Submitted:
16 October 2023
Posted:
16 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
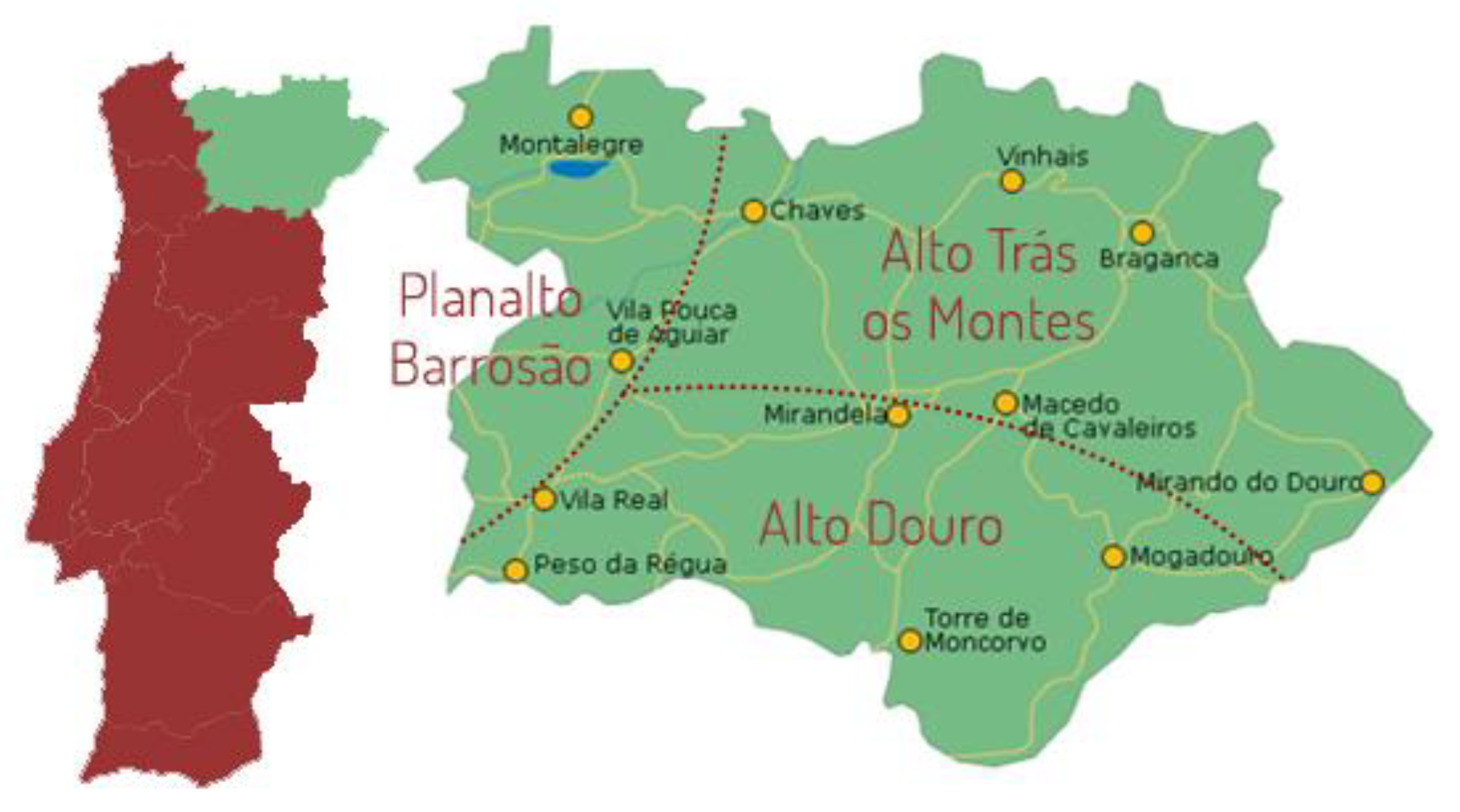
2.1. Sampling
2.2. Physicochemical and microbiological analysis of sausages
2.3. LAB isolation
2.4. Phenotypic characterization
2.5. Genotypic characterization
2.5.1. DNA extraction
2.5.2. 16S rRNA amplification
2.5.3. 16S rRNA sequencing
2.6. Data analysis
2.6.1. Species identification
2.6.2. Phylogenetic tree
