Submitted:
14 October 2023
Posted:
17 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Mouse and Tumor Models
2.3. Plasmid Transfection and Overexpression of EHF
2.4. Determination of Cell Growth
2.5. RNA Isolation and Quantitative Real-Time RT-PCR
2.6. Knockdown of EHF Expression by Small Interfering RNA (siRNA)
2.7. Western Blot
2.8. Wound Healing Assay
2.9. Transwell Assay
2.10. Senescence-Associated β-gal Assay
2.11. Annexin V/PI Apoptosis Assay
2.12. Survival Analysis
2.13. Dataset Analysis
2.14. Statistical Analysis
| Gene | Species | Sequence |
|---|---|---|
| GAPDH | Human | FW – CTGGGCTACACTGAGCACCA RV – CCAGCGTCAAAGGTGGAG |
| EHF | Human | FW – TGCAGCATCTGAAGTGGAAC RV – AGGAAGGTGACTGGTGGTTG |
| NDRG2 | Human | FW – TCTGTCACTTTCACTGTCTAR RV – CCAGAGATGGGTACTGATAT |
| N-CADHERIN | Human | FW – CCTCCAGAGTTTACTGCCATGAC RV – GTAGGATCTCCGCCACTGATTC |
| SLUG | Human | FW – ATCTGCGGCAAGGCGTTTTCCA RV – GAGCCCTCAGATTTGACCTGTC |
| CYCLOPHILIN | Mouse | FW – CATACAGGTCCTGGCATCTTGTC RV – AGACCACATGCTTGCCATCCAG |
| EHF | Mouse | FW – ATGCAATGTTTCCAGCGGTT RV – GTCGAACTCCTGGAAAGGGA |
| NDRG2 | Mouse | FW – CAGCATCACAGGGCACTTGA RV – CGCCGAGACCTGAACTTTG |
3. Results
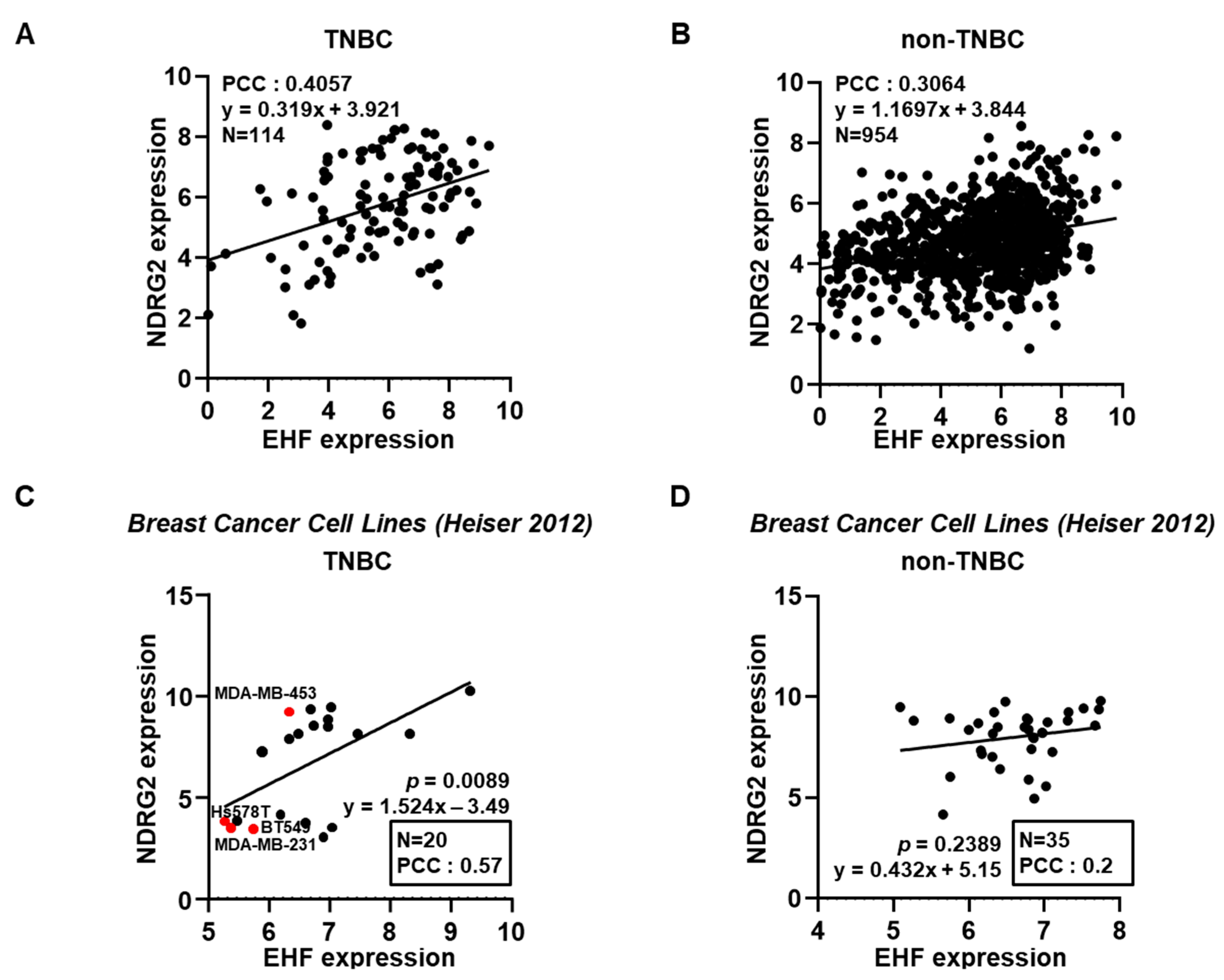
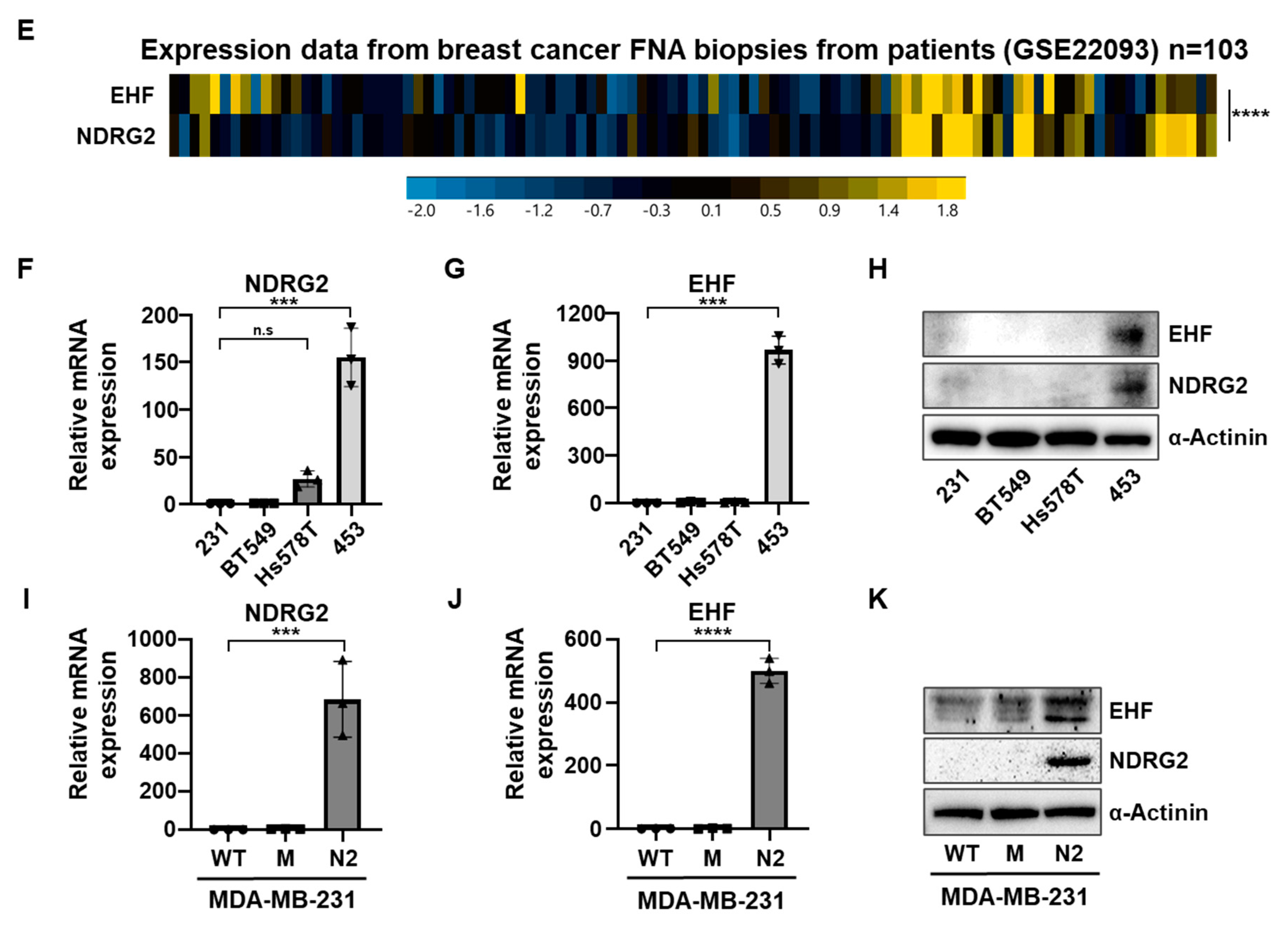
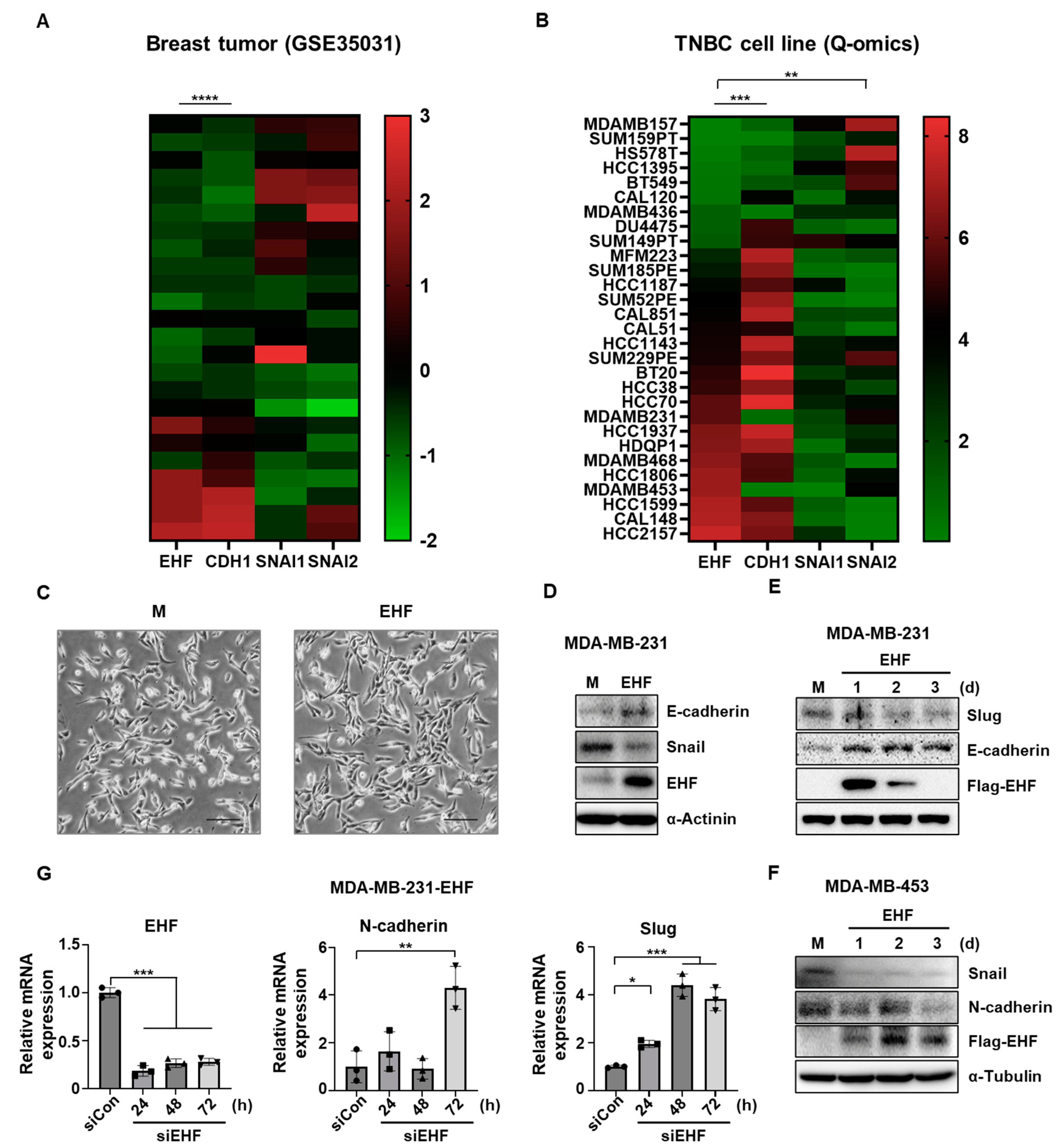
