Submitted:
27 September 2023
Posted:
30 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Traditional Approaches in Maize Crop Improvement
3. Molecular Breeding and Marker-Assisted Selection (MAS)
4. Genomic Selection
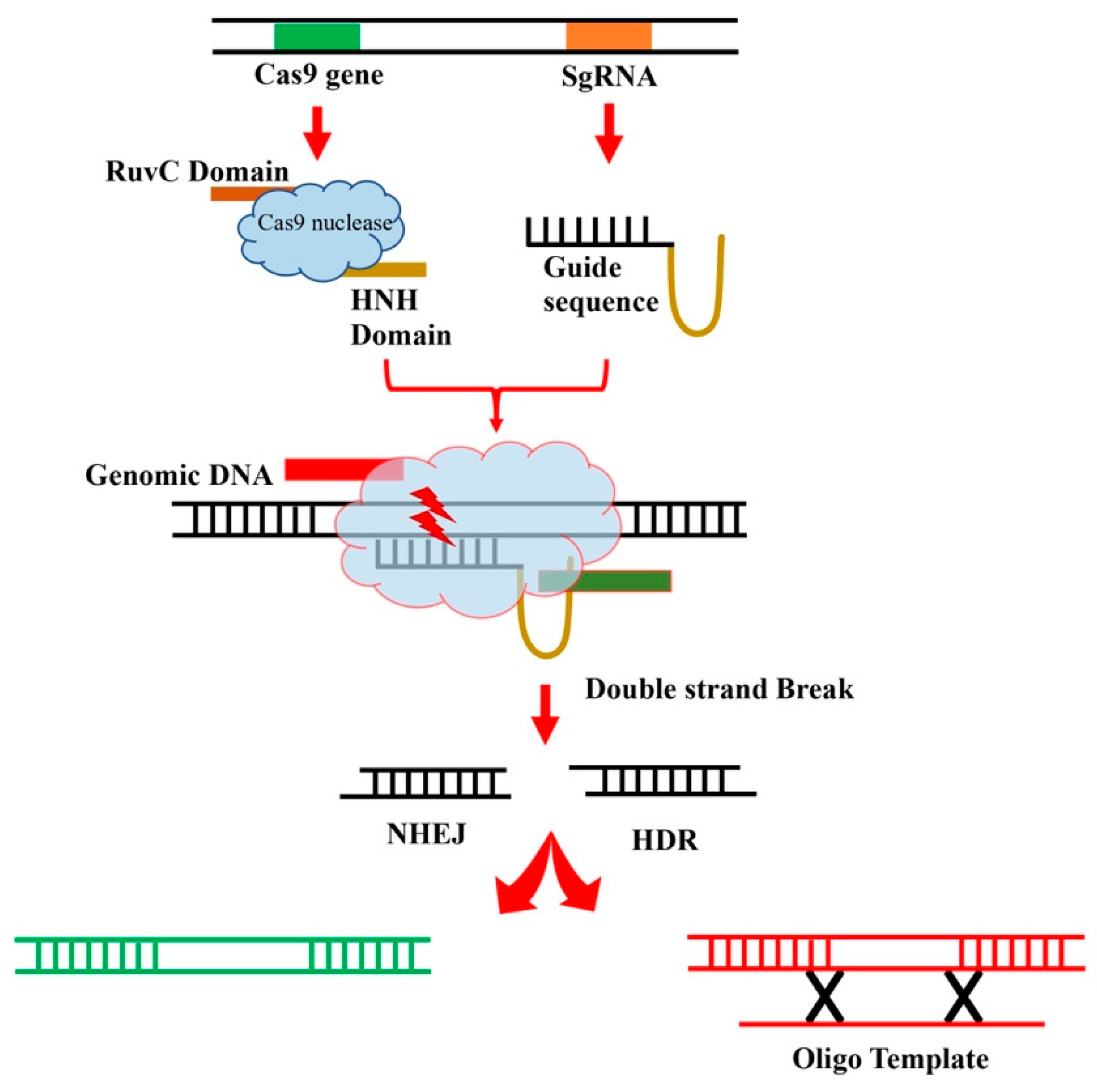
5. Genome Editing and CRISPR-Cas9
5.1. CRISPR-Cas9 in Maize
5.2. Potential Benefits of Genome Editing and Ethical Considerations
6. Use of Transgenic Approaches to Introduce Foreign Genes into Maize
6.1. Enhancing Traits in Genetically Modified (GM) Maize Varieties
6.2. Concerns and Regulations Related to GM Crops
