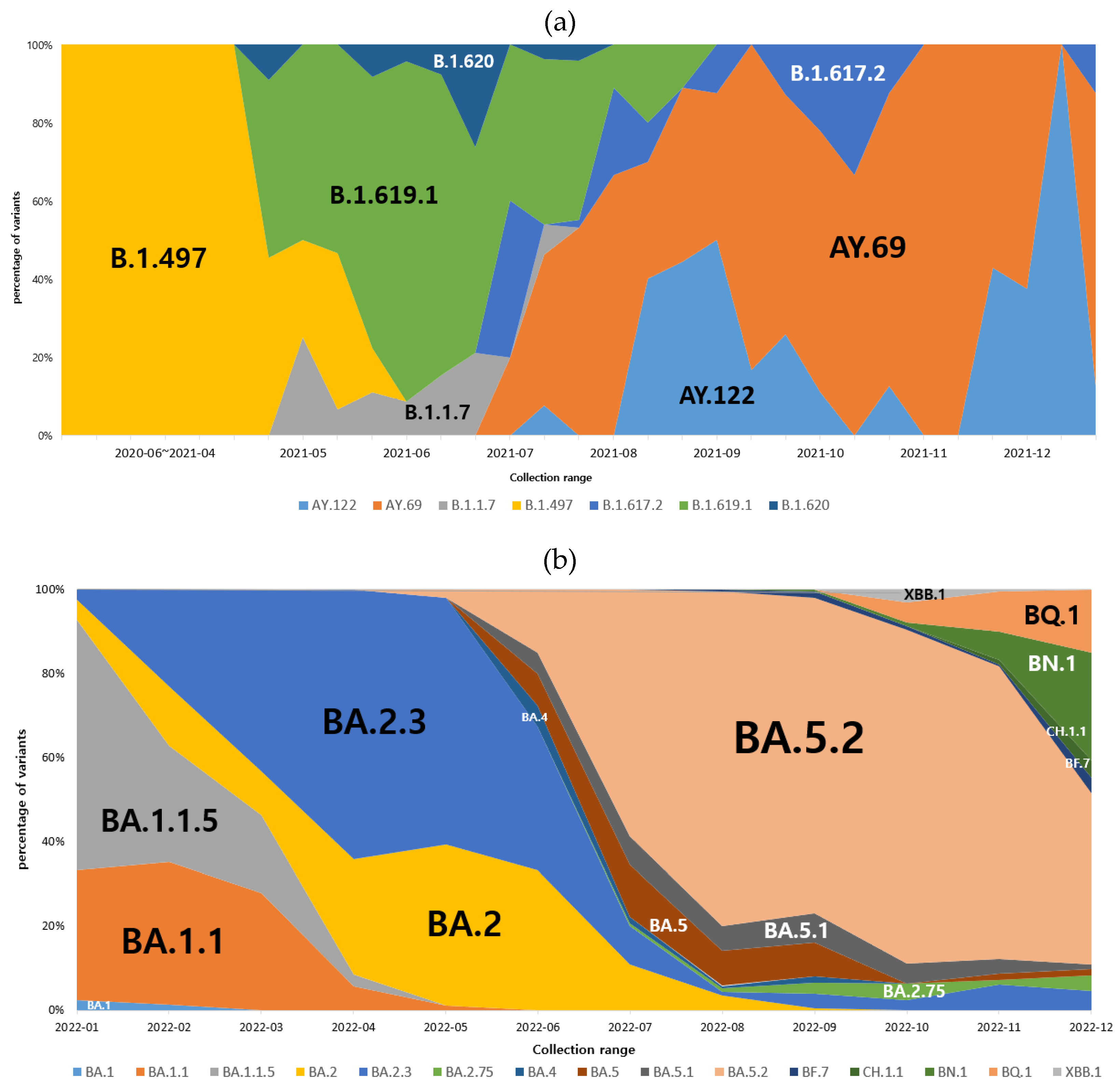
3.2. Dominant SARS-CoV-2 lineages during waves
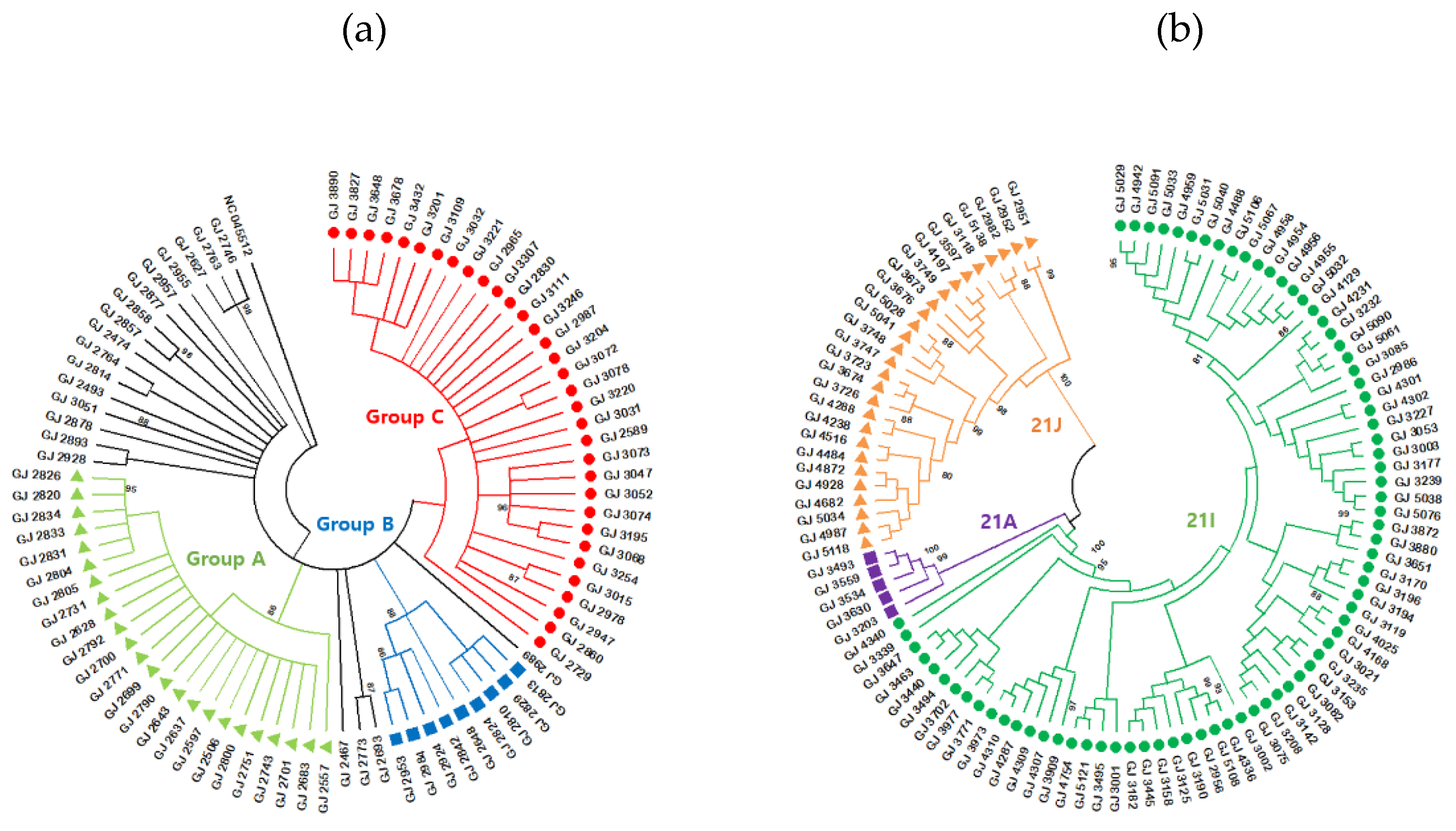
3.3. B.1.619.1 genome sequences
3.4. B.1.617.2 genome sequences
3.5. Omicron variant spread in Gwangju in 2022
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohan, B.S.; Vinod, N. COVID-19: An Insight into SARS-CoV2 Pandemic Originated at Wuhan City in Hubei Province of China. J. Infect. Dis. Epidemiol. 2020, 6, 146. [Google Scholar] [CrossRef]
- Kim, J.M., et al. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020, 11, 3–7. [CrossRef] [PubMed]
- Kim, M.J., et al. Isolation and Genomic Analyses of an Early SARS-CoV-2 Strains from the 2020 Epidemic in Gwangju, South Korea. J. Bacteriol. Virol. 2021, 51, 138–147. [CrossRef]
- Markov, P.V., et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [CrossRef] [PubMed]
- (WHO). W.H.O. Tracking SARS-CoV-2 variants. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants.