3.1. The Positive Correlation between NDRG2 and EHF Gene Expression Levels in Breast Cancer
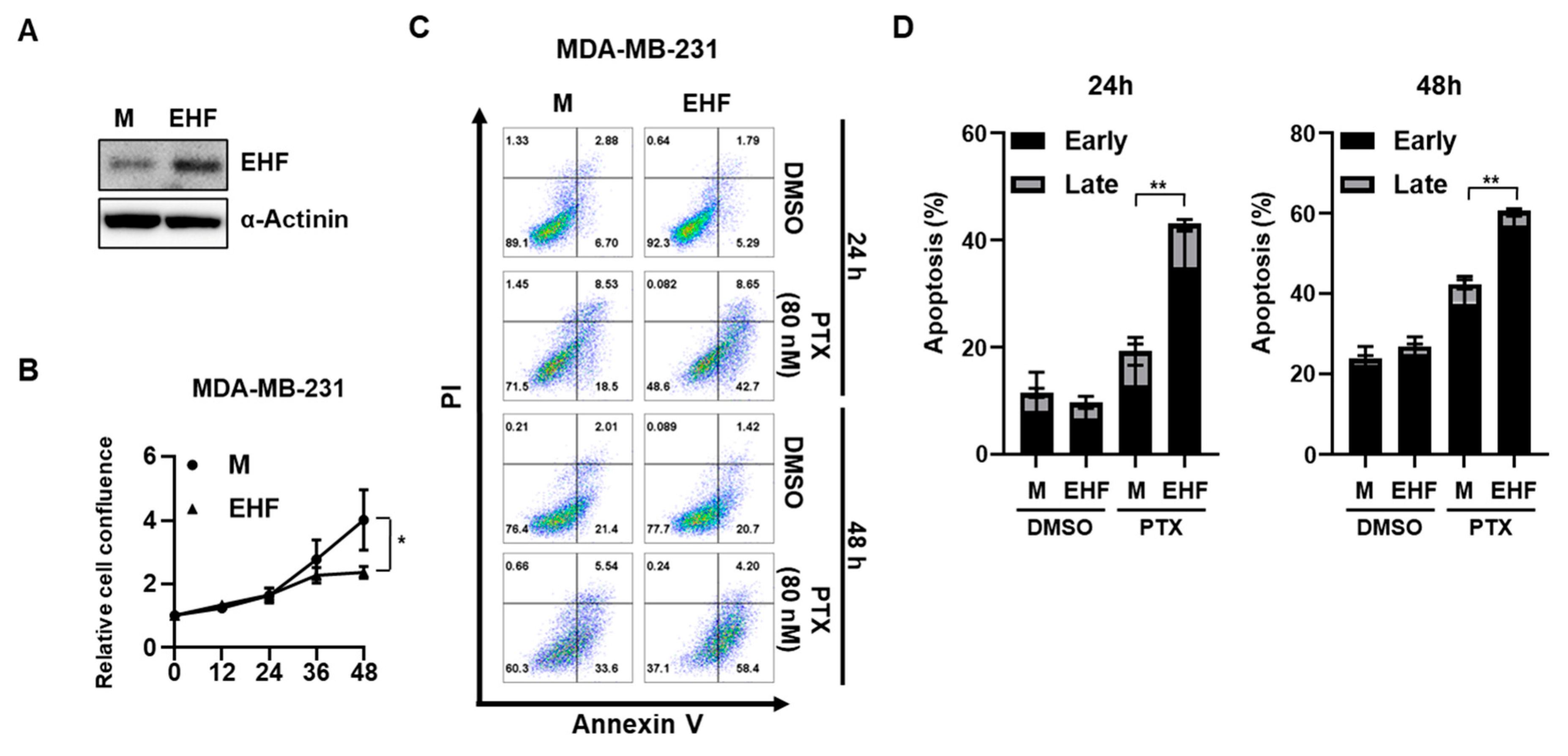
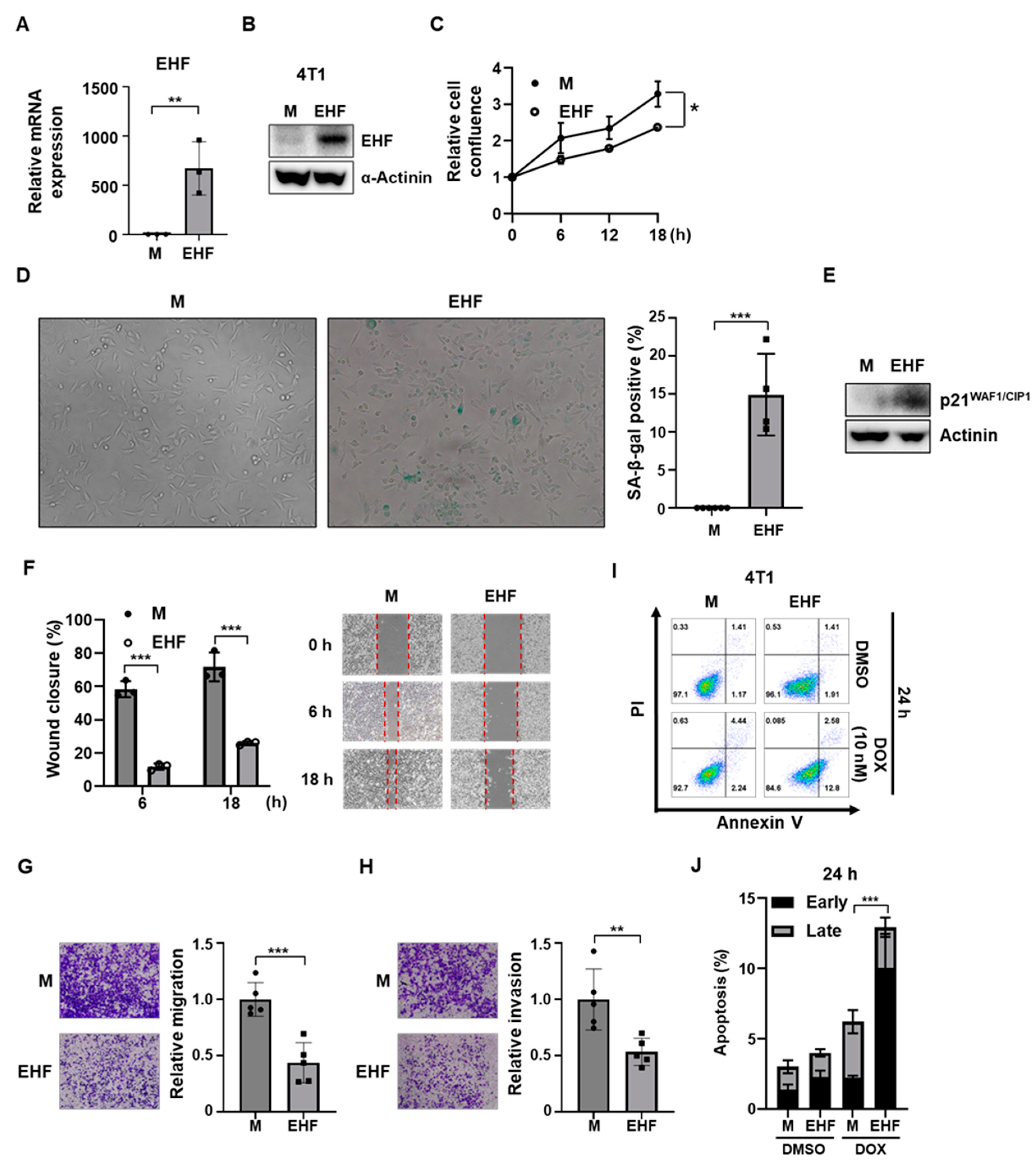
3.2. EHF Inhibited Cell Growth and Promoted Paclitaxel-Induced Apoptosis
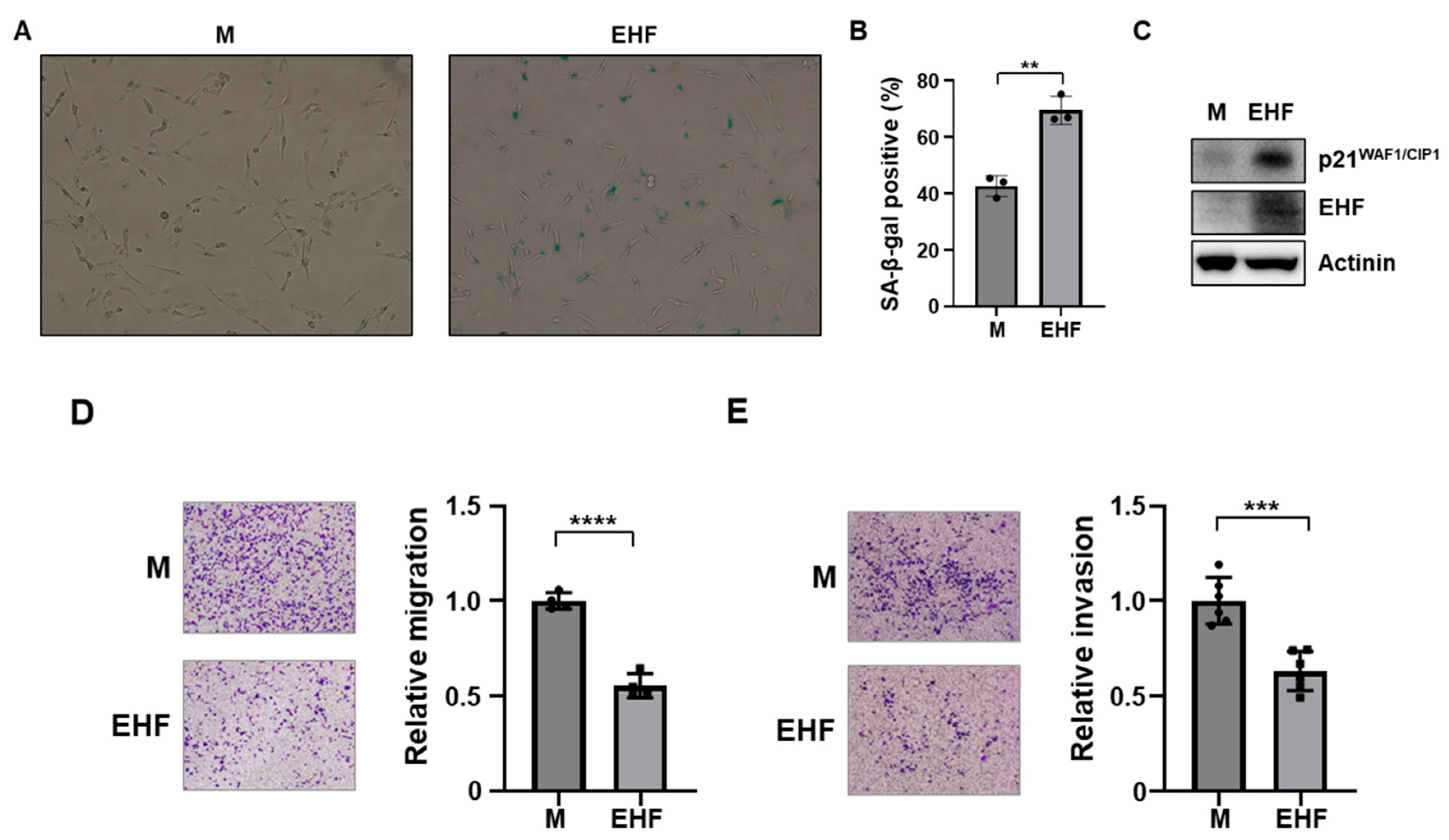
3.3. Induction of Senescence and Inhibition of Cell Migration by EHF Expression
3.4. EHF Inhibited EMT in TNBC Cells
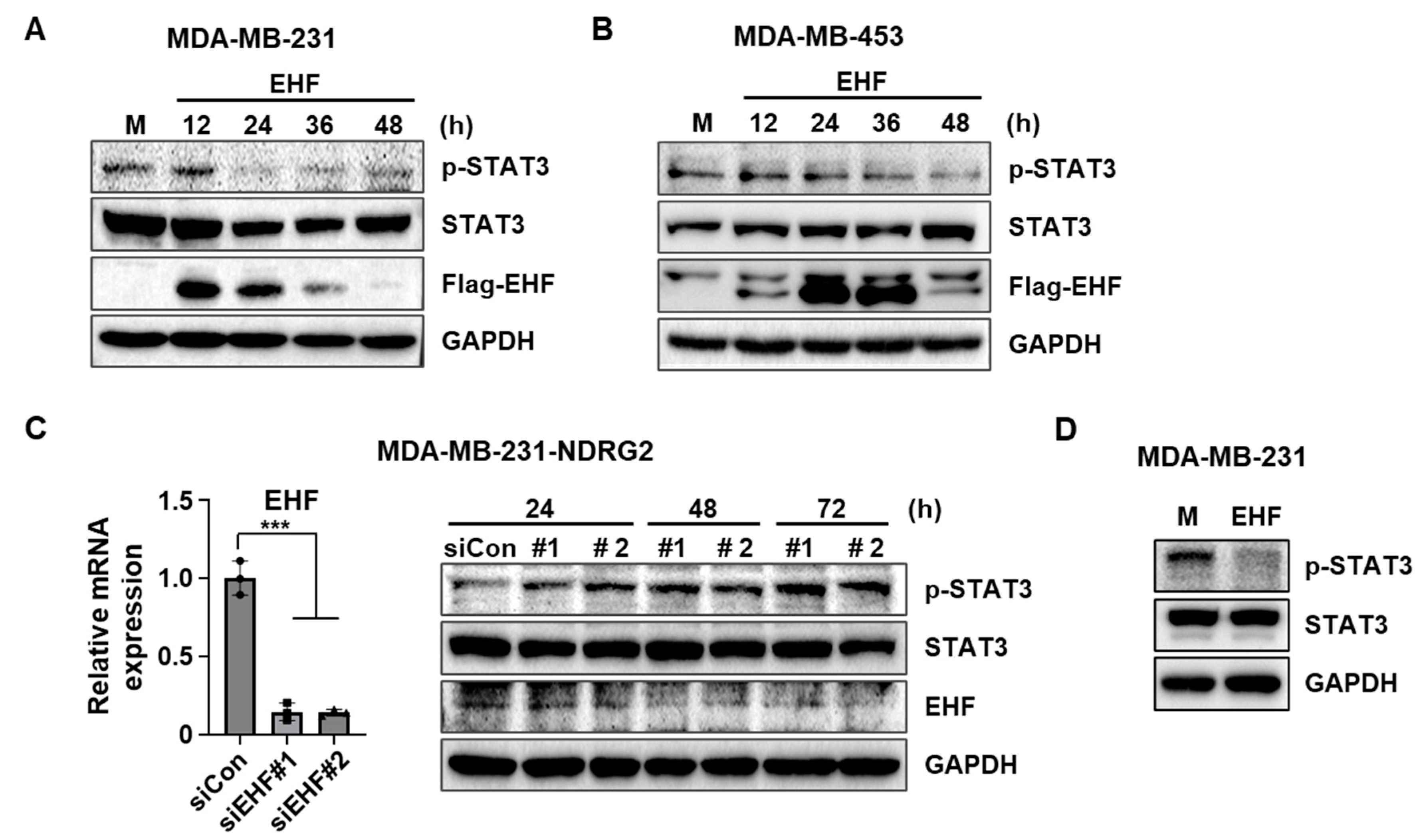
3.5. EHF Overexpression Inhibited the Phosphorylation of STAT3
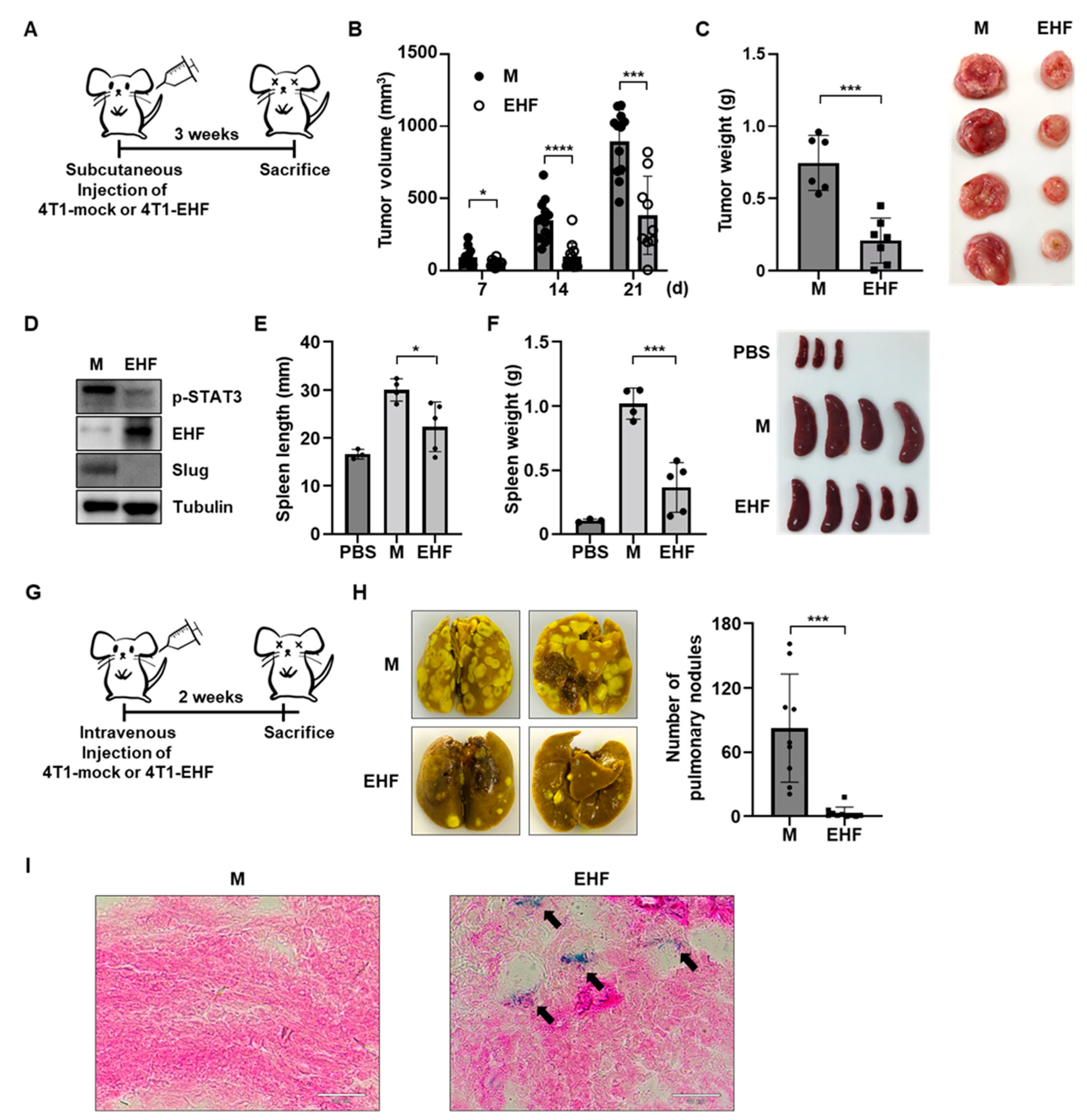
3.6. EHF Exhibited Antitumor Effects in Mouse Mammary Tumor Cells
3.7. EHF Suppressed Breast Cancer Progression
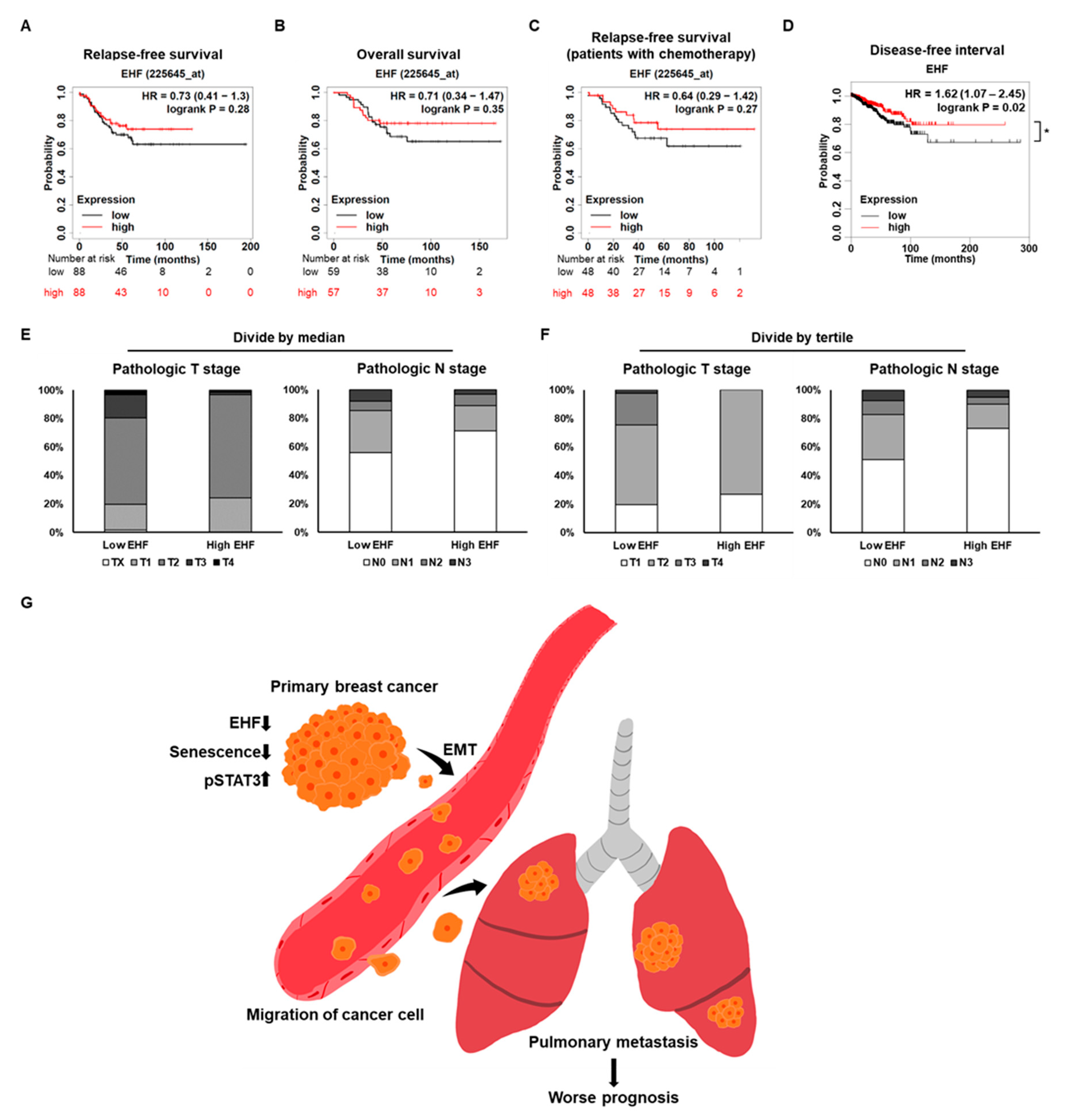
3.8. Breast Cancer Patients with High EHF Expression Showed an Increased Survival Rate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends in cell biology 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Calcinotto, A.; Kohli, J.; Zagato, E.; Pellegrini, L.; Demaria, M.; Alimonti, A. Cellular Senescence: Aging, Cancer, and Injury. Physiological reviews 2019, 99, 1047–1078. [Google Scholar] [CrossRef]
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular senescence: the good, the bad and the unknown. Nature reviews. Nephrology 2022, 18, 611–627. [Google Scholar] [CrossRef]
