Submitted:
08 October 2023
Posted:
09 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Viral Sequences and Metadata Information
2.2. Hierarchical Cluster Analysis
2.3. Phylogenetic Analysis
2.4. Evolutionary Analysis
3. Results
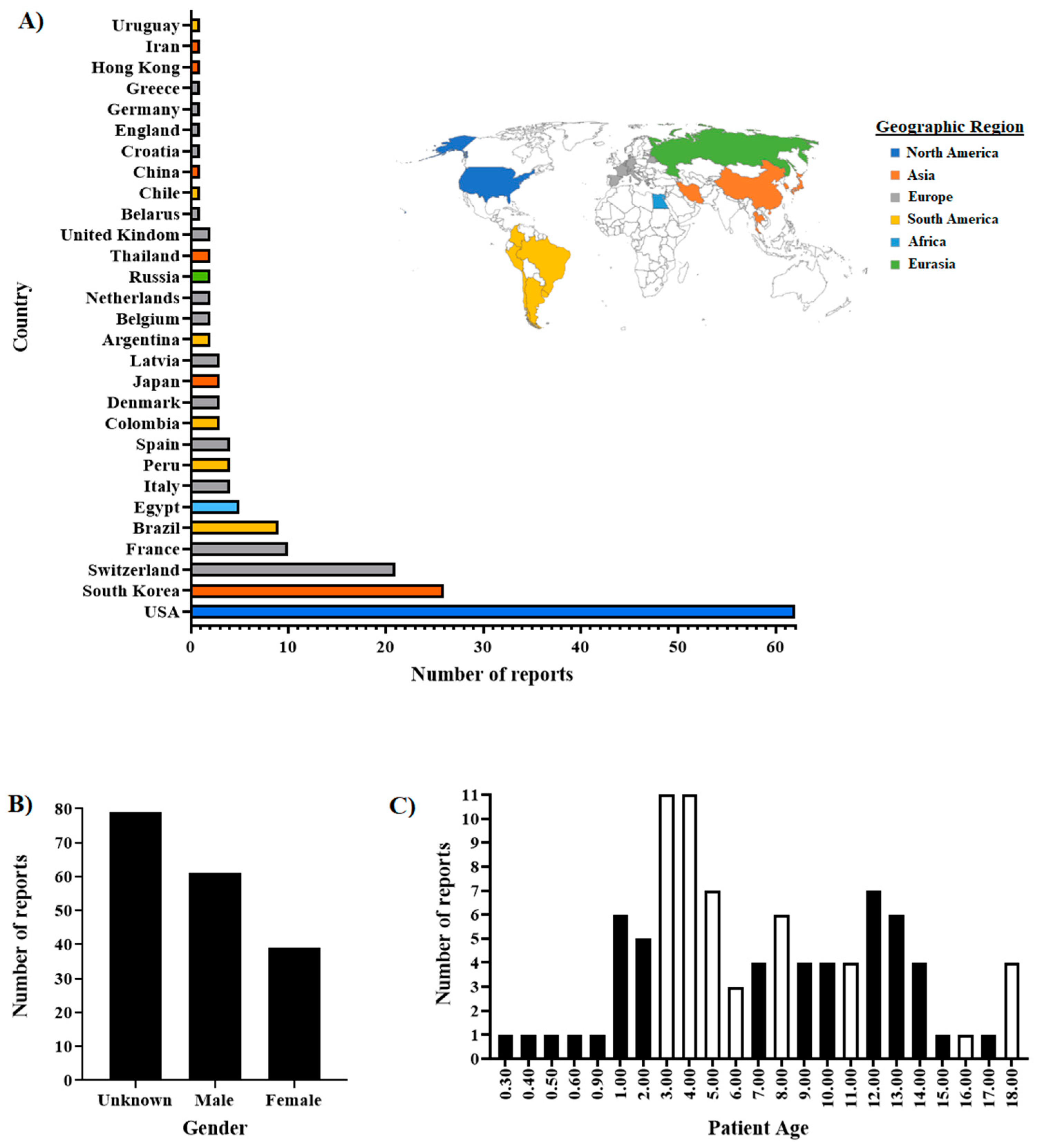
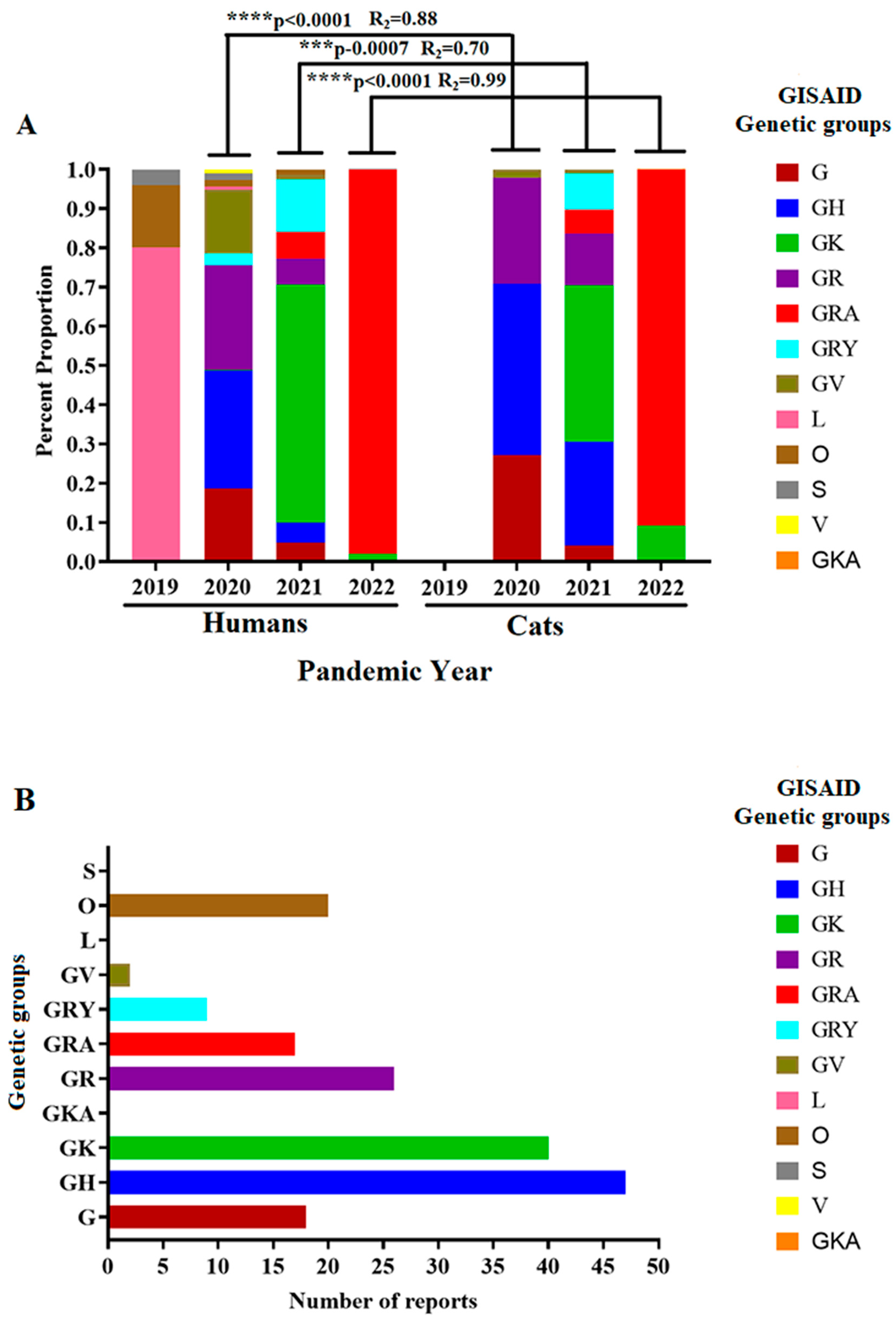
3.1. Overview of the SARS-CoV-2 Lineages Isolated from Cat Infections Around the World
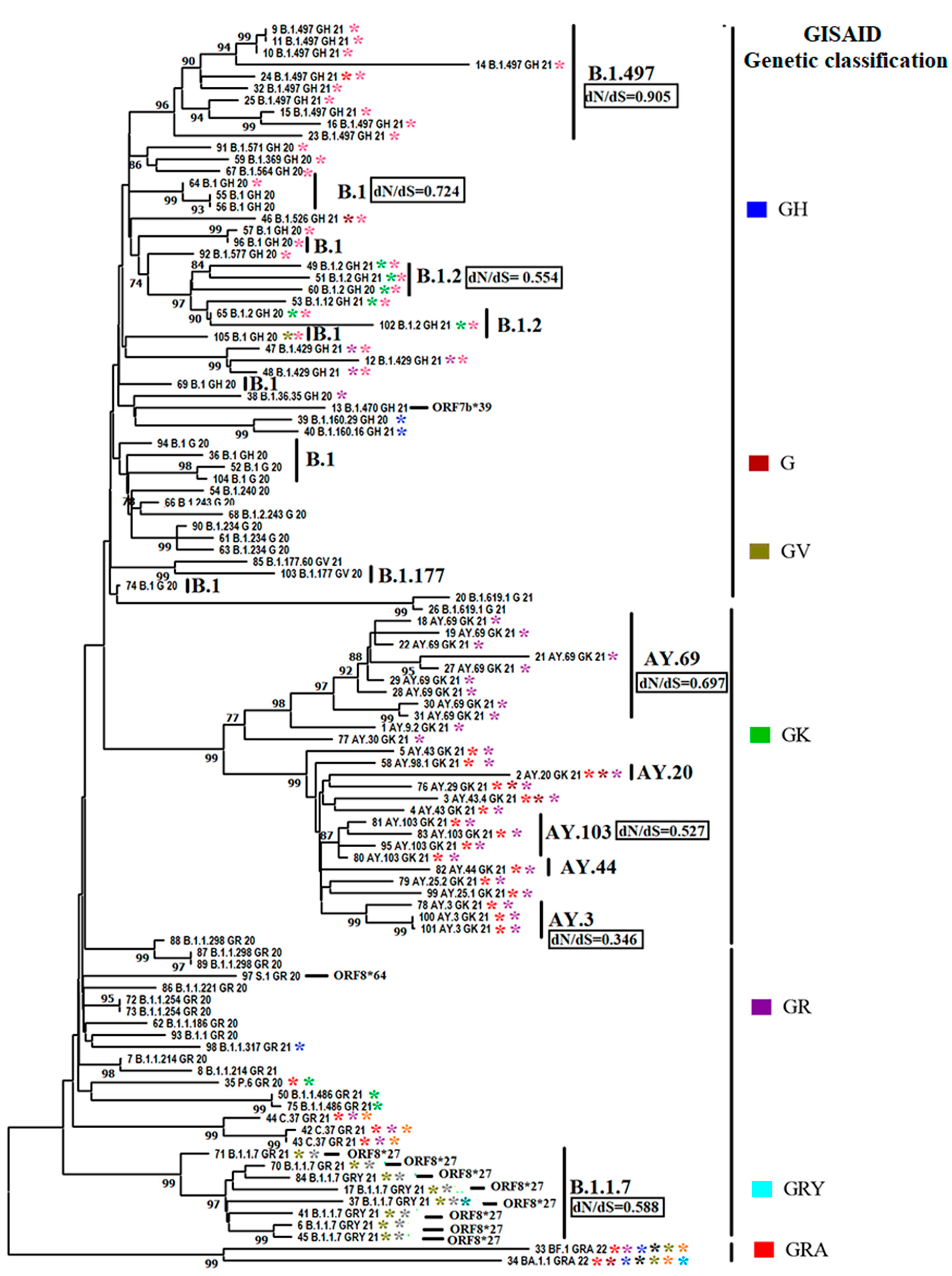
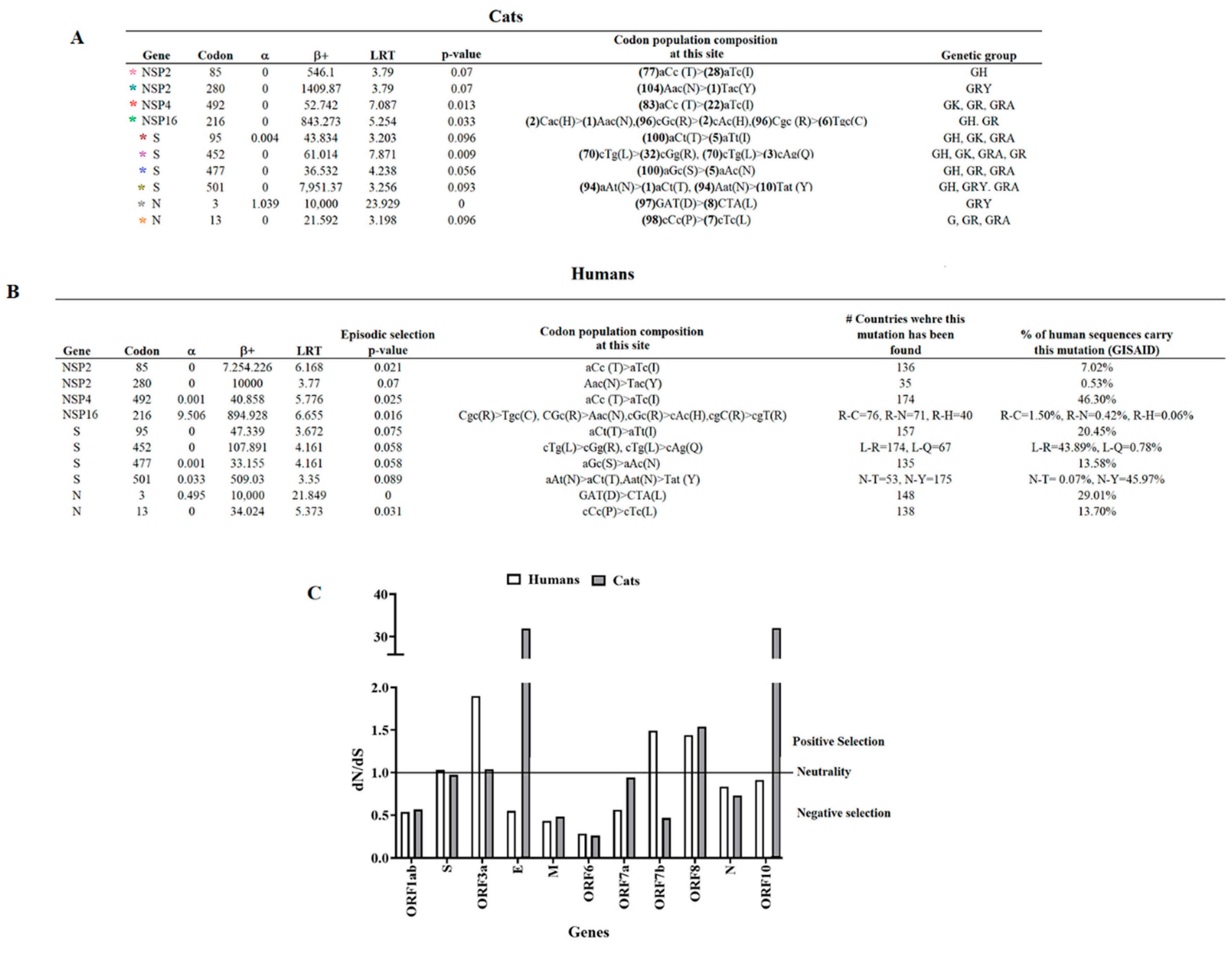
3.2. Evolutionary Characteristics of Viral Lineages Affecting Cat Populations
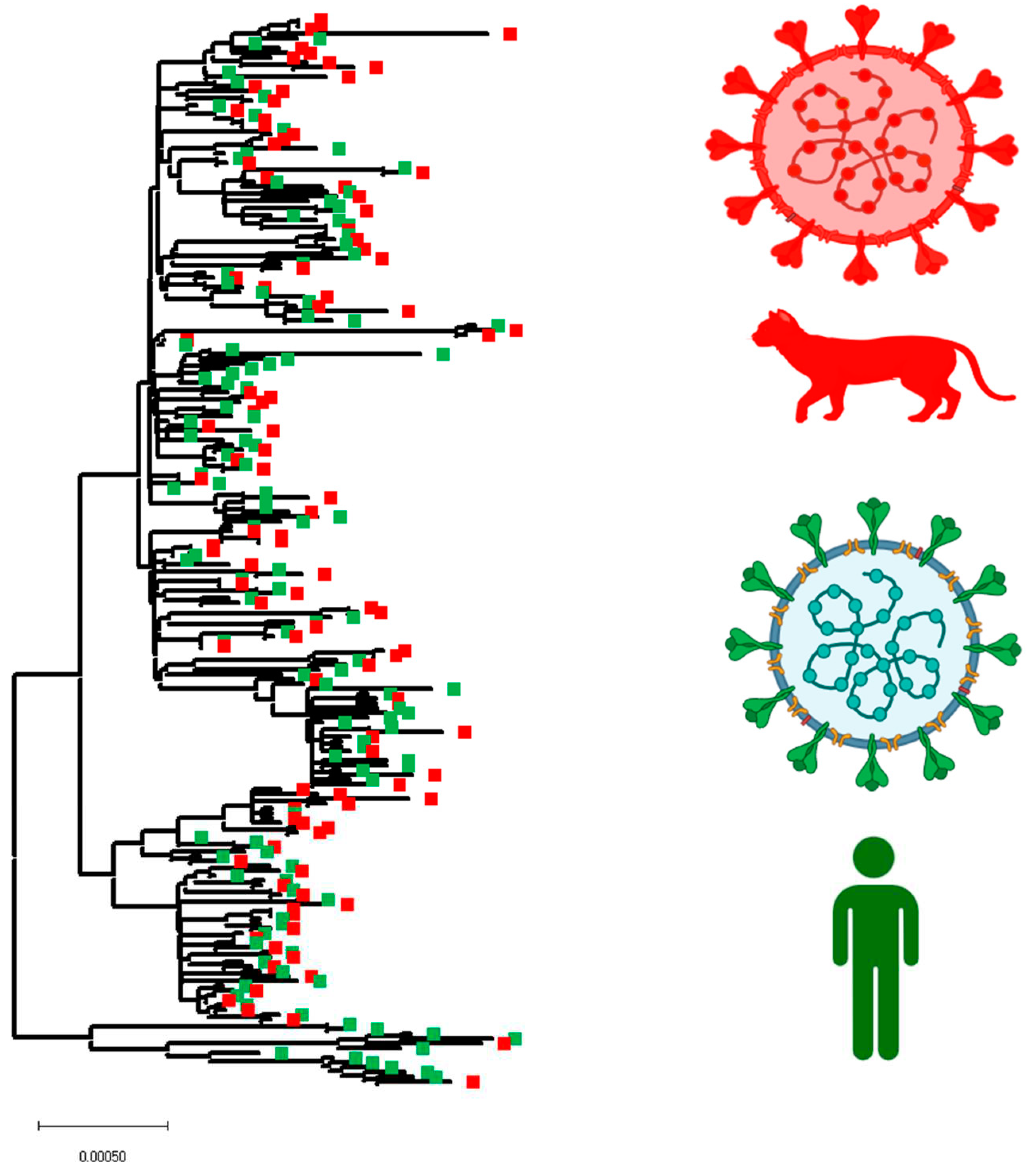
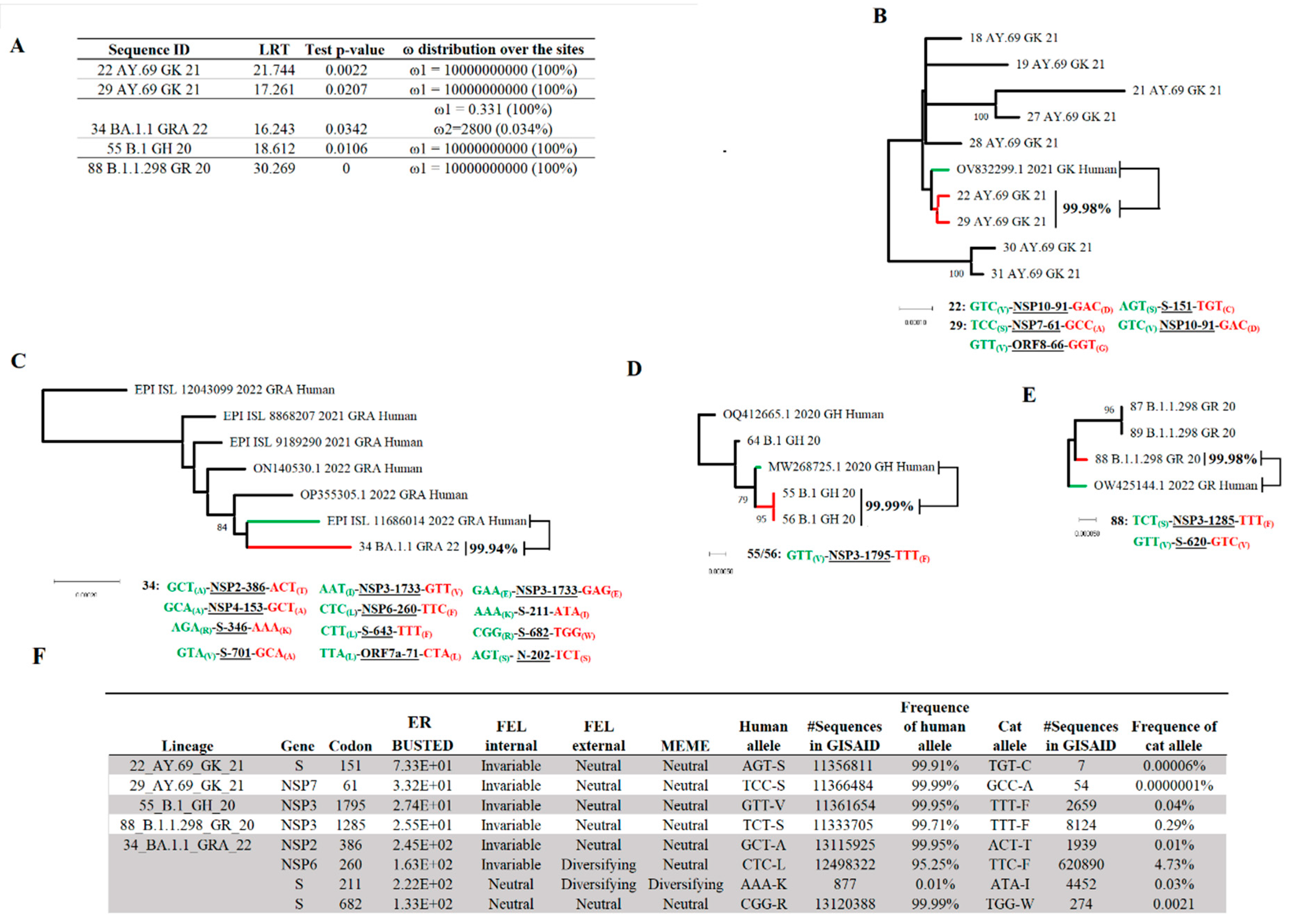
3.3. Evaluating the Hypothesis about Independent Evolution of SARS-CoV-2 in Cat Populations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velazquez-Salinas, L.; Zarate, S.; Eberl, S.; Gladue, D.P.; Novella, I.; Borca, M.V. Positive Selection of ORF1ab, ORF3a, and ORF8 Genes Drives the Early Evolutionary Trends of SARS-CoV-2 During the 2020 COVID-19 Pandemic. Front Microbial. 2020, 11, 550674. [Google Scholar] [CrossRef]
- Martin, D.P.; Weaver, S.; Tegally, H.; San, J.E.; Shank, S.D.; Wilkinson, E.; Lucaci, A.G.; Giandhari, J.; Naidoo, S.; Pillay, Y.; et al. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell 2021, 184, 5189–5200. [Google Scholar] [CrossRef]