- O'Toole, A., et al. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022, 23, 121.
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017, 22, 30494. [Google Scholar] [CrossRef]
- Flores-Vega, V.R., et al. SARS-CoV-2: Evolution and Emergence of New Viral Variants. Viruses 2022, 14, 653. [CrossRef] [PubMed]
- Rando, H.M., et al. Pathogenesis, Symptomatology, and Transmission of SARS-CoV-2 through Analysis of Viral Genomics and Structure. mSystems 2021, 6, e0009521. [CrossRef]
- Ha, J.H., et al. COVID-19 Waves and Their Characteristics in the Seoul Metropolitan Area (Jan 20, 2020-Aug 31, 2022). Public Health Wkly. Rep. 2023, 16, 111–136.
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Mishra, P.M., et al. SARS-CoV-2 Spike mutations modify the interaction between virus Spike and human ACE2 receptors. Biochem. Biophys. Res. Commun. 2022, 620, 8–14. [CrossRef] [PubMed]
- Carabelli, A.M., et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177.
- Cosar, B., et al. SARS-CoV-2 Mutations and their Viral Variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.R., et al. Structure genomics of SARS-CoV-2 and its Omicron variant: Drug design templates for COVID-19. Acta Pharmacol. Sin. 2022, 43, 3021–3033. [CrossRef] [PubMed]
- Huang, Y., et al. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [CrossRef]
- Brant, A.C., et al. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [CrossRef] [PubMed]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Lekana-Douki, S.E., et al. Screening and Whole Genome Sequencing of SARS-CoV-2 Circulating During the First Three Waves of the COVID-19 Pandemic in Libreville and the Haut-Ogooue Province in Gabon. Front. Med. 2022, 9, 877391. [CrossRef] [PubMed]
- Zhou, D., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361. [CrossRef] [PubMed]
- GISAID. GISAID Analysis Update (10/01/2021). 2021; Available from: http://gisaid.org.
- Yu, C.Y., et al. Whole genome sequencing analysis of SARS-CoV-2 from Malaysia: From alpha to Omicron. Front. Med. 2022, 9, 1001022. [CrossRef] [PubMed]
- Park, A.K., et al. SARS-CoV-2 B.1.619 and B.1.620 Lineages, South Korea, 2021. Emerg. Infect. Dis. 2022, 28, 415–419. [CrossRef] [PubMed]
- Park, A.K., et al. Genomic Surveillance of SARS-CoV-2: Distribution of Clades in the Republic of Korea in 2020. Osong Public Health Res. Perspect. 2021, 12, 37–43. [CrossRef] [PubMed]
- Harvey, W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [CrossRef]
- Wang, W.B., et al. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: Binding free energy calculation studies. J. Mol. Graph Model 2021, 109, 108035. [CrossRef] [PubMed]
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): A structural perspective. Mol. Biol. Rep. 2023, 50, 2713–2721. [Google Scholar] [CrossRef]
- Zhang, Y., et al. Immune Evasive Effects of SARS-CoV-2 Variants to COVID-19 Emergency Used Vaccines. Front. Immunol. 2021, 12, 771242. [CrossRef]