- Mayer, I.A.; Abramson, V.G.; Lehmann, B.D.; Pietenpol, J.A. New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clinical cancer research : an official journal of the American Association for Cancer Research 2014, 20, 782–790. [Google Scholar] [CrossRef]
- Melotte, V.; Qu, X.; Ongenaert, M.; van Criekinge, W.; de Bruine, A.P.; Baldwin, H.S.; van Engeland, M. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2010, 24, 4153–4166. [Google Scholar] [CrossRef]
- Hu, W.; Fan, C.; Jiang, P.; Ma, Z.; Yan, X.; Di, S.; Jiang, S.; Li, T.; Cheng, Y.; Yang, Y. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget 2016, 7, 209–223. [Google Scholar] [CrossRef]
- Nakahata, S.; Ichikawa, T.; Maneesaay, P.; Saito, Y.; Nagai, K.; Tamura, T.; Manachai, N.; Yamakawa, N.; Hamasaki, M.; Kitabayashi, I.; et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nature communications 2014, 5, 3393. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, A.; Lewinsky, R.H.; Bornholdt, J.; Vogel, L.K.; Mitchelmore, C. Expression profile of the N-myc Downstream Regulated Gene 2 (NDRG2) in human cancers with focus on breast cancer. BMC cancer 2011, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Moradi Monfared, M.; Alizadeh Zarei, M.; Rafiei Dehbidi, G.; Behzad