- O'Toole, A.; Pybus, O.G.; Abram, M.E.; Kelly, E.J.; Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genomics 2022, 23, 121. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L. The complex evolutionary dynamics of SARS-CoV-2, a big challenge to control the pandemic of COVID-19. J. Med. Virol. 2022, 94, 5082–5085. [Google Scholar] [CrossRef]
- Lenharo, M. WHO declares end to COVID-19's emergency phase. Nature 2023. [Google Scholar] [CrossRef]
- Mahdy, M.A.A.; Younis, W.; Ewaida, Z. An Overview of SARS-CoV-2 and Animal Infection. Front. Vet. Sci. 2020, 7, 596391. [Google Scholar] [CrossRef]
- Prince, T.; Smith, S.L.; Radford, A.D.; Solomon, T.; Hughes, G.L.; Patterson, E.I. SARS-CoV-2 Infections in Animals: Reservoirs for Reverse Zoonosis and Models for Study. Viruses 2021, 13, 494. [Google Scholar] [CrossRef]
- Navarro-Lopez, R.; Solis-Hernandez, M.; Rocha-Martinez, M.K.; Eberl, S.; Gomez-Romero, N.; Velazquez-Salinas, L.; Estrada-Franco, J.G. Near-Full-Length Genome Sequences Representing an Event of Zooanthroponotic Transmission of SARS-CoV-2 Lineage B.1.189 in Mexico during 2020. Microbiol Resour Announc 2022, 11, e0049722. [Google Scholar] [CrossRef]