- Winger, A. and T. Caspari, The Spike of Concern-The Novel Variants of SARS-CoV-2. Viruses 2021, 13. [CrossRef] [PubMed]
- Lee, S., et al., Phylodynamic analysis revealed that human mobility and vaccination were correlated to the local spread of SARS-CoV-2 in Republic of Korea. Emerg. Microbes Infect. 2023, 12, 2228934. [CrossRef]
- Klink, G.V. , et al., The rise and spread of the SARS-CoV-2 AY.122 lineage in Russia. Virus Evol. 2022, 8, veac017. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, K.; Zha, S.; Wang, L.; Chen, R. Epidemiological Characteristics of COVID-19 Resurgence in Areas Initially Under Control. Front. Public Health 2021, 9, 749294. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gong, D.; Xiao, J.; Hu, J.; He, G.; Rong, Z.; Ma, W. Cluster infections play important roles in the rapid evolution of COVID-19 transmission: A systematic review. Int. J. Infect. Dis. 2020, 99, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Yong, S.E.F.; Anderson, D.E.; E Wei, W.; Pang, J.; Ni Chia, W.; Tan, C.W.; Teoh, Y.L.; Rajendram, P.; Toh, M.P.H.S.; Poh, C.; et al. Connecting clusters of COVID-19: An epidemiological and serological investigation. Lancet Infect. Dis. 2020, 20, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Choi, J.H.; Cho, J.H. Estimation of the Effectiveness of a Tighter, Reinforced Quarantine for the Coronavirus Disease 2019 (COVID-19) Outbreak: Analysis of the Third Wave in South Korea. J. Pers. Med. 2023, 13, 402. [Google Scholar] [CrossRef] [PubMed]
- Organization, W.H. Strategy to Achieve Global Covid-19 Vaccination by mid-2022. Available from: https://cdn.who.int/media/docs/default-source/immunization/covid-19/strategy-to-achieve-global-covid-19-vaccination-by-mid-2022.pdf?sfvrsn=5a68433c_5.
- Chavda, V.P. , et al., The Delta and Omicron Variants of SARS-CoV-2: What We Know So Far. Vaccines (Basel), 2022. 10(11).
- Viana, R. , et al., Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature, 2022. 603, 679-686.
- Boudet, A. , et al., Limitation of Screening of Different Variants of SARS-CoV-2 by RT-PCR. Diagnostics (Basel), 2021. 11(7).
- Focosi, D. , et al., Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. Int J Mol Sci, 2023. 24(3).
- I.H Kim, A.K.P., J. I.H Kim, A.K.P., J.S. No, H.J. Lee, J.A Kim, C.Y. Lee, Y.J. Ahn, J.E. Rhee, E.J Kim, Omicron Subvariants (BQ.1, BQ.1.1, etc.) Outbreak Status. Public Health Weekly Report, 2022. 15, 2917-2924.
- Dijokaite-Guraliuc, A. , et al., Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses. Cell Rep, 2023. 42, 112271.
- Zhang, Y.; Zhang, H.; Zhang, W. SARS-CoV-2 variants, immune escape, and countermeasures. Front. Med. 2022, 16, 196–207. [Google Scholar] [CrossRef]