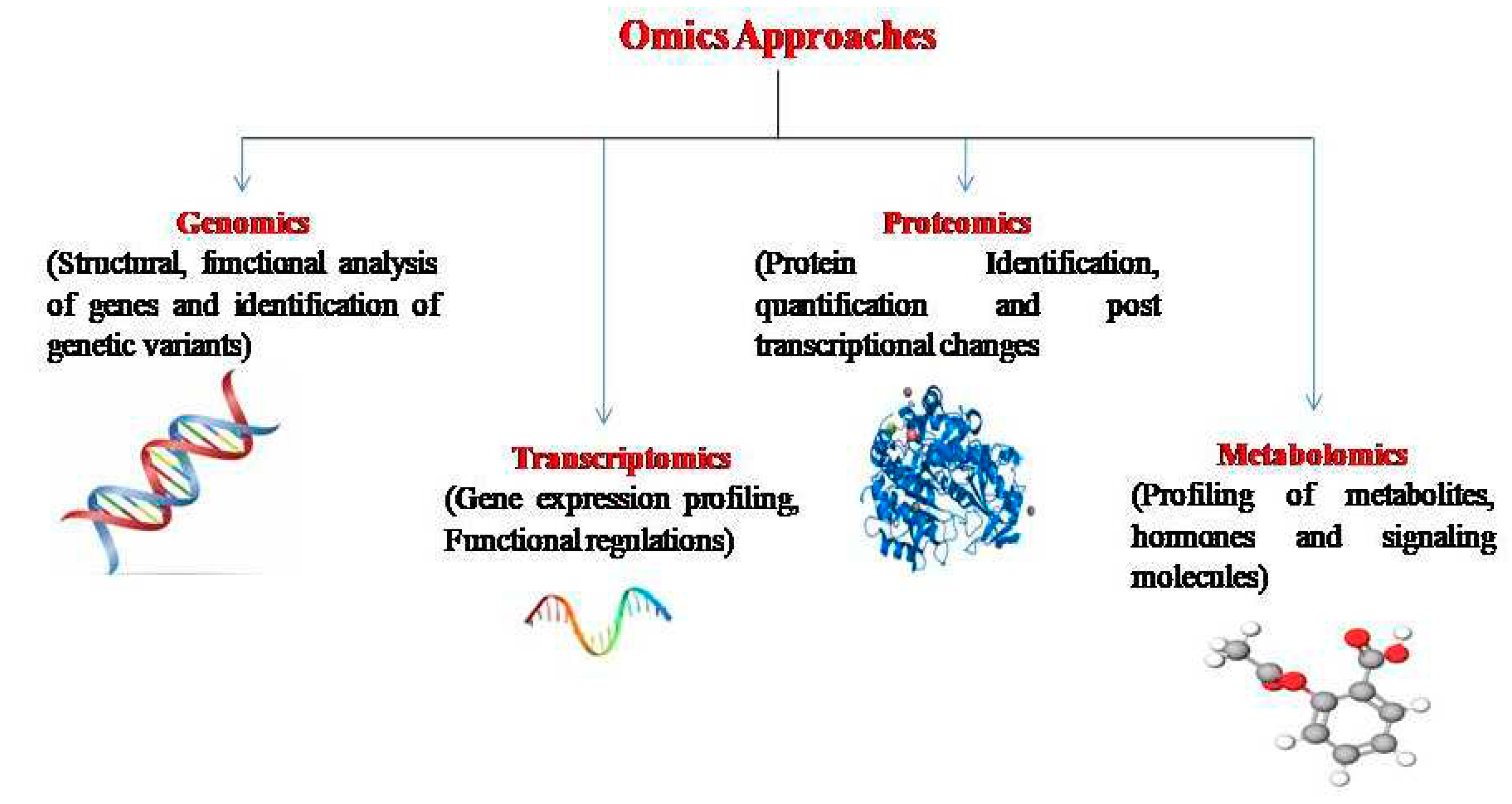
7. Omics Technologies in Maize Improvement
8. Conclusions
Conflicts of Interest
References
- Erenstein, O., Jaleta, M., Sonder, K., Mottaleb, K. and Prasanna, B.M. Global maize production, consumption and trade: Trends and R&D implications. Food Security 2022, 14, 1295–1319.
- Awika, J.M., 2011. Major cereal grains production and use around the world. In Advances in cereal science: implications to food processing and health promotion (pp. 1-13). American Chemical Society.
- FAOStat. (2022). FAO Stat. FAO, Rome. http:// www. fao. org/ faost at.