2.6.3. Phenotypic characterization of LAB
3. Results and Discussion
3.1. Genetic identification of Lactic Acid Bacteria Isolates
3.2. Alheira physicochemical and microbiological analysis
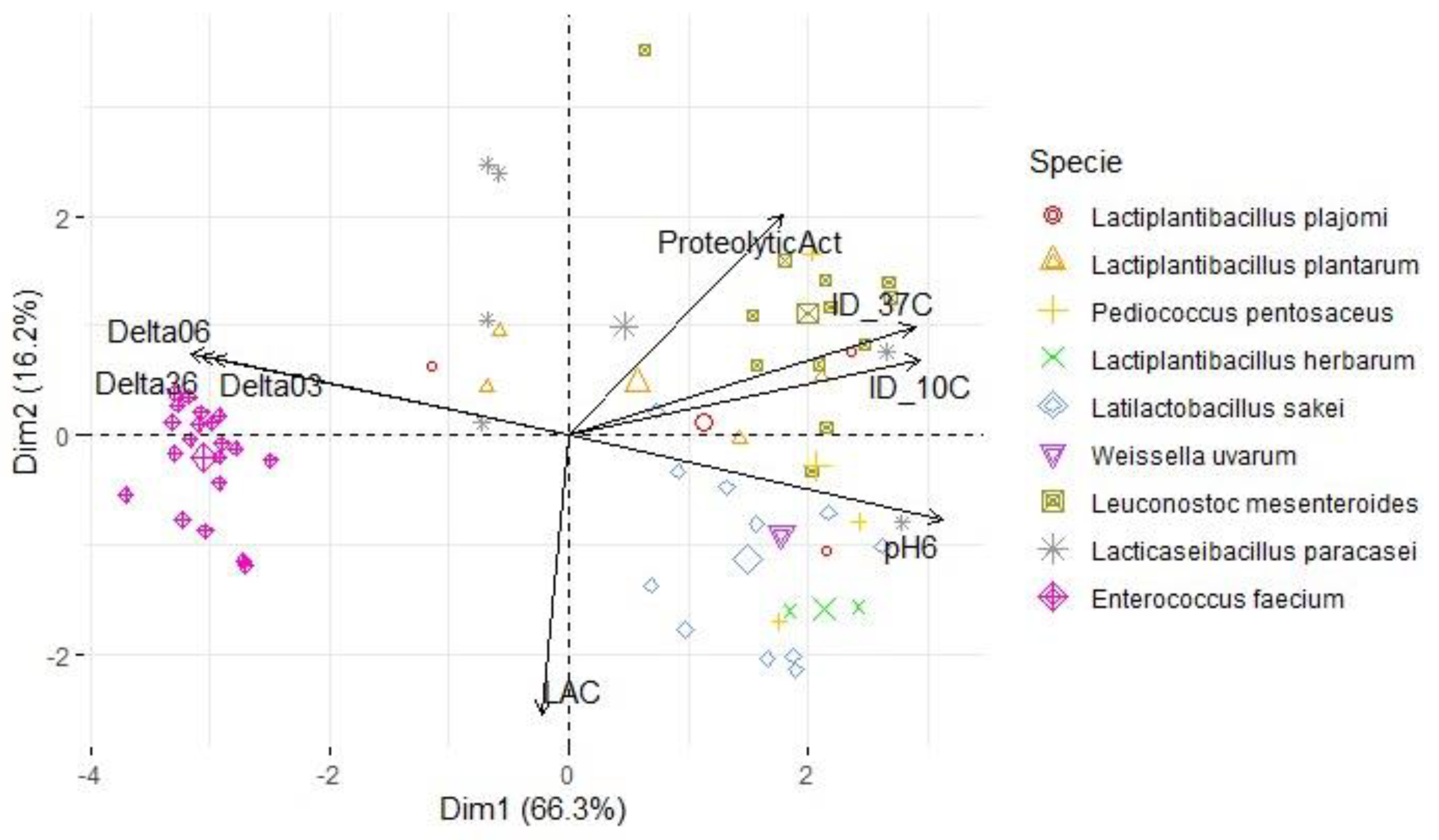
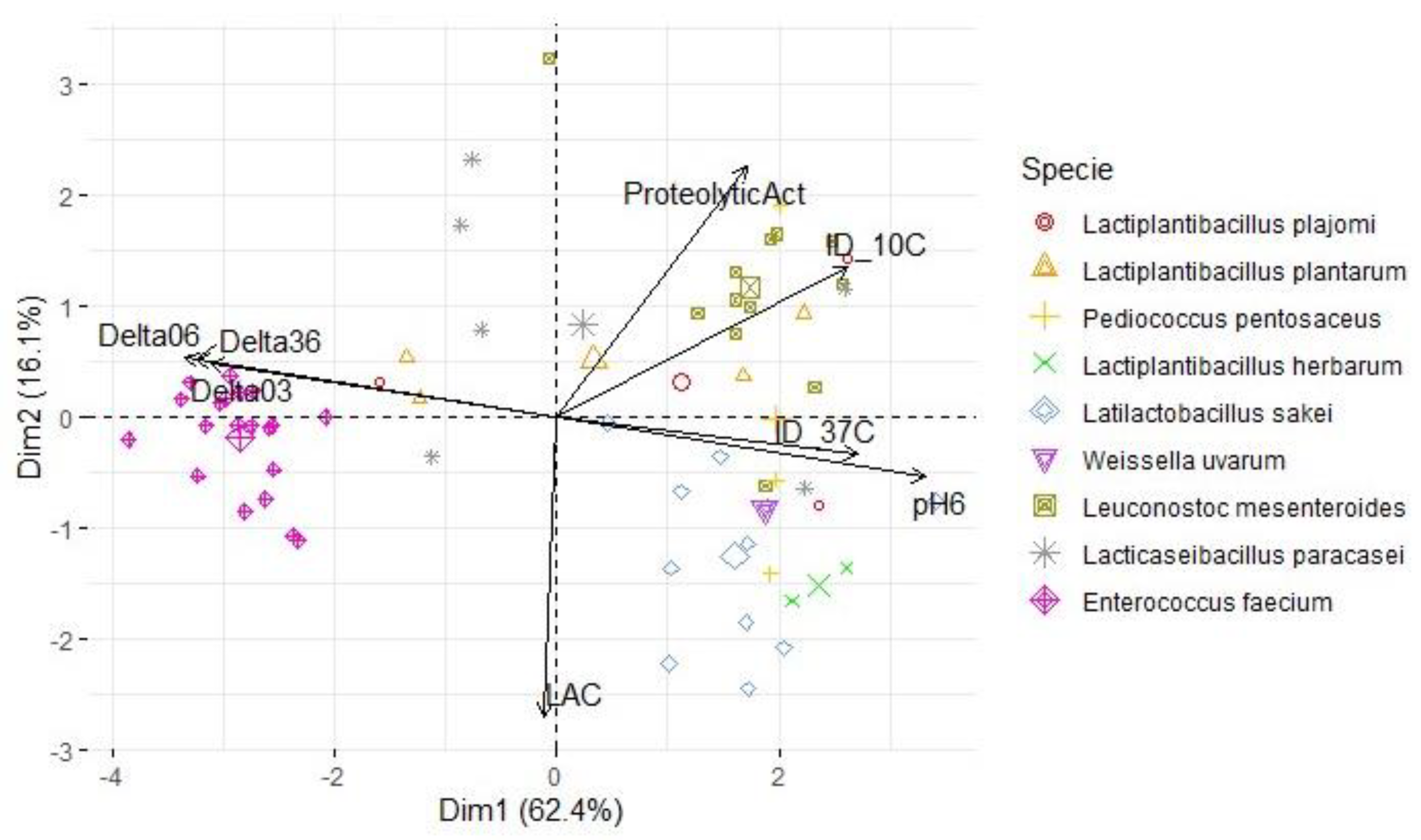
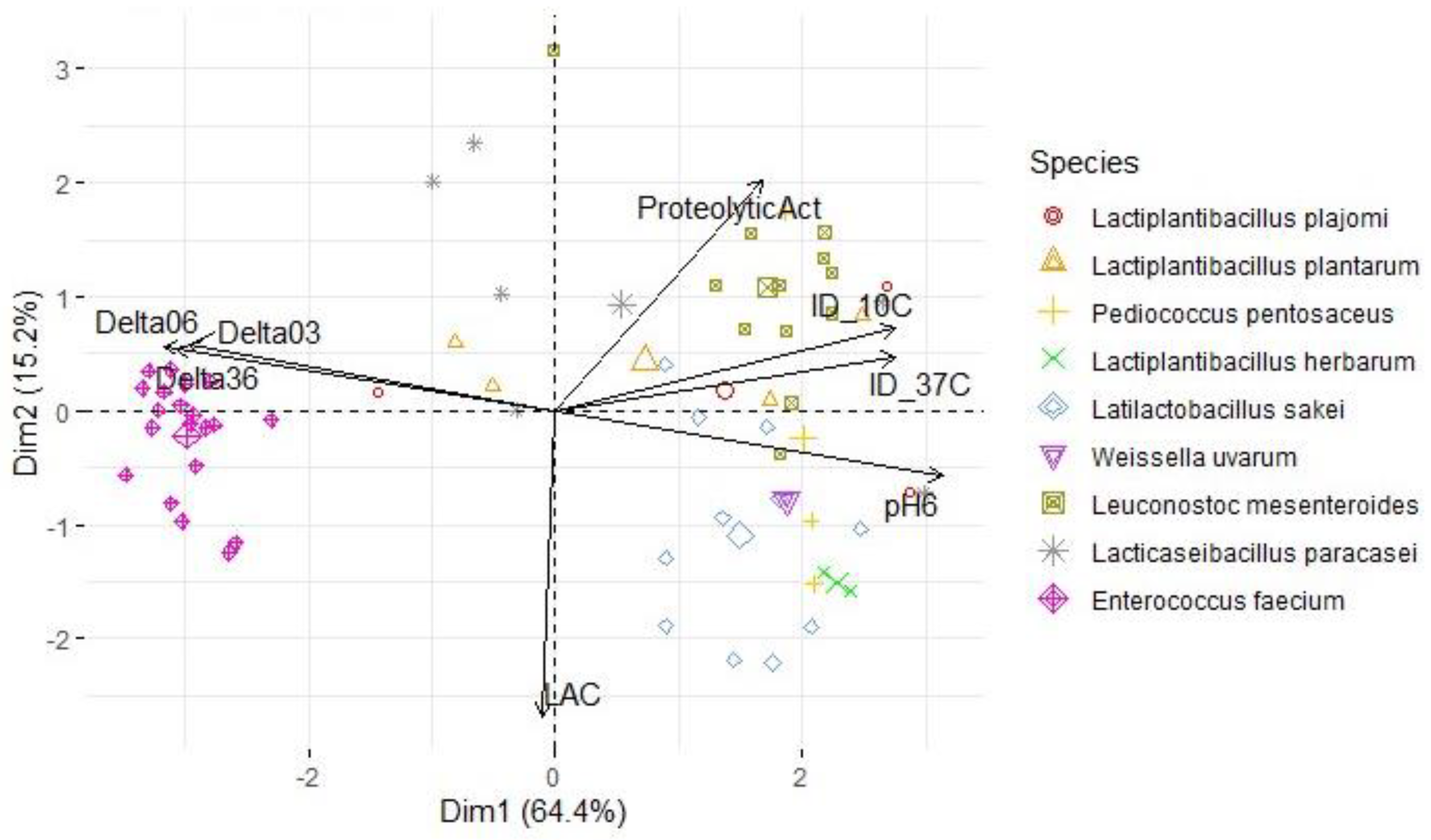
3.3. Lactic acid bacteria phenotypic and genetic analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferreira V, Barbosa J, Vendeiro S, Mota A, Silva F, Monteiro MJ, et al. Chemical and microbiological characterization of alheira: A typical Portuguese fermented sausage with particular reference to factors relating to food safety. Meat Sci. 2006;73(4):570–5. [CrossRef]
- Coelho-Fernandes S, Zefanias O, Rodrigues G, Faria AS, Fernandes Â, Barros L, et al. Microbiological and Physicochemical Assessment of Artisanally Produced “Alheira” Fermented Sausages in Northern Portugal. Proceedings. 2020;70(1):16.