- DeGrace, M.M. , et al. , Defining the risk of SARS-CoV-2 variants on immune protection. Nature 2022, 605, 640–652. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of the MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
| Period | Number of COVID-19 cases |
Number of sequencing cases |
|---|---|---|
| Total | 850,707 | 2399 |
| First Wave (Feb 2020 – May 2020) |
36 | 2 |
| Second Wave (Jun 2020 – Nov 2020) |
725 | 35 |
| Third Wave (Dec 2020 – Jun 2021) |
2285 | 165 |
| Fourth Wave (Jul 2021 – Jan 2022) |
17,123 | 312 |
| Fifth Wave (Feb 2022 – Jun 2022) |
518,055 | 757 |
| Sixth Wave (Jul 2022 - ) |
312,483 | 1128 |
| Pango lineage | Total | Sub-lineages (number of variants) |
|---|---|---|
| B.41 | 2 | B.41 (2) |
| B.1.497 | 97 | B.1.497 (97) |
| B.1.1.7(Alpha) | 17 | B.1.1.7 (17) |
| B.1.351(Beta) | 2 | B.1.351 (2) |
| B.1.619.1 | 119 | B.1.619.1 (119) |
| B.1.617.2(Delta) | 173 | B.1.617.2 (7), AY.20 (1), AY.43 (1), AY.69 (117), AY.75.2 (1), AY.106 (1), AY.122 (42), AY.122.5 (3) |
| BA.1(Omicron) | 270 | BA.1 (2), BA.1.1 (128), BA.1.1.5 (136), BA.1.9 (1), BA.1.15 (1), BA.1.17 (1), BA.1.17.2 (1) |
| BA.2(Omicron) | 697 | BA.2 (114), BA.2.1 (1), BA.2.2 (1), BA.2.3 (302), BA.2.3.1 (1), BA.2.3.2 (5), BA.2.3.7 (1), BA.2.3.8 (4), BA.2.3.10 (1), BA.2.3.12 (19), BA.2.3.13 (4), BA.2.3.14 (12), BA.2.3.20 (10), BA.2.5 (1), BA.2.9 (3), BA.2.10 (13), BA.2.12.1 (12), BA.2.17 (2), BA.2.18 (1), BA.2.38.1 (2), BA.2.40.1 (1), BA.2.51 (1), BA.2.56 (5), BA.2.65 (17), BA.2.68 (39), BA.2.74 (1), BA.2.75 (1), BA.2.75.2 (3), BA.2.75.3 (1), BA.2.75.4 (4), BA.2.75.5 (2), BA.2.76 (1), BG.5 (1), BH.1 (1), BL.1 (7), BM.1.1 (1), BM.1.1.3 (3), BM.4.1.1 (1), BN.1.1 (4), BN.1.2 (15), BN.1.3 (44), BN.1.4 (1), BN.4 (1), BS.1.1 (4), CH.1.1 (10), CM.1 (1), CM.3 (1), CM.4 (13), CM.5 (1), CM.6 (1), CM.8.1 (2) |
| BA.4(Omicron) | 17 | BA.4 (1), BA.4.1 (7), BA.4.1.1 (6), BA.4.1.8 (1), BA.4.4 (1), BA.4.6 (1) |
| BA.5(Omicron) | 997 | BA.5 (2), BA.5.1 (39), BA.5.1.1 (1), BA.5.1.3 (3), BA.5.1.5 (1), BA.5.1.10 (7), BA.5.1.22 (2), BA.5.1.23 (4), BA.5.1.28 (7), BA.5.2 (388), BA.5.2.1 (225), BA.5.2.2 (3), BA.5.2.3 (2), BA.5.2.6 (10), BA.5.2.9 (5), BA.5.2.12 (1), BA.5.2.16 (2), BA.5.2.19 (23), BA.5.2.20 (14), BA.5.2.22 (12), BA.5.2.25 (1), BA.5.2.26 (3), BA.5.2.27 (9), BA.5.2.28 (3), BA.5.2.32 (4), BA.5.2.34 (3), BA.5.2.44 (2), BA.5.5 (34), BA.5.6 (7), BA.5.6.1 (1), BA.5.8 (1), BA.5.9 (3), BA.5.10 (7), BA.5.10.1 (2), BE.1 (6), BE.1.1 (10), BE.1.1.2 (1), BE.1.4 (1), BE.4 (3), BE.4.1.1 (1), BF.3 (1), BF.5 (43), BF.7 (10), BF.7.4.2 (2), BF.7.5 (1), BF.10 (7), BF.11 (2), BF.21 (11), BF.24 (1), BF.26 (1), BF.27 (1), BF.28 (7), BK.1 (1), BQ.1 (1), BQ.1.1 (19), BQ.1.1.1 (1), BQ.1.1.4 (1), BQ.1.1.17 (7), BQ.1.1.18 (2), BQ.1.2 (9), BQ.1.3 (4), BQ.1.5 (5), BQ.1.11 (4), BQ.1.15 (1), BQ.1.23 (1), CK.3 (1) |
| recombinant | 8 | XAH (1), XAZ (1), XBB (1), XBB.1 (4), XBC.1 (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