- Grote, U., Fasse, A., Nguyen, T.T. and Erenstein, O. Food security and the dynamics of wheat and maize value chains in Africa and Asia. Frontiers in Sustainable Food Systems 2021, 4, 617009. [CrossRef]
- Poole N, Donovan J, Erenstein O. Agri-nutrition research: Revisiting the contribution of maize and wheat to human nutrition and health. Food Policy. 2021, 100, 101976. [Google Scholar] [CrossRef]
- Serna-Saldivar SO, Perez-Carrillo E, Heredia-Olea E. Soybean-fortified wheat flour tortillas. InFlour and breads and their fortification in health and disease prevention 2019 Jan 1 (pp. 291-306). Academic Press.
- Tiwari, A., Choudhary, S., Padiya, J., Ubale, A., Mikkilineni, V. and Char, B., 2022. Recent Advances and Applicability of GBS, GWAS, and GS in Maize. Genotyping by Sequencing for Crop Improvement, pp.188-217.
- Walbot, V., 2008. Maize genome in motion.
- Schnable, P.S., Ware, D., Fulton, R.S., Stein, J.C., Wei, F., Pasternak, S., Liang, C., Zhang, J., Fulton, L., Graves, T.A. and Minx, P. The B73 maize genome: complexity, diversity, and dynamics. Science 2009, 326, 1112–1115.
- Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M.C., Wang, B., Campbell, M.S., Stein, J.C., Wei, X., Chin, C.S. and Guill, K. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527.
- Chapman, M.A., He, Y. and Zhou, M. Beyond a reference genome: pangenomes and population genomics of underutilized and orphan crops for future food and nutrition security. New Phytologist 2022, 234, 1583–1597. [CrossRef]
- Voss-Fels, K.P., Cooper, M. and Hayes, B.J. Accelerating crop genetic gains with genomic selection. Theoretical and Applied Genetics 2019, 132, 669–686. [CrossRef] [PubMed]
- Liu C, Li X, Meng D, Zhong Y, Chen C, Dong X, Xu X, Chen B, Li W, Li L, Tian X. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Molecular plant. 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium BP maize from the Central Balsas River Valley, Mexico. Proceedings of the National Academy of Sciences. 2009, 106, 5019–5024. [Google Scholar] [CrossRef]
- Ranere AJ, Piperno DR, Holst I, Dickau R, Iriarte J. The cultural and chronological context of early Holocene maize and squash domestication in the Central Balsas River Valley, Mexico. Proceedings of the National Academy of Sciences. 2009, 106, 5014–5018. [Google Scholar] [CrossRef] [PubMed]
- Alene AD, Menkir A, Ajala SO, Badu-Apraku B, Olanrewaju AS, Manyong VM, Ndiaye A. The economic and poverty impacts of maize research in West and Central Africa. Agricultural Economics. 2009, 40, 535–550. [Google Scholar] [CrossRef]
- Gedil M, Menkir A. An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Frontiers in Plant Science. 2019, 10, 1430. [Google Scholar] [CrossRef]
- Lamichhane S, Thapa S. Advances from conventional to modern plant breeding methodologies. Plant breeding and biotechnology. 2022, 10, 1–4. [Google Scholar] [CrossRef]
- Johannsen, W. Heredity in populations and pure lines. Classic papers in genetics. 1903, 20–26. [Google Scholar]
- Poehlman, JM. Breeding field crops. Springer Science & Business Media; 2013 Apr 17.
- Begna, T. Conventional Breeding Methods Widely used to Improve Self-Pollinated Crops. International Journal of Research. 2021, 7, 1–6. [Google Scholar]
- Brown J, Caligari P. An introduction to plant breeding. John Wiley & Sons; 2011 Aug 26.
- Aleksoski, J. The effect of backcross method in tobacco breeding. Journal of Agriculture and Plant Sciences 2018, 16, 9–19. [Google Scholar]
- Vogel, KE. Backcross breeding. Transgenic Maize: Methods and Protocols. 2009, 161–169.
- Singh, SP. Alternative methods to backcross breeding. Annu. Rep. Bean Improv. Coop. 1982, 25, 11–12. [Google Scholar]
- Hull, FH. Recurrent selection for specific combining ability in corn 1. Agronomy Journal. 1945, 37, 134–145. [Google Scholar] [CrossRef]
- Ramya P, Singh GP, Jain N, Singh PK, Pandey MK, Sharma K, Kumar A, Prabhu KV. Effect of recurrent selection on drought tolerance and related morpho-physiological traits in bread wheat. PloS one. 2016, 11, e0156869. [Google Scholar]
- Bangarwa, S. 2021. Recurrent Selection - Definition and Types. Biotecharticles.com. 19 October.
