1. Introduction
Two key processes guiding tissue and organ development both during embryogenesis and in various pathophysiological contexts are epithelial-to-mesenchymal transition (EMT) and its reverse mesenchymal-to-epithelial transition (MET)1. Epithelial cells form stationary barriers delineating tissues and organs2, create lumen and tube morphologies (e.g., via ‘hollowing’ and ‘cavitation’3), maintain basal–apical cell polarity by basement membrane (BM) anchoring, and ensure strict cell–cell contact through desmosomes, gap-, adherens-, and tight-junctions4. Mesenchymal cells, in contrast, are spindle shaped, reside within, and remodel the 3D fibrous extracellular matrix (ECM), and can travel long distances while invading tissues within the body2,5. The epithelial-to-mesenchymal spectrum is wide, and contains many intermediate phenotypes6.
Cell state transitioning (both EMT and MET) is crucial for tissue and organ development and is vital for almost every step of embryogenesis, including extraembryonic trophectoderm specification, embryonic gastrulation, body-plan patterning and, ultimately, organogenesis (establishment and diversification of body organs)5,7-9. For example, neural crest cells (NCCs) delaminate via EMT from the neural plate border and migrate to distant embryonic destinations7,10,11 including the pharyngeal arches12, head and neck mesenchyme11, second heart field13, and adrenal gland medulla14. Upon reaching their destinations, NCCs undergo at least one more MET to assume parenchymal differentiated cell states and morphologies9.
Moreover, EMT and MET are also of paramount importance for adult tissue homeostasis and regeneration15, and are involved in numerous pathological conditions including cancer metastasis16, fibrosis during aging2, cellular senescence17,18 and MET mediated escape from senescence19. In essence, most tissue formation and remodeling processes necessitate at least one EMT-MET cycle, making the study of these processes critical not only for developmental biology and tissue engineering purposes but also for better understanding of pathologies and possible therapy development.
Global, often antagonistic, cellular programs ensure the protection of epithelial-mesenchymal homeostasis—including specific splicing, miRNA regulatory networks and other epigenetic mechanisms, which are still not completely understood (particularly, for MET)1,5. Direct human organogenesis embryo models, however, are notoriously difficult to generate (also limited by the ‘14-day’ ethical rule)20, and EMT/MET regulatory network manipulation is, in many cases, lethal. Hence, current EMT/MET knowledge is mostly deduced from animal models (i.e., drosophila, chick, zebrafish, and murine)9, and by employing human cancer cell spheroids and/or human primary and stem cell-derived models21-23.
One possible downstream common mediator of MET may be heart and neural crest derivatives 2 (HAND2), a type B basic helix-loop-helix (bHLH) transcription factor (TF)24, and its associated anti sense long non-coding RNA (lncRNA, HAND2-AS1). Embryonically, HAND2 induces type 1 (embryonic) MET patterning of NCCs to branching aortic vessels and of mesodermal second heart field (SHF) cells to the right ventricle (RV), parts of the atria, interventricular septum, and outflow tract25-27. HAND2 murine knockout (KO) is lethal at E9.5-10.5, yielding RV hypoplasia, single chambered hearts, and vascular malformations25,28. Molecularly, HAND2 is a crucial downstream regulator of the endocardial VEGF-Notch signaling pathway during cardiogenesis and coronary vasculogenesis27. HAND2 also instructs type 1 MET in anterior-posterior limb-bud digit patterning (initiating a SHH/FGF feedback loop)29 and induces embryonic mesothelium progenitor formation delineating internal organs and contributing to organogenesis, tissue homeostasis and regeneration, through unclear signaling30. HAND2 was, thus, suggested as a “regulator of tissue morphogenesis and patterning through a mechanism independent of direct DNA binding”29, indicating tissue context specificity during normal development, as further supported by the evolutionary conserved roles of bHLH TFs31.