- Koeppel, K.N.; Mendes, A.; Strydom, A.; Rotherham, L.; Mulumba, M.; Venter, M. SARS-CoV-2 Reverse Zoonoses to Pumas and Lions, South Africa. Viruses 2022, 14, 120. [Google Scholar] [CrossRef]
- Cool, K.; Gaudreault, N.N.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; McDowell, C.; Carossino, M.; Bold, D.; Mitzel, D.; Kwon, T.; et al. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg. Microbes Infect. 2022, 11, 95–112. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Neira, V.; Brito, B.; Agüero, B.; Berrios, F.; Valdés, V.; Gutierrez, A.; Ariyama, N.; Espinoza, P.; Retamal, P.; Holmes, E.C.; et al. A household case evidences shorter shedding of SARS-CoV-2 in naturally infected cats compared to their human owners. Emerg. Microbes Infect. 2021, 10, 376–383. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Martins, M.; do Nascimento, G.M.; Nooruzzaman, M.; Yuan, F.; Chen, C.; Caserta, L.C.; Miller, A.D.; Whittaker, G.R.; Fang, Y.; Diel, D.G. The Omicron Variant BA.1.1 Presents a Lower Pathogenicity than B.1 D614G and Delta Variants in a Feline Model of SARS-CoV-2 Infection. J. Virol. 2022, 96, e0096122. [Google Scholar] [CrossRef]