Behbahani, A.; Arabsolghar, R.; Takhshid, M.A. NDRG2 Regulates the Expression of Genes Involved in Epithelial Mesenchymal Transition of Prostate Cancer Cells. Iranian journal of medical sciences 2019, 44, 118–126. [Google Scholar] [PubMed]
- Li, S.J.; Wang, W.Y.; Li, B.; Chen, B.; Zhang, B.; Wang, X.; Chen, C.S.; Zhao, Q.C.; Shi, H.; Yao, L. Expression of NDRG2 in human lung cancer and its correlation with prognosis. Medical oncology 2013, 30, 421. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.Q.; Wang, C.G.; Sun, J.C.; Gao, J.J.; Tao, L.; Du, X.L.; Zhao, H.D.; Yang, J.D.; Li, Y. Overexpression of NDRG2 Increases Iodine Uptake and Inhibits Thyroid Carcinoma Cell Growth In Situ and In Vivo. Oncol Res 2015, 23, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lim, J.; Yang, Y.; Lee, M.S.; Lim, J.S. N-myc downstream-regulated gene 2 (NDRG2) suppresses the epithelial-mesenchymal transition (EMT) in breast cancer cells via STAT3/Snail signaling. Cancer letters 2014, 354, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Yamada, T. Molecular biology of the Ets family of transcription factors. Gene 2003, 303, 11–34. [Google Scholar] [CrossRef] [PubMed]