Our lab’s research focuses on the intersection of tissue engineering (TE32) and developmental biology (DB) that is often termed developmental engineering (DE)33-36. While traditional TE uses biomaterial scaffolds and signaling molecules to guide (top-down) cellular production of biological tissue substitutes, DB studies the basic science principles of tissue, organ and whole organism formation (bottom-up)37,38. Developmental Engineering (DE) aims, therefore, to identify, study and mimic nature’s building blocks and key developmental processes to produce in vitro human tissues and organ models36.
We came across the HAND2-HAND2-AS1 genomic locus by chance as part of an experiment detailed in this manuscript, involving human mesenchymal stem cells (hMSCs) (as model cells) that were seeded on a bioinductive porcine cardiac extracellular matrix (pcECM) scaffold. We believe accidental discoveries made through this work are of interest and possibly with wide implications. We, therefore, hope that this publication would provide an impetus for other researchers that might help elucidate the enigma involving HAND2-HAND2-AS1 regulation of MET in both tissue development, homeostasis, regeneration, and pathogenesis.
2. Materials and Methods
2.1. Decellularization and pcECM Characterization
Acellular pcECM samples (15×10×1.5 mm) were produced as we previously reported39-41. Briefly, native porcine left ventricular slices were sequentially immersed in alternating hyper-hypotonic solutions (1.1% and 0.7% NaCl, respectively), a trypsin (0.05%w/v)-EDTA (0.02%w/v) solution and 1% (v/v) Triton™-x-100 (in 0.1%v/v ammonium hydroxide) PBS solution. All decellularization reagents were purchased from Sigma-Aldrich (St. Louis, MO). Prior to experimentation, sliced matrices were disinfected with 70% v/v ethanol and washed with PBS containing double antibiotic-antimycotic concentration (2%v/v, Gibco™, TermoFisher Scientifc, Waltham, MA) followed by immersion in cell culture media overnight in 37 °C and 5% CO2.
For histological analyses, three representative samples of each experimental group were fixated in fresh paraformaldehyde (4% w/v solution, PFA, Sigma-Aldrich, St. Lous, MO) for 2 hrs, and were then processed for paraffin blocks. Native tissue of similar dimensions was used as the positive control. Paraffin sections (5 µm) were processed for hematoxylin and eosin (H&E) and Masson’s trichrome (MTC) stains. For immunohistochemical stains we used routine IHC staining protocols. H&E, MTC and IHC protocols used were the same as we as previously published42 . For IHC stains we used the following primary antibodies: rabbit IgG-anti epidermal growth factor (EGF, abcam ab9695), mouse IgG anti basic fibroblast growth factor (bFGF, Mareck-Millipore, 05-118), and mouse IgG anti vascular endothelial growth factor A (VEGF, abcam, ab1316). For all staining primary antibody dilution was 1:100, antigen retrieval steps were performed in a pressure cooker for 40 min, and secondary antibody staining was performed with the Novolink polymer detection system (RE7290-K, Leica Biosystems, Germany), according to the manufacturer instructions.
2.2. hMSC culture and characterization
Human bone marrow mesenchymal stem cells (hMSCs) were commercially purchased (PoieticsTM, Lonza, Switzerland, PT-2501) and are guaranteed by the supplier to be CD105+CD166+CD29+CD44+ CD14-CD34-CD45- (by flow cytometry) and of being capable to differentiate down the adipogenic, chondrogenic, and osteogenic lineages when cultured in the recommended differentiation medium43. Cells were cultured in standard cell culture dishes using α-MEM with 10% ml FBS, 1% (v/v) antibiotic-antimycotic solution, and 1% (v/v) of 200 mM L-glutamine solution. bFGF (5·10-3 µg/ml) was supplemented every other day. Cells were split at 70-80% confluency at a 1:4 ratio and were used up until passage 4-5 in all experiments described herein.