- Zoccola, R.; Beltramo, C.; Magris, G.; Peletto, S.; Acutis, P.; Bozzetta, E.; Radovic, S.; Zappulla, F.; Porzio, A.M.; Gennero, M.S.; et al. First detection of an Italian human-to-cat outbreak of SARS-CoV-2 Alpha variant - lineage B.1.1.7. One Health 2021, 100295. [Google Scholar] [CrossRef]
- Schiaffino, F.; Ferradas, C.; Jara, L.M.; Salvatierra, G.; Dávila-Barclay, A.; Sanchez-Carrion, C.; Ulloa, A.; Mascaro, L.; Pajuelo, M.J.; Guevara Sarmiento, L.; et al. First Detection and Genome Sequencing of SARS-CoV-2 Lambda (C.37) Variant in Symptomatic Domestic Cats in Lima, Peru. Front. Vet. Sci. 2021, 8, 737350. [Google Scholar] [CrossRef]
- Kuhlmeier, E.; Chan, T.; Agüí, C.V.; Willi, B.; Wolfensberger, A.; Beisel, C.; Topolsky, I.; Beerenwinkel, N.; Stadler, T. ; Swiss Sars-CoV-Sequencing Consortium; Jones, S.; et al. Detection and Molecular Characterization of the SARS-CoV-2 Delta Variant and the Specific Immune Response in Companion Animals in Switzerland. Viruses. [CrossRef]
- Ferasin, L.; Fritz, M.; Ferasin, H.; Becquart, P.; Corbet, S.; Ar Gouilh, M.; Legros, V.; Leroy. E.M. Infection with SARS-CoV-2 variant B.1.1.7 detected in a group of dogs and cats with suspected myocarditis. Vet. Rec. 2021, 189, e944. [Google Scholar] [CrossRef]