- Schlegel, RH. History of plant breeding. CRC press; 2017 Dec 15.
- Acquaah, G. Principles of plant genetics and breeding. John Wiley & Sons; 2009 Mar 12.
- Bharti G, Chimata M K. Review on New Plant Breeding Techniques. International Journal of Science and Research 2019, 8, 723–730. [Google Scholar]
- Lema, M. Marker assisted selection in comparison to conventional plant breeding. Agric Res Technol. 2018, 14, 555914. [Google Scholar]
- Muntean L, Ona A, Berindean I, Racz I, Muntean S. Maize Breeding: From Domestication to Genomic Tools. Agronomy. 2022, 12, 2365. [Google Scholar] [CrossRef]
- Allard, RW. Principles of plant breeding. Soil Science. 1961, 91, 414. [Google Scholar] [CrossRef]
- Breseghello F, Coelho AS. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). Journal of agricultural and food chemistry. 2013, 61, 8277–8286. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R. Prospects for using marker-assisted breeding to improve maize production in Africa. Journal of the Science of Food and Agriculture. 2008, 88, 745–755. [Google Scholar] [CrossRef]
- Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, Van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Molecular breeding. 1997, 3, 381–390. [Google Scholar] [CrossRef]
- Winter P, Kahl G. Molecular marker technologies for plant improvement. World Journal of Microbiology and Biotechnology. 1995, 11, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Mohan M, Nair S, Bhagwat A, Krishna TG, Yano M, Bhatia CR, Sasaki T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Molecular breeding. 1997, 3, 87–103. [Google Scholar] [CrossRef]
- Bhullar NK, Zhang Z, Wicker T, Keller B. Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC plant biology. 2010, 10, 1–3. [Google Scholar]
- Kumawat G, Kumawat CK, Chandra K, Pandey S, Chand S, Mishra UN, Lenka D, Sharma R. Insights into marker assisted selection and its applications in plant breeding. In Plant Breeding-Current and Future Views 2020 Nov 30. Intechopen.
- Ribaut JM, Hoisington D. Marker-assisted selection: new tools and strategies. Trends in Plant Science. 1998, 3, 236–239. [Google Scholar] [CrossRef]
- Hasan MM, Rafii MY, Ismail MR, Mahmood M, Rahim HA, Alam MA, Ashkani S, Malek MA, Latif MA. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnology & Biotechnological Equipment. 2015, 29, 237–254. [Google Scholar]
- Ribaut JM, Ragot M. Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives. Journal of experimental botany. 2007, 58, 351–360. [Google Scholar]
- Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Yang X, Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nature genetics. 2016, 48, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Prasanna BM, Palacios-Rojas N, Hossain F, Muthusamy V, Menkir A, Dhliwayo T, Ndhlela T, San Vicente F, Nair SK, Vivek BS, Zhang X. Molecular breeding for nutritionally enriched maize: status and prospects. Frontiers in genetics. 2020, 10, 1392. [Google Scholar] [CrossRef]
- Bhatt V, Muthusamy V, Jha S, Zunjare RU, Baveja A, Dosad S, Hossain F. Development of low phytic acid maize through marker assisted introgression of lpa1-1 and lpa2-1 genes. In Abstracts: 13th Asian Maize Conference on and Expert Consultation on Maize for Food, Feed, Nutrition and Environmental Security, Ludhiana, India, -10 2018 Oct 8 (pp. 143-144). 8 October.