To visualize the cell expression of characteristic markers we performed immunofluorescent staining on glass cover slides for representative positive and negative marker. Briefly, cells were cultured on round 18mm (0.2mm thick) cover slides in standard tissue culture plate. When reaching 70% confluence cells were fixed to the slides using ice cold methanol for 5 min followed by partial brief air drying. Subsequently slides were blocked with 3% FBS containing PBS and incubated in a humidity chamber for 30 min at RT with the following mouse anti human primary antibodies: CD90 (BD Biosciences, 555593), CD105 (BD Biosciences, 555690), CD73 (Santa Cruz Biotechnology, Inc., 32299) and CD31 used as isotype negative control (R&D systems, BBA7) all at a 1:50 dilution in 3% FBS containing PBS. Following primary antibody washes (2x) slides were incubated with a PE anti-mouse (BD Biosciences 550589, dilution 1:100) secondary antibody for 30 min RT. Subsequently slides were washed in the same serum containing wash buffer and mounted with aqueous mounting medium containing DAPI (DAPI Fluromount-G®, SouthernBiotech, Birmingham, AL). Images were acquired using the Nikon Eclipse TE2000-U equipped with a fluorescent camera at 400x magnification.
To assess the cells differentiation capacity at higher passages (passage 5), we performed cell differentiation using commercial differentiation kits. A cell solution of 1.6×107 cells/mL, 5×103 cells/mL and 1×104 cells/mL of hMSC were prepared with complete α-MEM media, for chondrogenesis, osteogenesis and adipogenesis differentiation, respectively (as recommended by the manufacturer protocols). 10µL droplets of cells of each cell concentrations were added to the centre of each well of 12-well plates and incubated in a standard 37oC 5% CO2 incubator for 2 hrs. The α-MEM media was then replaced with the respective differentiation media and replenished every other day: StemPro® Chondrogenesis Differentiation Kit, StemPro® Osteogenesis Differentiation Kit, and StemPro® Adipogenesis Differentiation Kit (Gibco™, TermoFisher Scientifc, Waltham, MA). Staining with respective dye indicators were used after 14 days of differentiation: Alcian Blue for chondrogenesis, Alizarin Red S for osteogenesis and Oil Red O for adipogenesis (all purchased from ThermoFisher Scientific, Waltham, MA).
2.3. pcECM induced MET in hMSCs
Seeding of hMSC on the pcECM was performed on pre-cut disinfected and culture media incubated pcECM (96 well plate format) at a density of 3x10
5 cells/cm
2 as previously published
44. hMSC seeded pcECM constructs were cultured for 30 days. At designated time points throughout the culture mRNA was extracted (phenol and guanidine thiocyanate, qiasol
TM, Qiagen, Germany) from at least n=3 samples per time point (t=4, 23 and 30 days post seeding). Extracted mRNA integrity was evaluated using the 2100 Bioanalyzer (Agilent, Santa Clara, CA), and RIN values of >7 were used for subsequent steps. mRNA was reverse transcribed to cDNA using the RT
2 First Strand Kit (330401, Qiagen) followed by real-time qPCR analyses using two separate Qiagen RT
2 PCR arrays custom designed with Qiagen for specific lineage identification of endothelial and cardiomyocyte cell phenotypes (given the cardio-vascular origin of the pcECM). Lists of gene array composition and identities appear in Supplementary
Tables S1-S2.
Qualitative detection of HAND2 and CD31 expression in hMSCs at the protein level in situ (on the pcECM) was performed by immunohistochemistry (IHC) and immunofluorescence (IF), respectively. HAND2 IHC was performed using a rabbit IgG anti human HAND2 (abcepta, AP17008C, 1:100) as per the protocol described above for pcECM growth factors staining with slight modifications that include antigen retrieval at pH=9, and 0.4% Triton-X-100 perforation for intracellular staining. CD31 IF was similarly performed on fixated tissue sections using the R&D systems, BBA7 mouse IgG anti human primary antibody diluted 1:50 in 3% FBS containing PBS, and a secondary PE anti-mouse (BD Biosciences 550589, dilution 1:100) for 30 min RT. Dapi was used for counter staining. Images were acquired using a fluorescent and brightfield inverted microscope (Nikon eclipse TE-2000U).