- Bashor, L.; Gagne, R.B.; Bosco-Lauth, A.; Stenglein, M.; VandeWoude, S. Rapid evolution of SARS-CoV-2 in domestic cats. Virus Evol. 2022, 8, veac092. [Google Scholar] [CrossRef]
- Bashor, L.; Gagne, R.B.; Bosco-Lauth, A.M.; Bowen, R.A.; Stenglein, M.; VandeWoude, S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl. Acad. Sci. USA 2021, 118, e2105253118. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W. GISAID's Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Smith, M.D.; Wertheim, J.O.; Weaver, S.; Murrell, B.; Scheffler, K.; Kosakovsky Pond, S.L. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015, 32, 1342–1353. [Google Scholar] [CrossRef]
- Murrell, B.; Weaver, S.; Smith, M.D.; Wertheim, J.O.; Murrell, S.; Aylward, A.; Eren, K.; Pollner, T.; Martin, D.P.; Smith, D.M.; et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015, 32, 1365–1371. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Gonzales, J.L.; de Jong, M.C.M.; Gerhards, N.M.; Van der Poel, W.H.M. The SARS-CoV-2 Reproduction Number R0 in Cats. Viruses 2021, 13, 2480. [Google Scholar] [CrossRef]
- Gerhards, N.M.; Gonzales, J.L.; Vreman, S.; Ravesloot, L.; van den Brand, J.M.A.; Doekes, H.P.; Egberink, H.F.; Stegeman, A.; Oreshkova, N.; van der Poel, W.H.M.; et al. Efficient Direct and Limited Environmental Transmission of SARS-CoV-2 Lineage B.1.22 in Domestic Cats. Microbiol. Spectr. 2023, 11, e0255322. [Google Scholar] [CrossRef]
- Han, A.X.; Parker, E.; Scholer, F.; Maurer-Stroh, S.; Russell, C.A. Phylogenetic Clustering by Linear Integer Programming (PhyCLIP). Mol. Biol. Evol. 2019, 36, 1580–1595. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L. Deciphering the evolutionary mechanisms of SARS-CoV-2: Absence of ORF8 protein and its potential advantage in the emergence of viral lineages. J. Med. Virol. 2023, 95, e29002. [Google Scholar] [CrossRef]
- Braun, K.M.; Moreno, G.K.; Halfmann, P.J.; Hodcroft, E.B.; Baker, D.A.; Boehm, E.C.; Weiler, A.M.; Haj, A.K.; Hatta, M.; Chiba, S.; et al. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog. 2021, 17, e1009373. [Google Scholar] [CrossRef]
- Lin, X.; Sha, Z.; Trimpert, J.; Kunec, D.; Jiang, C.; Xiong, Y.; Xu, B.; Zhu, Z.; Xue, W.; Wu, H. The NSP4 T492I mutation increases SARS-CoV-2 infectivity by altering non-structural protein cleavage. Cell Host Microbe 2023, 31, 1170–1184. [Google Scholar] [CrossRef]