- Goswami R, Zunjare R, Khan S, Baveja A, Muthusamy V, Hossain F. Marker-assisted introgression of rare allele of crtRB1 gene into elite quality protein maize inbred for combining high lysine, tryptophan and provitamin A in maize. Plant Breed. 2019, 138, 174–183. [Google Scholar] [CrossRef]
- Mehta BK, Muthusamy V, Zunjare RU, Baveja A, Chauhan HS, Chhabra R, Singh AK, Hossain F. Biofortification of sweet corn hybrids for provitamin-A, lysine and tryptophan using molecular breeding. Journal of Cereal Science. 2020, 96, 103093. [Google Scholar] [CrossRef]
- Prasanna BM, Palacios-Rojas N, Hossain F, Muthusamy V, Menkir A, Dhliwayo T, Ndhlela T, San Vicente F, Nair SK, Vivek BS, Zhang X. Molecular breeding for nutritionally enriched maize: status and prospects. Frontiers in genetics. 2020, 10, 1392. [Google Scholar] [CrossRef]
- Baveja A, Muthusamy V, Panda KK, Zunjare RU, Das AK, Chhabra R, Mishra SJ, Mehta BK, Saha S, Hossain F. Development of multinutrient-rich biofortified sweet corn hybrids through genomics-assisted selection of shrunken2, opaque2, lcyE and crtRB1 genes. Journal of Applied Genetics. 2021, 62, 419–429. [Google Scholar] [CrossRef]
- Das AK, Gowda MM, Muthusamy V, Zunjare RU, Chauhan HS, Baveja A, Bhatt V, Chand G, Bhat JS, Guleria SK, Saha S. Development of maize hybrids with enhanced vitamin-E, vitamin-A, lysine, and tryptophan through molecular breeding. Frontiers in plant science. 2021, 12, 659381. [Google Scholar] [CrossRef]
- Singh J, Sharma S, Kaur A, Vikal Y, Cheema AK, Bains BK, Kaur N, Gill GK, Malhotra PK, Kumar A, Sharma P. Marker-assisted pyramiding of lycopene-ε-cyclase, β-carotene hydroxylase1 and opaque2 genes for development of biofortified maize hybrids. Scientific Reports. 2021, 11, 12642. [Google Scholar] [CrossRef]
- Zhao X, Tan G, Xing Y, Wei L, Chao Q, Zuo W, Lübberstedt T, Xu M. Marker-assisted introgression of qHSR1 to improve maize resistance to head smut. Molecular breeding. 2012, 30, 1077–1088. [Google Scholar] [CrossRef]
- Li, P. Understanding Maize Biology for Better Crop Improvement. Molecular Plant. 2017, 10, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Yang Q, He Y, Kabahuma M, Chaya T, Kelly A, Borrego E, Bian Y, El Kasmi F, Yang L, Teixeira P, Kolkman J. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nature Genetics. 2017, 49, 1364–1372. [Google Scholar] [CrossRef]
- Xu Z, Hua J, Wang F, Cheng Z, Meng Q, Chen Y, Han X, Tie S, Liu C, Li X, Wang Z. Marker-assisted selection of qMrdd8 to improve maize resistance to rough dwarf disease. Breeding Science. 2020, 70, 183–192. [Google Scholar] [CrossRef]
- Hossain F, Muthusamy V, Bhat JS, Zunjare RU, Kumar S, Prakash NR, Mehta BK. Maize Breeding. InFundamentals of Field Crop Breeding 2022 (pp. 221-258). Singapore: Springer Nature Singapore. 6 May.
- Wang X, Xu Y, Hu Z, Xu C. Genomic selection methods for crop improvement: Current status and prospects. The Crop Journal. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Sun Q, Wang P, Li W, Li W, Lu S, Yu Y, Zhao M, Meng Z. Genomic selection on shelling percentage and other traits for maize. Breeding science. 2019, 69, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen TH, Hayes BJ, Goddard M. Prediction of total genetic value using genome-wide dense marker maps. genetics. 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Bernardo R, Yu J. Prospects for genomewide selection for quantitative traits in maize. Crop Science. 2007, 47, 1082–1090. [Google Scholar] [CrossRef]
- Madhusudhana, R. Marker-assisted breeding in sorghum. InBreeding sorghum for diverse end uses 2019 Jan 1 (pp. 93-114). Woodhead Publishing.