In addition, and as a more quantitative measure that avoids potential background fluorescence from the ECM, hMSCs were also harvested at designated culture time points from the pcECM and subsequently FACS analysed for quantification of CD31. For cell harvesting, hMSC seeded pcECM matrices were washed twice with 0.625 mM EDTA in PBS and transferred into 0.5 ml of Tryple 10X (ThermoFisher Scientific, Waltham, MA) in a 1.5-ml eppendorf tube. After 8 min incubation, pcECMs were removed and 0.5 ml of complete culture media (containing 10% FBS) was added in each eppendorf before centrifugation at 300 g for 5 min. All cell pellets were collected in one single tube, washed twice with cold (4°C) PBS and followed immunofluorescence staining protocol with formaldehyde against CD31 (Biolegend 303102, dilution 1:50) and PE anti-mouse (BD Biosciences 550589, dilution 1:100) for flow cytometry analysis using a BD FACS CaliburTM machine. Tissue culture plate cells served as control for this experiment. Representative results are shown out of at least n=3 samples at each time point and experimental condition as indicated.
2.4. Bioinformatic Analyses
Bioniformatic analyses for HAND2 and HAND2-AS1 expression levels was performed using several separate datasets: The National Center for Biotechnology Information (NCBI) bioproject #PRJNA280600 (“RNA sequencing of total RNA from 20 human tissues”,
45); NCBI bioproject #PRJNA270632 (“Tissue-specific circular RNA induction during human fetal development”,
46); The national institute of health (NIH) and national cancer institute (NCI) supported cancer genome atlas program (TCGA,
https://www.cancer.gov/tcga); and the Genotype-Tissue Expression (GTEx) project database (dbGaP accession #phs000424.vN.pN,
47) supported by the Common Fund of the Office of the Director of the NIH , and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Datasets from the NCBI bioprojects were copied from the NCBI Gene resource for HAND2 (Gene ID #9464) and HAND2-AS1 (Gene ID #79804). TCGA and GTEx data were obtained through the gene expression profiling interactive analysis 2 (GEPIA2) server
48. All data obtained was further statistically analysed and presented using GraphPad Prism version 10.0.2 (232) for Windows (GraphPad Software, Boston, Massachusetts USA,
www.graphpad.com).
2.5. CRISPR-Cas9 Knockout of HAND2-HAND2-AS1 in hMSCs
In the interest of space detailed CRISPR-Cas9 protocol and design employed appears in the supplementary information.
2.6. Statistical Analyses
Unless otherwise stated results indicate mean ± standard deviation of at least n=3 biological repetitions per sample group and time point. For statistical analyses results were first tested for normality using the Shapiro-Wilk test criteria. All results obeyed gaussian normal distribution and were therefore analysed using one-way analysis of variance (ANOVA) with Tukey’s honest significant difference (HSD) post hoc correction for multiple comparisons. Adjusted p values tested are indicated by (*, p<0.05), (**, p<0.01), (***, p<0.001) and (****, p<0.0001) in the appropriate figures. (ns) indicated non-significant difference. All statistical tests and graphical results were analysed and displayed using the GraphPad Prism software version 10.0.2.