- Esteves A, Patarata L, Saraiva C, Martins C. Assessment of the Microbiological Characteristics of Industrially Produced Alheira, with Particular Reference to Foodborne Pathogens. J Food Saf. 2008 Feb;28(1):88–102. [CrossRef]
- Mokoena MP. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules. 2017 Aug;22(8):1255. [CrossRef]
- 5. Doyle MP, Steenson LR, Meng J. Bacteria in Food and Beverage Production. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Applied Bacteriology and Biotechnology [Internet]. Berlin, Heidelberg: Springer; 2013 [cited 2023 Aug 21]. p. 241–56. [CrossRef]
- Khalid K. An overview of lactic acid bacteria. IJB. 2011;V1(N3):P1-13.
- 14:00-17:00. ISO. [cited 2023 Oct 9]. ISO 1442:2023. Available from: https://www.iso.org/standard/82664.html.
- 14:00-17:00. ISO. [cited 2023 Oct 9]. ISO 937:2023. Available from: https://www.iso.org/standard/82663.html.
- 14:00-17:00. ISO. [cited 2023 Oct 9]. ISO 936:1998. Available from: https://www.iso.org/standard/24783.html.
- 14:00-17:00. ISO. [cited 2023 Oct 9]. ISO 6888-1:2021. Available from: https://www.iso.org/standard/76672.html.
- Dwivedi HP, Smiley RD, Pincus DH. 11. Rapid Methods for the Detection and Identification of Foodborne Pathogens. In: Compendium of Methods for the Microbiological Examination of Foods [Internet]. American Public Health Association; 2013 [cited 2023 Oct 9]. Available from: https://ajph.aphapublications.org/doi/full/10.2105/MBEF.0222.016.
- Nutrition C for FS and A. BAM Chapter 5: Salmonella. FDA [Internet]. 2023 Sep 18 [cited 2023 Oct 9]; Available from: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella.
- Faria AS, Fernandes N, Cadavez V, Gonzales-Barron U. Screening of Lactic Acid Bacteria Isolated from Artisanally Produced Alheira Fermented Sausages as Potential Starter Cultures. 2021.
- Mohania D, Nagpal R, Kumar M, Bhardwaj A, Yadav M, Jain S, et al. Molecular approaches for identification and characterization of lactic acid bacteria. J Dig Dis. 2008;9(4):190–8. [CrossRef]
- Hou Q, Bai X, Li W, Gao X, Zhang F, Sun Z, et al. Design of Primers for Evaluation of Lactic Acid Bacteria Populations in Complex Biological Samples. Front Microbiol [Internet]. 2018 [cited 2022 Mar 28];9. [CrossRef]
- Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022 Jan 7;50(D1):D20–6.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–10.
- Wilhelm H. Holzapfel, Brian J.B. Wood. Lactic Acid Bacteria : Biodiversity and Taxonomy. Chichester, West Sussex, UK: Wiley-Blackwell; 2014.
- R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2021. Available from: https://www.R-project.org/.
- Bonatesta E, Horejs-Kainrath C, Bodenhofer U. msa: Multiple Sequence Alignment [Internet]. Bioconductor version: Release (3.17); 2023 [cited 2023 Oct 10]. Available from: https://bioconductor.org/packages/msa/.
- Charif D, Clerc O, Frank C, Lobry JR, Necşulea A, Palmeira L, et al. seqinr: Biological Sequences Retrieval and Analysis [Internet]. 2023 [cited 2023 Oct 10]. Available from: https://cran.r-project.org/web/packages/seqinr/index.html.
- Paradis E, Blomberg S, Bolker [aut B, cph, Brown J, Claramunt S, et al. ape: Analyses of Phylogenetics and Evolution [Internet]. 2023 [cited 2023 Oct 10]. Available from: https://cran.r-project.org/web/packages/ape/index.html.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul 1;4(4):406–25. [CrossRef]
- Yu G, Lam TTY, Xu S, Li L, Jones B, Silverman J, et al. ggtree: an R package for visualization of tree and annotation data [Internet]. Bioconductor version: Release (3.17); 2023 [cited 2023 Oct 10]. Available from: https://bioconductor.org/packages/ggtree/.
- Dinov ID, Rubin D, Lorensen W, Dugan J, Ma J, Murphy S, et al. iTools: A Framework for Classification, Categorization and Integration of Computational Biology Resources. PLOS ONE. 2008 May 28;3(5):e2265. [CrossRef]
- Kolde R. pheatmap: Pretty Heatmaps [Internet]. 2019 [cited 2023 Aug 31]. Available from: https://cran.r-project.org/web/packages/pheatmap/index.html.