- Jannink JL, Lorenz AJ, Iwata H. Genomic selection in plant breeding: from theory to practice. Briefings in functional genomics. 2010, 9, 166–177. [Google Scholar] [CrossRef]
- Heffner EL, Sorrells ME, Jannink JL. Genomic selection for crop improvement. Crop Science. 2009, 49, 1–2. [Google Scholar] [CrossRef]
- Mayor PJ, Bernardo R. Genome wide selection and marker-assisted recurrent selection in doubled haploid versus F2 populations. Crop Science. 2009, 49, 1719–1725. [Google Scholar] [CrossRef]
- Massman JM, Jung HJ, Bernardo R. Genomewide selection versus marker-assisted recurrent selection to improve grain yield and stover-quality traits for cellulosic ethanol in maize. Crop Science. 2013, 53, 58–66. [Google Scholar] [CrossRef]
- Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D, De Los Campos G, Burgueño J, González-Camacho JM, Pérez-Elizalde S, Beyene Y, Dreisigacker S. Genomic selection in plant breeding: methods, models, and perspectives. Trends in plant science. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Wang T, Ma X, Li Y, Bai D, Liu C, Liu Z, Tan X, Shi Y, Song Y, Carlone M, Bubeck D. Changes in yield and yield components of single-cross maize hybrids released in China between 1964 and 2001. Crop Science. 2011, 51, 512–525. [Google Scholar] [CrossRef]
- Sun, G.; Yu, H.; Wang, P.; Guerrero, M.L.; Mural, R.V.; Mizero, O.N.; Grzybowski, M.; Song, B.; van Dijk, K.; Schachtman, D.P.; et al. A role for heritable transcriptomic variation in maize adaptation to temperate environments. BioRxiv 2022, 39. [Google Scholar] [CrossRef] [PubMed]
- Zaidi SK, Frietze SE, Gordon JA, Heath JL, Messier T, Hong D, Boyd JR, Kang M, Imbalzano AN, Lian JB, Stein JL. Bivalent epigenetic control of oncofetal gene expression in cancer. Molecular and cellular biology. 2017, 37, e00352–17. [Google Scholar]
- Osakabe K, Osakabe Y, Toki S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proceedings of the National Academy of Sciences. 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS biology. 2014, 12, e1001877. [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Pegoraro C, Mertz LM, da Maia LC, Rombaldi CV, de Oliveira AC. Importance of heat shock proteins in maize. J Crop Sci Biotechnol. 2011, 14, 85–95. [Google Scholar] [CrossRef]
- Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucl Acids Res. 1993, 21, 5034–5040. [Google Scholar] [CrossRef]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun. 2016, 7, 13274. [Google Scholar] [CrossRef] [PubMed]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013, 154, 442–451. [Google Scholar] [CrossRef]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Newman M, Ausubel FM. Introduction to gene editing and manipulation using CRISPR/Cas9 technology. Curr Protoc Mol Biol. 2016, 115, 1–6. [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef]
- Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, Becraft PW, Meyers BC, Walbot V, Wang K, Yang B. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2017, 15, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J. L., et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 2018, 13, 413–430. [CrossRef]
- Pineda, M., Lear, A., Collins, J. P., Kiani, S. Safe CRISPR: challenges and possible solutions. Trends Biotechnol. 2019, 37, 389–401. [CrossRef] [PubMed]
- Garcia Ruiz, M. T., Knapp, A. N., Garcia-Ruiz, H. Profile of genetically modified plants authorized in Mexico. GM Crops Food 2018, 9, 152–168. [CrossRef] [PubMed]
- Eckerstorfer, M. F., Engelhard, M., Heissenberger, A., Simon, S., Teichmann, H. Plants developed by new genetic modification techniques—comparison of existing regulatory frameworks in the EU and non-EU countries. Front. In Bioeng. Biotechnol. 2019, 7, 26. [CrossRef]
- Brookes, G., and Barfoot, P. GM Crops: Global Socio-economic and Environmental Impacts 1996–2018. 2020. Available online: https://pgeconomics.co.uk/ pdf/globalimpactfinalreportJuly2020.pdf (accessed on 28 August 2021).
- ISAAA Brief (2019). Global Status of Commercialized Biotech/GM Crops in 2019. ISAAA Ithaca: NY.
- ISAAA database (2021). GM Approval Database. Available online: https://www. isaaa.org/gmapprovaldatabase/default.asp (accessed on 10 August 2021).