3. Results
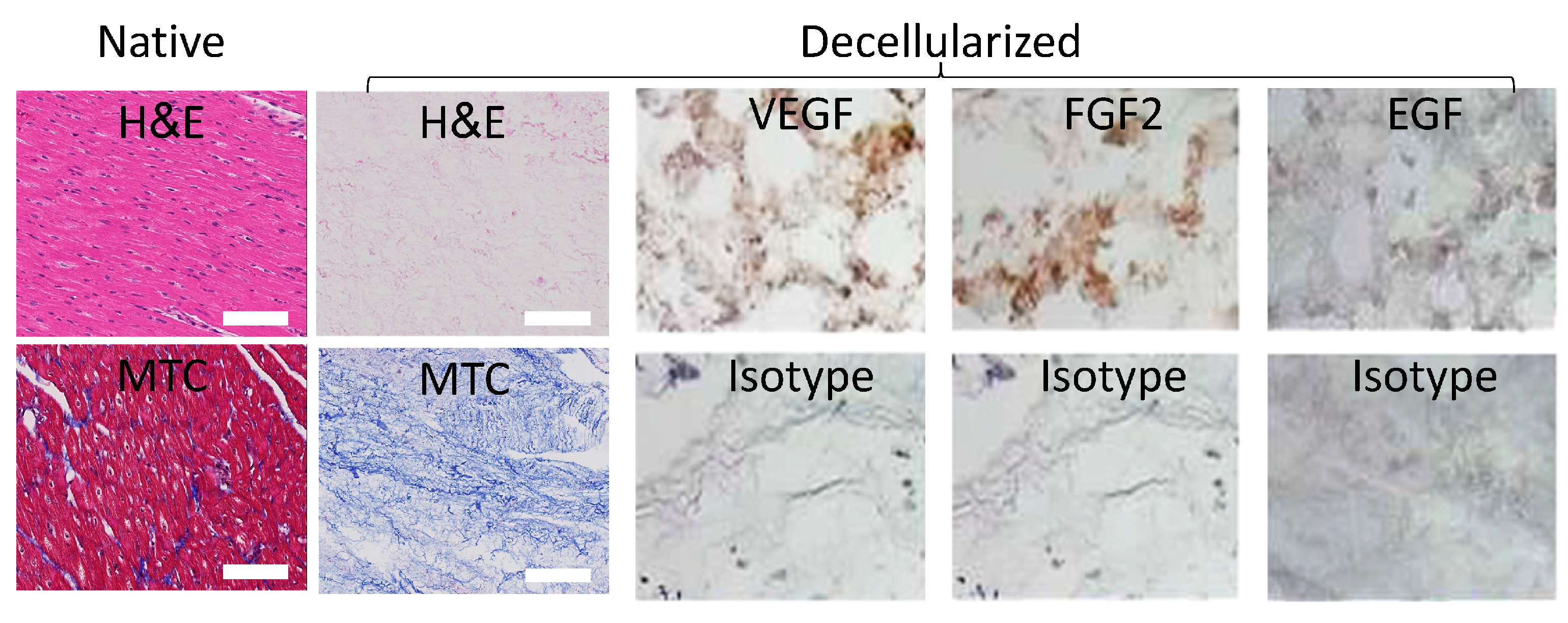
We began our work with the decellularization of porcine hearts as we previously published
39,40 to obtain slabs of bioinductive pcECM (
Figure 1). To demonstrate the preservation of ECM fibers and structure we stained paraffin sections of the native tissue (as control) and decellularized slabs with hematoxylin and eosin (H&E) and masson’s trichrome (MTC) reagents. Additional immunohistochemical (IHC) stains were consequently performed for two cardio-vascular lineage specific representative growth factors (FGF2 and VEGF, respectively), and an epidermal growth factor (EGF) serving as negative control (
Figure 1 as indicated). These stains confirmed retention of cardio-vascular growth factor pockets within the pcECM following decellularization, which is in accordance with previously described mechanisms of ECM biological composition and function
42,49-51.