- Correia Santos S, Fraqueza MJ, Elias M, Salvador Barreto A, Semedo-Lemsaddek T. Traditional dry smoked fermented meat sausages: Characterization of autochthonous enterococci. LWT - Food Sci Technol. 2017 Jun 1;79:410–5. [CrossRef]
- Fraqueza MJ. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int J Food Microbiol. 2015 Nov 6;212:76–88. [CrossRef]
- Moracanin SV, Turubatovic L, Skrinjar M, Obradovic D. Antilisterial Activity of Bacteriocin Isolated from Leuconostoc mesenteroides ssp mesenteroides IMAU:10231 in the Production of Sremska Sausages: Lactic Acid Bacteria Isolation, Bacteriocin Identification and Meat Application Experiments. FOOD Technol Biotechnol. 2013 Jun;51(2):247–56.
- Franciosa I, Ferrocino I, Giordano M, Mounier J, Rantsiou K, Cocolin L. Specific metagenomic asset drives the spontaneous fermentation of Italian sausages. Food Res Int. 2021 Jun 1;144:110379. [CrossRef]
- Albano H, van Reenen CA, Todorov SD, Cruz D, Fraga L, Hogg T, et al. Phenotypic and genetic heterogeneity of lactic acid bacteria isolated from “Alheira”, a traditional fermented sausage produced in Portugal. Meat Sci. 2009;82(3):389–98.