- Abduljaleel, Z.; Melebari, S.; Athar, M.; Dehlawi, S.; Udhaya Kumar, S.; Aziz, S.A.; Dannoun, A.I.; Malik, S.M.; Thasleem, J.; George Priya Doss, C. SARS-CoV-2 vaccine breakthrough infections (VBI) by Omicron variant (B.1.1.529) and consequences in structural and functional impact. Cell Signal 2023, 109, 110798. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H.D.; et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Crawford, K.H.D.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; Veesler, D.; Bloom, J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Guo, H.; Fan, Q.; Song, S.; Shen, S.; Zhou, B.; Wang, H.; Cheng, L.; Ge, X.; Ju, B.; Zhang, Z. Increased resistance of SARS-CoV-2 Lambda variant to antibody neutralization. J. Clin. Virol. 2022, 150–151, 105162. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, L.; Zhang, Y.; Wang, H.; Ding, R.; Nie, J.; Li, Q.; Liu, S.; Yu, Y.; Yang, X.; et al. The Antigenicity of Epidemic SARS-CoV-2 Variants in the United Kingdom. Front. Immunol. 2021, 12, 687869. [Google Scholar] [CrossRef]
- Wang, H.X.; Zhang, L.; Liang, Z.T.; Nie, J.H.; Wu, J.J.; Li, Q.Q.; Ding, R.X.; Zhang, Y.; Chen, G.Q.; Wang, Y.C.; et al. Infectivity and antigenicity of pseudoviruses with high-frequency mutations of SARS-CoV-2 identified in Portugal. Arch. Virol. 2022, 167, 459–470. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef]