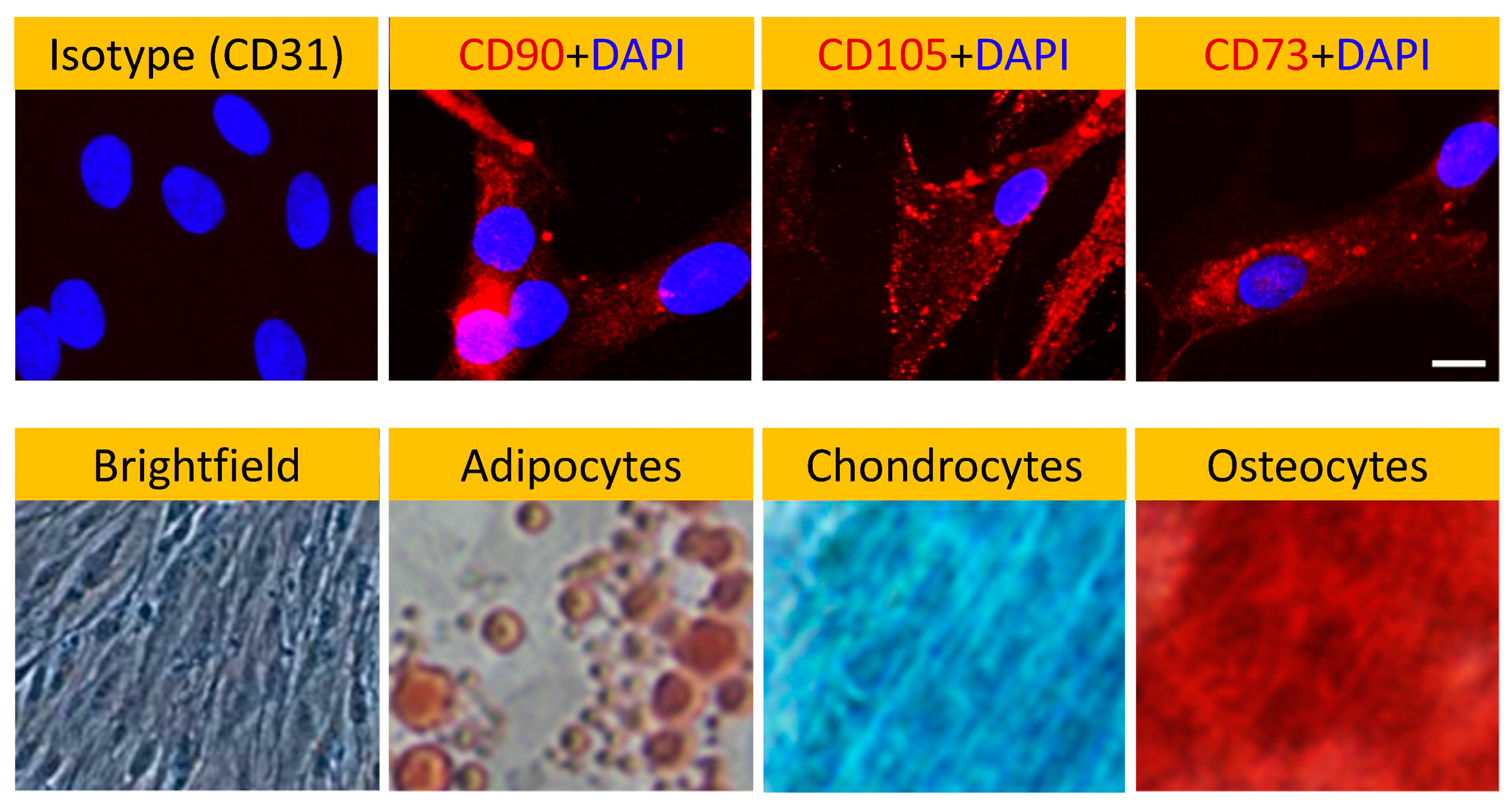
Prior to repopulating our pcECM scaffold with hMSCs (commercially purchased from Lonza, Switzerland) we performed characterization of cell morphology, marker expression profile and differentiation potency (
Figure 2). These analyses demonstrated tissue-culture plastic adherence, characteristic spindle shaped morphology, consensus marker expression (CD73
+CD90
+CD105
+CD31
-), and multipotency following standard protocols for adipose, bone and cartilage-like cell differentiation. Of note was the absence of CD31 expression by the hMSCs (used as iso-type negative control), which suggested the lack of endothelial commitment by the cells used.