- Patarata L, Judas I, Silva JA, Esteves A, Martins C. A comparison of the physicochemical and sensory characteristics of alheira samples from different-sized producers. Meat Sci. 2008 May 1;79(1):131–8. [CrossRef]
- Joint FAO/WHO Expert Consultation on. 2001;
- Fernandes N, Faria AS, Carvalho L, Choupina A, Rodrigues C, Cadavez V, et al. Molecular Identification of Lactic Acid Producing Bacteria Isolated from Alheira, a Traditional Portuguese Fermented Sausage. Biol Life Sci Forum. 2022;18(1):73.
- Albano H, Henriques I, Correia A, Hogg T, Teixeira P. Characterization of microbial population of ‘Alheira’(a traditional Portuguese fermented sausage) by PCR-DGGE and traditional cultural microbiological methods. J Appl Microbiol. 2008;105(6):2187–94. [CrossRef]
- Albano H, van Reenen CA, Todorov SD, Cruz D, Fraga L, Hogg T, et al. Phenotypic and genetic heterogeneity of lactic acid bacteria isolated from “Alheira”, a traditional fermented sausage produced in Portugal. Meat Sci. 2009 Jul;82(3):389–98.
- Franz CMAP, Stiles ME, Schleifer KH, Holzapfel WH. Enterococci in foods—a conundrum for food safety. Int J Food Microbiol. 2003 Dec 1;88(2):105–22. [CrossRef]