- Luk, I.Y.; Reehorst, C.M.; Mariadason, J.M. ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Choi, Y.S.; Sinha, S. Determination of the consensus DNA-binding sequence and a transcriptional activation domain for ESE-2. The Biochemical journal 2006, 398, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Sui, F.; Ma, J.; Ren, X.; Yang, Q.; Zhang, Y.; Guan, H.; Shi, B.; Hou, P.; Ji, M. Increased expression of EHF contributes to thyroid tumorigenesis through transcriptionally regulating HER2 and HER3. Oncotarget 2016, 7, 57978–57990. [Google Scholar] [CrossRef]
- Brenne, K.; Nymoen, D.A.; Hetland, T.E.; Trope, C.G.; Davidson, B. Expression of the Ets transcription factor EHF in serous ovarian carcinoma effusions is a marker of poor survival. Human pathology 2012, 43, 496–505. [Google Scholar] [CrossRef]
- Albino, D.; Civenni, G.; Rossi, S.; Mitra, A.; Catapano, C.V.; Carbone, G.M. The ETS factor ESE3/EHF represses IL-6 preventing STAT3 activation and expansion of the prostate cancer stem-like compartment. Oncotarget 2016, 7, 76756–76768. [Google Scholar] [CrossRef]
- Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational and structural biotechnology journal 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.; Jin, S.; Yoo, H.; Jeong, S.; Jeong, E.; Yoon, S. Q-omics: Smart Software for Assisting Oncology and Cancer Research. Molecules and cells 2021, 44, 843–850. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nature biotechnology 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Wong, A.K.; Krishnan, A.; Aure, M.R.; Tadych, A.; Zhang, R.; Corney, D.C.; Greene, C.S.; Bongo, L.A.; Kristensen, V.N.; et al. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nature methods 2015, 12, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Golestan, A.M.; Mojtahedi, Z.P.; Ghalamfarsa, G.P.; Hamidinia, M.M.; Takhshid, M.A.P. The Effects of NDRG2 Overexpression on Cell Proliferation and Invasiveness of SW48 Colorectal Cancer Cell Line. Iranian journal of medical sciences 2015, 40, 430–439. [Google Scholar] [PubMed]
- Kim, M.J.; Kim, H.S.; Lee, S.H.; Yang, Y.; Lee, M.S.; Lim, J.S. NDRG2 controls COX-2/PGE(2)-mediated breast cancer cell migration and invasion. Molecules and cells 2014, 37, 759–765. [Google Scholar] [CrossRef]
- Ma, J.; Liu, W.; Yan, X.; Wang, Q.; Zhao, Q.; Xue, Y.; Ren, H.; Wu, L.; Cheng, Y.; Li, S.; et al. Inhibition of endothelial cell proliferation and tumor angiogenesis by up-regulating NDRG2 expression in breast cancer cells. PloS one 2012, 7, e32368. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Endo, K.; Sakamoto, K.; Kayamori, K.; Ehata, S.; Ichikawa, J.; Ando, T.; Nakamura, R.; Kimura, Y.; Yoshizawa, K.; et al. EHF suppresses cancer progression by inhibiting ETS1-mediated ZEB expression. Oncogenesis 2021, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Jiang, W.; Wang, X.; Wang, H.; Zheng, C.; Li, Y.; Sun, Y.; Huang, C.; Han, Z.B.; Yang, S.; et al. ESE3 Inhibits Pancreatic Cancer Metastasis by Upregulating E-Cadherin. Cancer research 2017, 77, 874–885. [Google Scholar] [CrossRef]
- Segatto, I.; Baldassarre, G.; Belletti, B. STAT3 in Breast Cancer Onset and Progression: A Matter of Time and Context. International journal of molecular sciences 2018, 19. [Google Scholar] [CrossRef]
- Albino, D.; Civenni, G.; Dallavalle, C.; Roos, M.; Jahns, H.; Curti, L.; Rossi, S.; Pinton, S.; D'Ambrosio, G.; Sessa, F.; et al. Activation of the Lin28/let-7 Axis by Loss of ESE3/EHF Promotes a Tumorigenic and Stem-like Phenotype in Prostate Cancer. Cancer research 2016, 76, 3629–3643. [Google Scholar] [CrossRef]
- Wang, L.; Xing, J.; Cheng, R.; Shao, Y.; Li, P.; Zhu, S.; Zhang, S. Abnormal Localization and Tumor Suppressor Function of Epithelial Tissue-Specific Transcription Factor ESE3 in Esophageal Squamous Cell Carcinoma. PloS one 2015, 10, e0126319. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yoon, S.Y.; Kim, J.T.; Song, E.Y.; Lee, H.G.; Son, H.J.; Kim, S.Y.; Cho, D.; Choi, I.; Kim, J.H.; et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis 2009, 30, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.S.; Kim, D.; Kim, D.H.; Chang, H.H.; Sohn, K.C.; Kim, K.H.; Jung, S.H.; Lee, B.K.; Kim, J.H.; Kim, K.D. NDRG2 correlated with favorable recurrence-free survival inhibits metastasis of mouse breast cancer cells via attenuation of active TGF-beta production. Carcinogenesis 2012, 33, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, N.; Kedage, V.; Hollenhorst, P.C. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell communication and signaling : CCS 2015, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Shon, S.K.; Kim, A.; Kim, K.I.; Yang, Y.; Cho, D.H.; Lee, M.S.; Lim, J.S. SOCS1 induced by NDRG2 expression negatively regulates STAT3 activation in breast cancer cells. Biochemical and biophysical research communications 2007, 363, 361–367. [Google Scholar] [CrossRef]
- Li, Q.J.; Yang, S.H.; Maeda, Y.; Sladek, F.M.; Sharrocks, A.D.; Martins-Green, M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. The EMBO journal 2003, 22, 281–291. [Google Scholar] [CrossRef]
- Campisi, J. Cellular senescence as a tumor-suppressor mechanism. Trends in cell biology 2001, 11, S27–31. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