- . Simmons, C. R., Lafitte, H. R., Reimann, K. S., Brugière, N., Roesler, K., Albertsen, M. C., et al. Successes and insights of an industry biotech program to enhance maize agronomic traits. Plant Sci. 2021, 307, 110899. [CrossRef]
- Wang A, Shu X, Jing X, Jiao C, Chen L, Zhang J, Ma L, Jiang Y, Yamamoto N, Li S, Deng Q. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnology Journal. 2021, 19, 1553–1566. [Google Scholar] [CrossRef]
- Kumar V, Singh A, Mithra SA, Krishnamurthy SL, Parida SK, Jain S, Tiwari KK, Kumar P, Rao AR, Sharma SK, Khurana JP. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA research. 2015, 22, 133–145. [Google Scholar] [CrossRef]
- Alam MA, Rahman M, Ahmed S, Jahan N, Khan MA, Islam MR, Alsuhaibani AM, Gaber A, Hossain A. Genetic variation and genotype by environment interaction for agronomic traits in maize (Zea mays L.) hybrids. Plants. 2022, 11, 1522. [Google Scholar] [CrossRef]
- Tabashnik, B. E., and Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [CrossRef]
- Pellegrino, E. , Bedini, S., Nuti, M., and Ercoli, L. Impact of genetically engineered maize on agronomic, environmental and toxicological traits: a metaanalysis of 21 years of field data. Sci. Rep. 2018, 8, 3113. [CrossRef]
- Schellenberger, U., Oral, J., Rosen, B. A., Wei, J. Z., Zhu, G., Xie, W., et al. A selective insecticidal protein from pseudomonas for controlling corn rootworms. Science 2016, 354, 634–637. [CrossRef]
- Moar, W., Khajuria, C., Pleau, M., Ilagan, O., Chen, M., Jiang, C., et al. Cry3Bb1-resistant western corn rootworm, diabrotica virgifera virgifera (LeConte) does not exhibit cross-resistance to DvSnf7 dsRNA. PLoS One 2017, 12, 1–15. [CrossRef]
- Yin, Y., Flasinski, S., Moar, W., Bowen, D., Chay, C., Milligan, J., et al. A new bacillus thuringiensis protein for Western corn rootworm control. PLoS One 2020, 15, 1–16. [CrossRef]
- Wu, J., Lawit, S. J., Weers, B., Sun, J., Mongar, N., Van Hemert, J., et al. Overexpression of zmm28 increases maize grain yield in the field. Proc. Natl. Acad. Sci. 2019, 116, 23850–23858. [CrossRef]
- Simmons, C. R., Weers, B. P., Reimann, K. S., Abbitt, S. E., Frank, M. J., Wang, W., et al. Maize BIG GRAIN1 homolog overexpression increases maize GRAIN yield. Plant Biotechnol. J. 2020, 18, 2304–2315. [CrossRef]
- Shi, J., Habben, J. E., Archibald, R. L., Drummond, B. J., Chamberlin, M. A., Williams, R. W., et al. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in Both Arabidopsis and maize. Plant Physiol. 2015, 69, 266–282. [CrossRef]
- Woźniak, E., Tyczewska, A., and Twardowski, T. A shift towards biotechnology: social opinion in the EU. Trends Biotechnol. 2021, 39, 214–218. [CrossRef]
- Cardi, T. Cisgenesis and genome editing: combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 2016, 135, 139–147. [Google Scholar] [CrossRef]
- Harfouche, A. L., Petousi, V., Meilan, R., Sweet, J., Twardowski, T., and Altman, A. Promoting ethically responsible use of agricultural biotechnology. Trends Plant Sci. 2021, 26, 546–559. [CrossRef]
- Li, Q.; Yan, J. Sustainable agriculture in the era of omics: Knowledge-driven crop breeding. Genome Biol. 2020, 21, 154. [Google Scholar] [CrossRef] [PubMed]
- Somegowda, V.K.; Rayaprolu, L.; Rathore, A.; Deshpande, S.P.; Gupta, R. Genome-Wide Association Studies (GWAS) for Traits Related to Fodder Quality and Biofuel in Sorghum: Progress and Prospects. Protein Pept. Lett. 2021, 28, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Al-Qurainy, F.; Alshameri, A.; Gaafar, A.-R.; Khan, S.; Nadeem, M.; Alameri, A.A.; Tarroum, M.; Ashraf, M. Comprehensive Stress-Based De Novo Transcriptome Assembly and Annotation of Guar (Cyamopsis tetragonoloba (L. ) Taub.): An Important Industrial and Forage Crop. Int. J. Genomics 2019, 2019, 7295859. [Google Scholar] [PubMed]
- Abdurakhmonov, I.Y. Plant Genomics; InTech: London, UK, 2016; ISBN 978-953-51-2455-9. [Google Scholar]
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of genomic tools in plant breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Vinayan, M.T.; Seetharam, K.; Babu, R.; Zaidi, P.H.; Blummel, M.; Nair, S.K. Genome wide association study and genomic prediction for stover quality traits in tropical maize (Zea mays L.). Sci. Rep. 2021, 11, 686. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X. Transcriptome and metabolomic analyses reveal regulatory networks controlling maize stomatal development in response to blue light. Int. J. Mol. Sci. 2021, 22, 5393. [Google Scholar] [CrossRef]