- Liu Y, Wan Z, Yohannes KW, Yu Q, Yang Z, Li H, et al. Functional Characteristics of Lactobacillus and Yeast Single Starter Cultures in the Ripening Process of Dry Fermented Sausage. Front Microbiol [Internet]. 2021 [cited 2023 Sep 1];11. [CrossRef]
- Worsztynowicz P, Schmidt AO, Białas W, Grajek W. Identification and partial characterization of proteolytic activity of Enterococcus faecalis relevant to their application in dairy industry. Acta Biochim Pol. 2019 Feb 3;66(1):61–9. [CrossRef]
- Hugas M. Bacteriocinogenic lactic acid bacteria for the biopreservation of meat and meat products. Meat Sci. 1998 Jan 1;49:S139–50.
- O’Sullivan L, Ross RP, Hill C. Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie. 2002 May 1;84(5):593–604. [CrossRef]
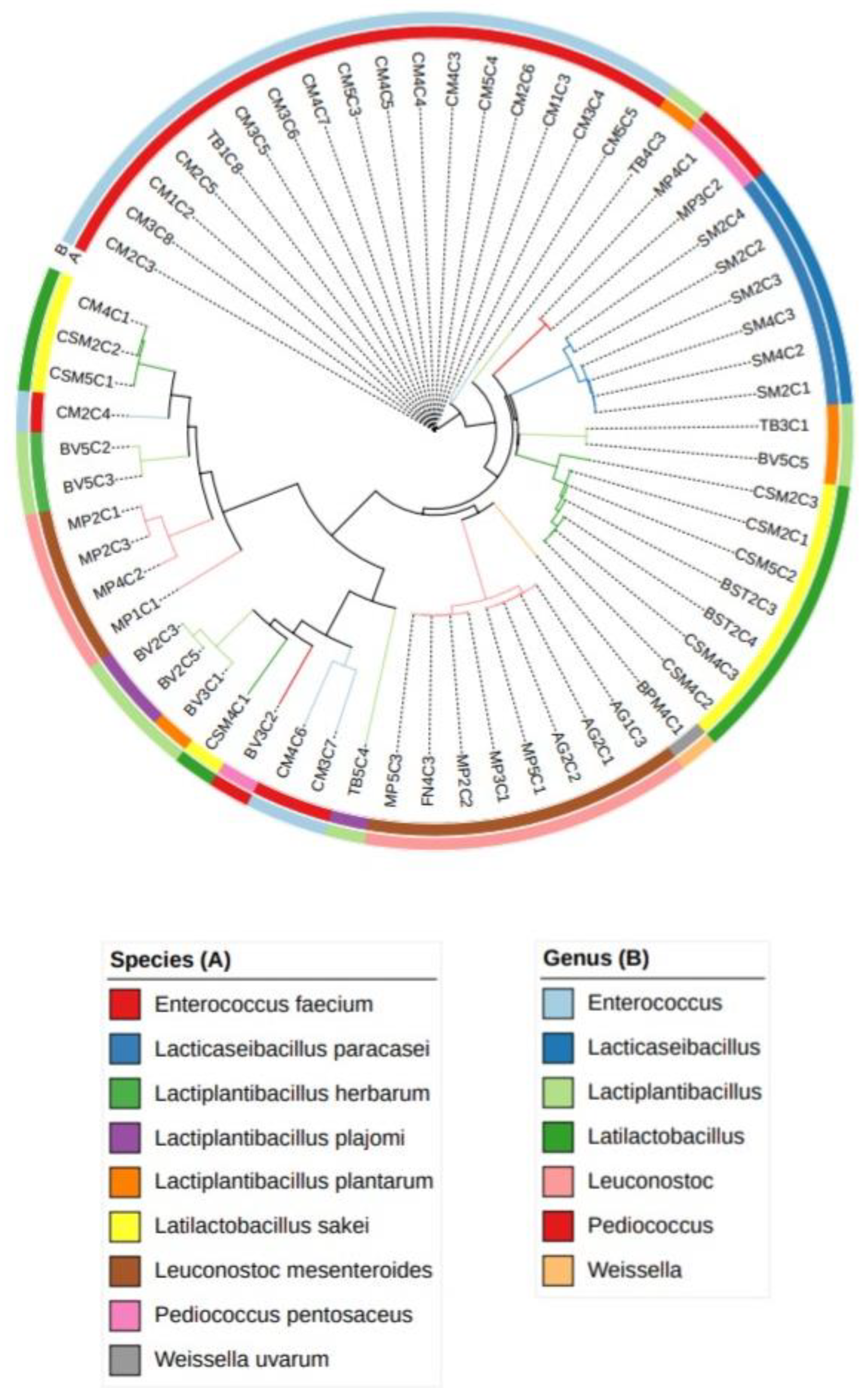
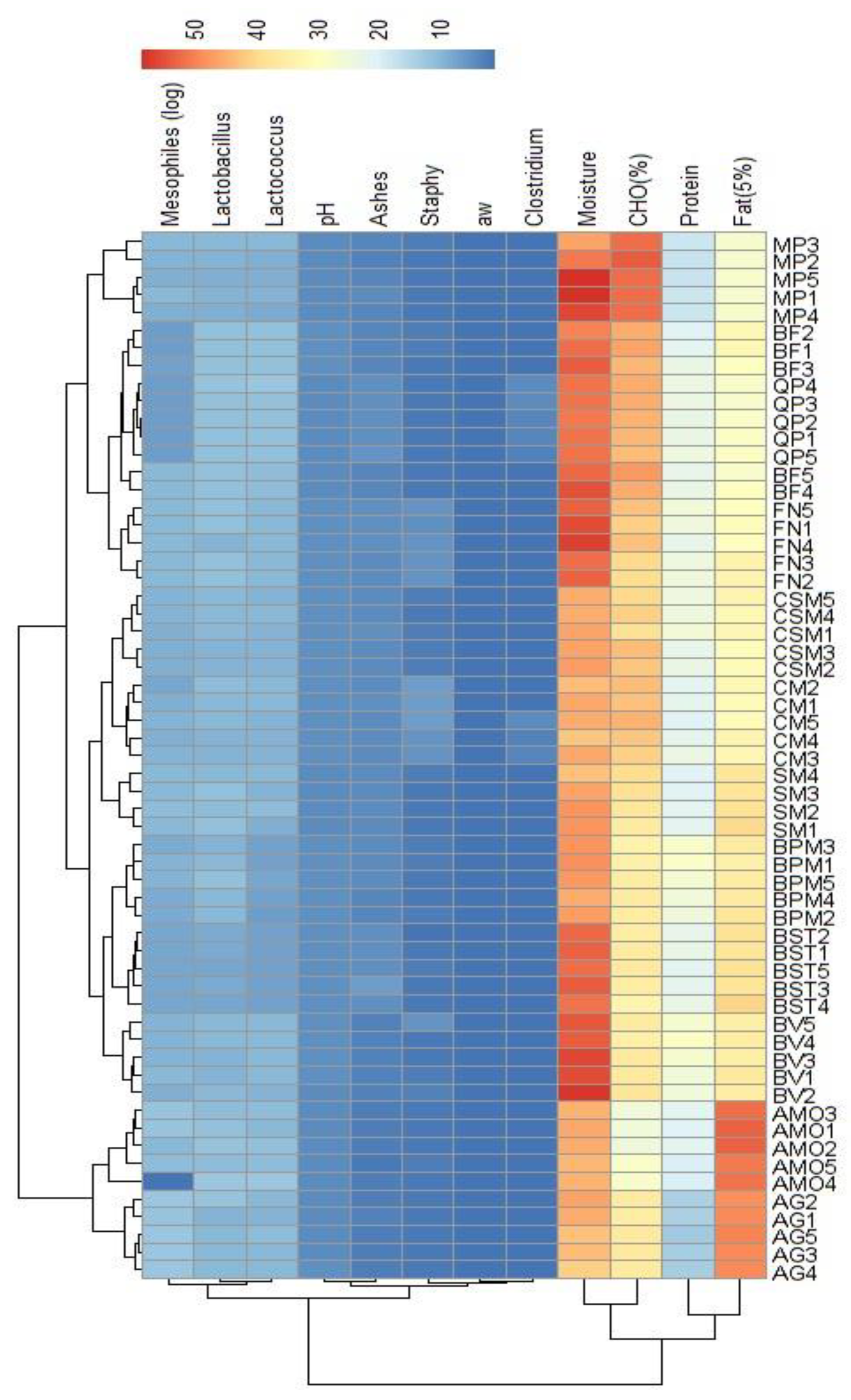
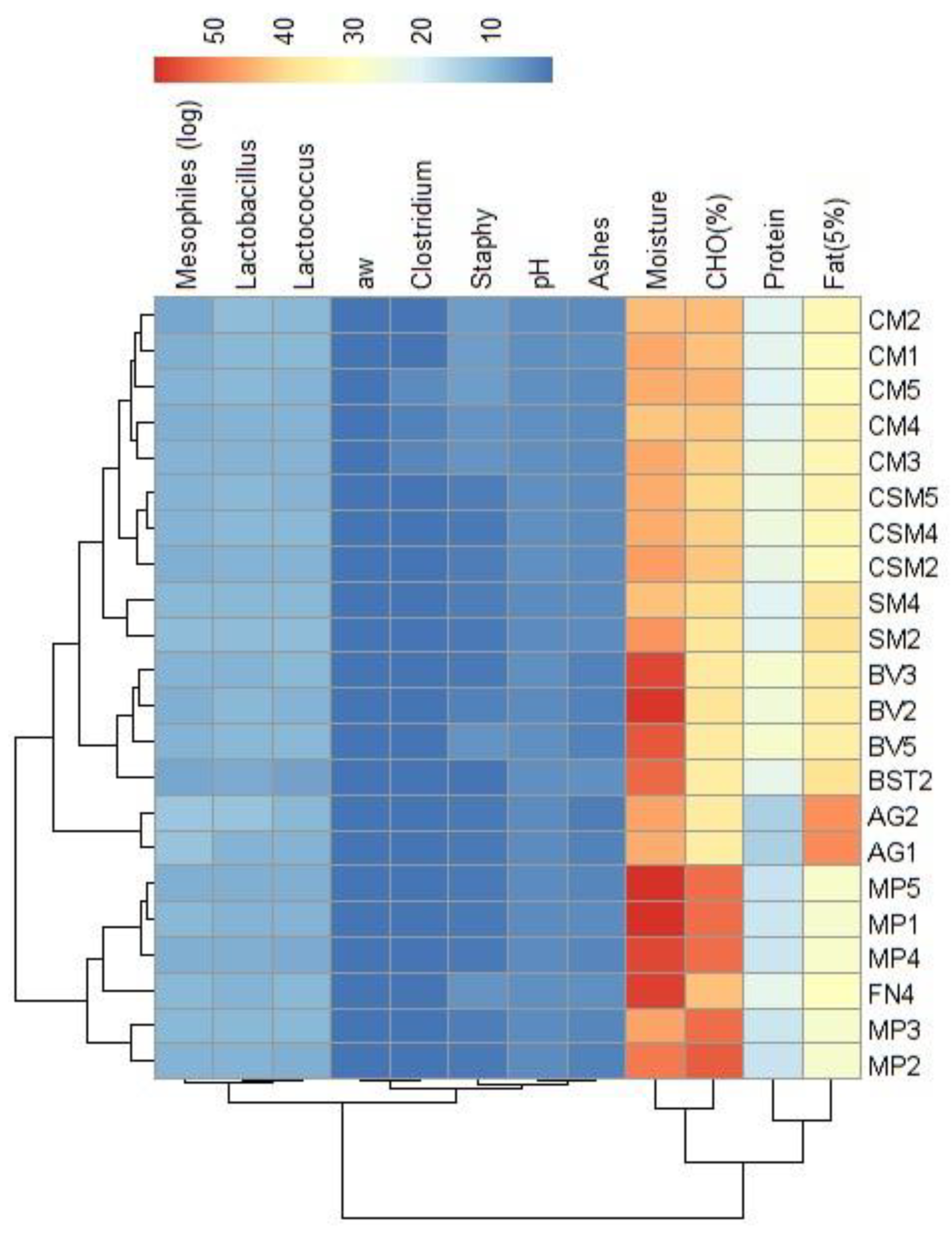
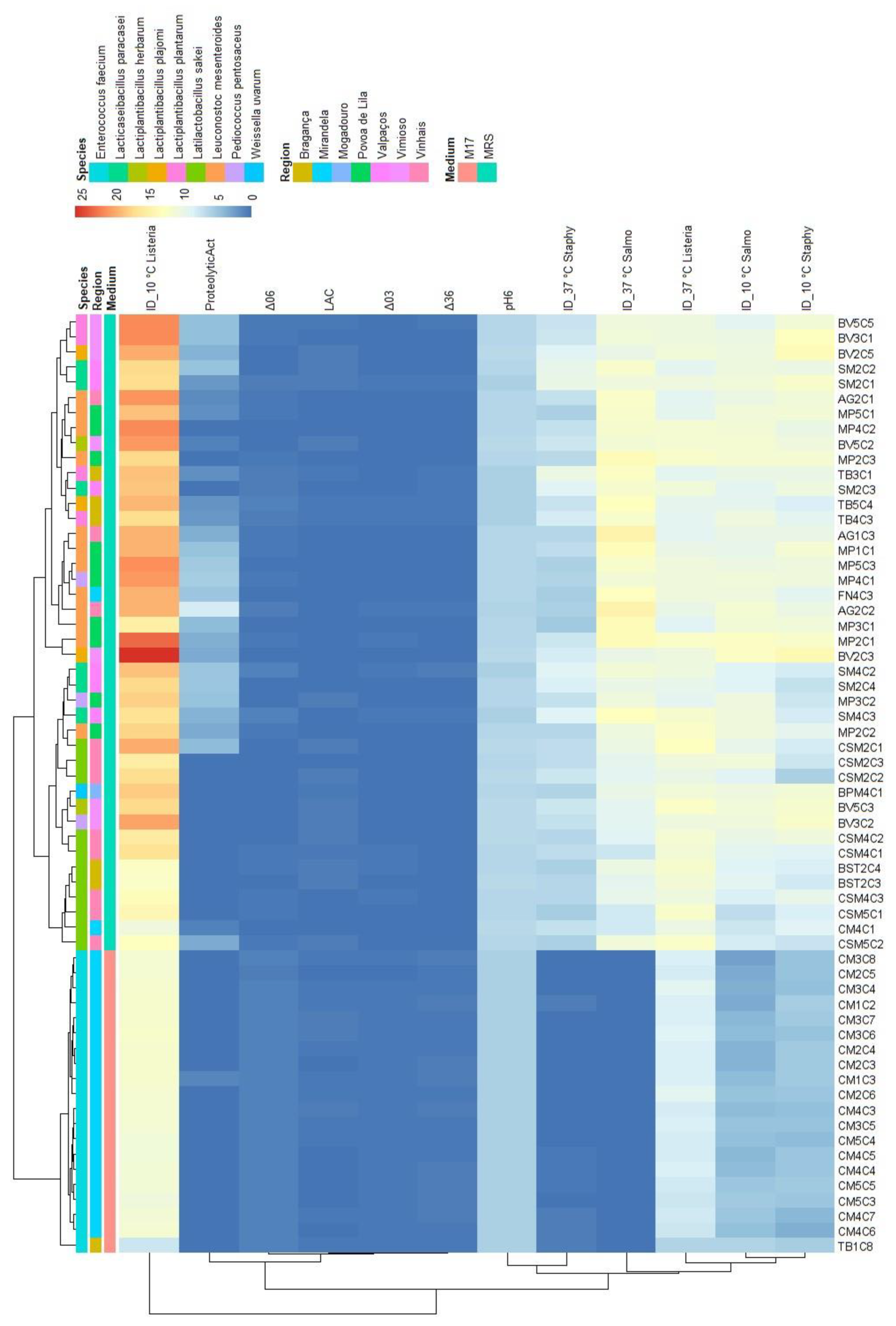
| N. isolates | Species | GenBank_ID | Identity (%) |
|---|---|---|---|
| 20 | Enterococcus faecium | NR_115764.1 | 99.2 [98.4; 100.0] |
| 6 | Lacticaseibacillus paracasei | NR_025880.1 | 100.0 [100; 100.0] |
| 2 | Lactiplantibacillus herbarum | NR_145899.1 | 99.5 [99.3; 99.6] |