- de Silva, T.I.; Liu, G.; Lindsey, B.B.; Dong, D.; Moore, S.C.; Hsu, N.S.; Shah, D.; Wellington, D.; Mentzer, A.J.; Angyal, A.; et al. COVID-19 Genomics UK (COG-UK) Consortium; Maini, M.K.; Ogg. G.; Knight, J.C.; ISARIC4C Investigators; Peng, Y.; Rowland-Jones, S.L.; Dong, T. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience 2021, 24, 103353. [Google Scholar] [CrossRef]
- Yalcinkaya, M.; Liu, W.; Islam, M.N.; Kotini, A.G.; Gusarova, G.A.; Fidler, T.P.; Papapetrou, E.P.; Bhattacharya, J.; Wang, N.; Tall, A.R. Modulation of the NLRP3 inflammasome by Sars-CoV-2 Envelope protein. Sci. Rep. 2021, 11, 24432. [Google Scholar] [CrossRef]
- Li, X.; Hou, P.; Ma, W.; Wang, X.; Wang, H.; Yu, Z.; Chang, H.; Wang, T.; Jin, S.; Wang, X.; et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol. Immunol. 2022, 19, 67–78. [Google Scholar] [CrossRef]
- Zhou, S.; Lv, P.; Li, M.; Chen, Z.; Xin, H.; Reilly, S.; Zhang, X. SARS-CoV-2 E protein: Pathogenesis and potential therapeutic development. Biomed. Pharmacother. 2023, 159, 114242. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