- Pan Y, Zhao SW, Tang XL, Wang S, Wang X, Zhang XX, Zhou JJ, Xi JH. Transcriptome analysis of maize reveals potential key genes involved in the response to belowground herbivore Holotrichia parallela larvae feeding. Genome. 2020, 63, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Pan Y, Zhao S, Wang Z, Wang X, Zhang X, Lee Y, Xi J. Quantitative proteomics suggests changes in the carbohydrate metabolism of maize in response to larvae of the belowground herbivore Holotrichia parallela. PeerJ. 2020, 8, e9819. [Google Scholar] [CrossRef]
- Zhou, K.; Zeng, X.; Zhang, B.; Aslam, M.; Xin, H.; Liu, W.; Zou, H. Bulk segregant transcriptome analysis based differential expression of drought response genes in maize. Pak. J. Agric. Sci. 2020, 57, 909–923. [Google Scholar]
- Kebede, A.Z.; Johnston, A.; Schneiderman, D.; Bosnich, W.; Harris, L.J. Transcriptome profiling of two maize inbreds with distinct responses to Gibberella ear rot disease to identify candidate resistance genes. BMC Genom. 2018, 19, 131. [Google Scholar] [CrossRef]
- He, W.; Zhu, Y.; Leng, Y.; Yang, L.; Zhang, B.; Yang, J.; Zhang, X.; Lan, H.; Tang, H.; Chen, J.; et al. Transcriptomic analysis reveals candidate genes responding maize gray leaf spot caused by Cercospora zeina. Plants 2021, 10, 2257. [Google Scholar] [CrossRef]
- Wu, X.; Wang, B.; Xie, F.; Zhang, L.; Gong, J.; Zhu, W.; Li, X.; Feng, F.; Huang, J. QTL mapping and transcriptome analysis identify candidate genes regulating pericarp thickness in sweet corn. BMC Plant Biol. 2020, 20, 117. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Komatsu, S. Plant proteomic research for improvement of food crops under stresses: A review. Mol. Omics 2021, 17, 860–880. [Google Scholar] [CrossRef]
- Guo, J.; Li, C.; Zhang, X.; Li, Y.; Zhang, D.; Shi, Y.; Song, Y.; Li, Y.; Yang, D.; Wang, T. Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 2020, 292, 110380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Zhang, Y.L.; Chen, S.X.; Yin, G.H.; Yang, Z.Z.; Lee, S.; Liu, C.G.; Zhao, D.D.; Ma, Y.K.; Song, F.Q.; et al. Proteomics of methyl jasmonate induced defense response in maize leaves against Asian corn borer. BMC Genom. 2015, 16, 224. [Google Scholar] [CrossRef]
- Wang J, Lin Z, Zhang X, Liu H, Zhou L, Zhong S, Li Y, Zhu C, Lin Z. krn1, a major quantitative trait locus for kernel row number in maize. New Phytologist. 2019, 223, 1634–1646. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Yang, Y.; Liu, S.; Zenda, T.; Liu, X.; Wang, Y.; Li, J.; Duan, H. Comparative proteomics analysis of two maize hybrids revealed drought-stress tolerance mechanisms. Biotechnol. Biotechnol. Equip. 2020, 34, 763–780. [Google Scholar] [CrossRef]
- Führs, H.; Götze, S.; Specht, A.; Erban, A.; Gallien, S.; Heintz, D.; Van Dorsselaer, A.; Kopka, J.; Braun, H.-P.; Horst, W.J. Characterization of leaf apoplastic peroxidases and metabolites in Vigna unguiculata in response to toxic manganese supply and silicon. J. Exp. Bot. 2009, 60, 1663–1678. [Google Scholar] [CrossRef]
- Kaur, B.; Sandhu, K.S.; Kamal, R.; Kaur, K.; Singh, J.; Röder, M.S.; Muqaddasi, Q.H. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: Applications, challenges, and prospects. Plants 2021, 10, 1989. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.Y.; Wang, L.H.; Dou, X.T.; Wang, Y.J.; Wang, H.Z. Comparative metabolomic profiling in the roots of salt-tolerant and salt-intolerant maize cultivars treated with NaCl stress. Biol. Plant. 2020, 64, 569–577. [Google Scholar] [CrossRef]
- Begcy, K.; Nosenko, T.; Zhou, L.-Z.; Fragner, L.; Weckwerth, W.; Dresselhaus, T. Male sterility in maize after transient heat stress during the tetrad stage of pollen development. Plant Physiol. 2019, 181, 683–700. [Google Scholar] [CrossRef]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite Profiling of Low-P Tolerant and Low-P Sensitive Maize Genotypes under Phosphorus Starvation and Restoration Conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