| 3 | Lactiplantibacillus plajomi | NR_136785.1 | 98.9 [98; 99.8] |
| 4 | Lactiplantibacillus plantarum | NR_042394.1 | 98.5 [97; 100.0] |
| 11 | Latilactobacillus sakei | NR_115172.1 | 99.8 [99.6; 100.0] |
| 4 8 |
Leuconostoc mesenteroides | NR_074957.1 NR_157602.1 |
98.5 [97; 100.0] 99.9 [99.8;100.0] |
| 3 | Pediococcus pentosaceus | NR_042058.1 | 99.5 [99.4; 99.6] |
| 1 | Weissela uvarum | NR_134229.1 | 100.0 |
| Variables | S. Typhimurium | L. monocytogenes | S. aureus | |||
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |
| ProteolyticAct | 0.538 | 0.609 | 0.494 | 0.653 | 0.508 | 0.612 |
| pH6 | 0.942 | -0.232 | 0.960 | -0.159 | 0.950 | -0.175 |
| ΔpH03 | -0.896 | 0.212 | -0.903 | 0.137 | -0.901 | 0.173 |
| ΔpH06 | -0.956 | 0.225 | -0.968 | 0.152 | -0.961 | 0.173 |
| ΔpH36 | -0.927 | 0.218 | -0.940 | 0.149 | -0.931 | 0.162 |
| LAC | -0.068 | -0.771 | -0.032 | -0.783 | -0.032 | -0.812 |
| ID_10C | 0.884 | 0.209 | 0.758 | 0.387 | 0.834 | 0.218 |
| ID_37C | 0.872 | 0.299 | 0.784 | -0.099 | 0.834 | 0.143 |
| % of variance | 66.3 | 16.2 | 62.4 | 16.1 | 64.4 | 15.2 |
| Cumulative % of var. | 66.3 | 82.4 | 62.4 | 78.5 | 64.4 | 79.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