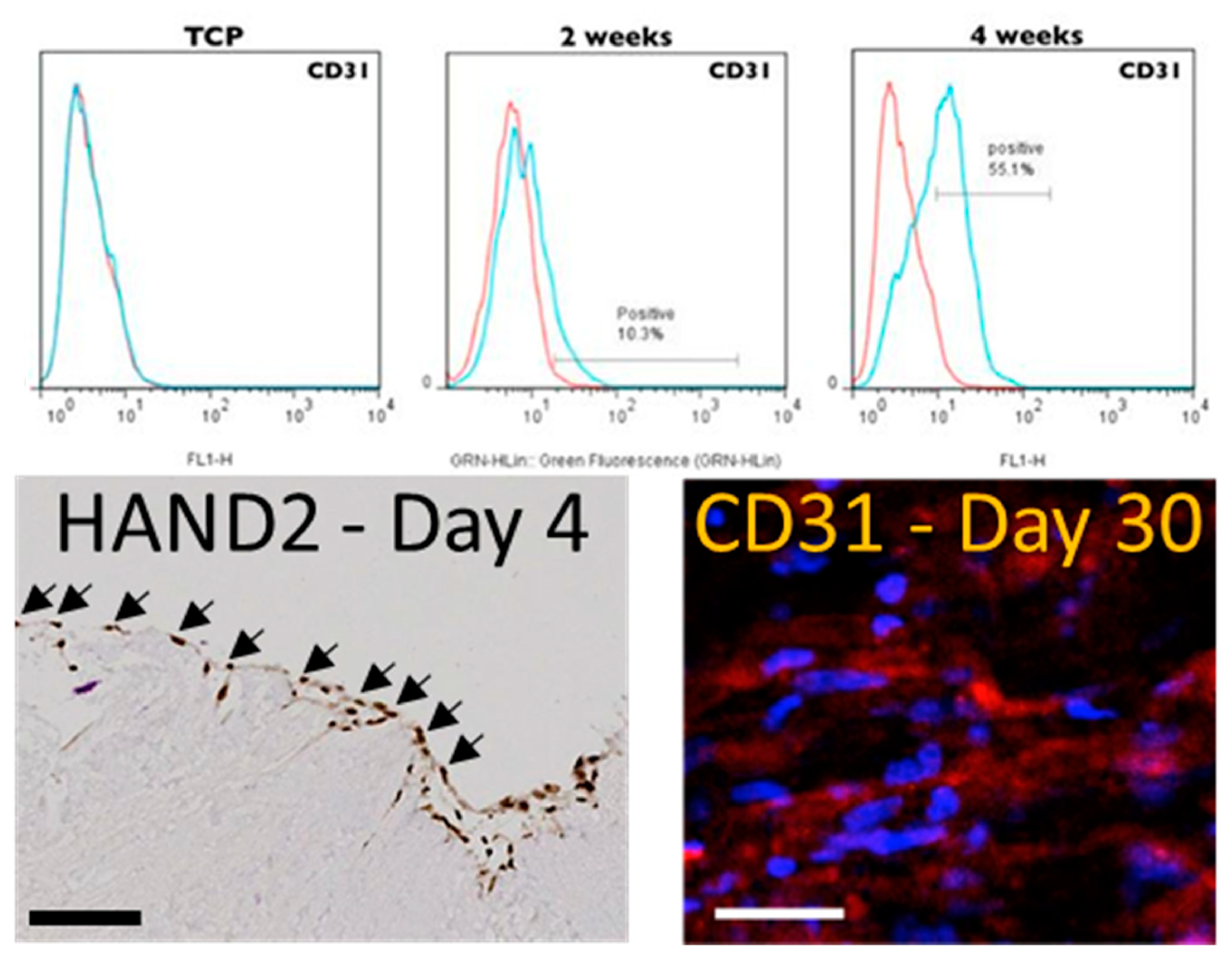
At distinct time points (t=3, 23 and 30 days) during the culture we isolated the hMSCs (through trypsinization) and obtained their mRNA. To identify possible induction pathways that may be triggered by the pcECM, the mRNA was then subject to reverse transcribed quantitative (realtime) polymerase chain reaction (RT-qPCR) arrays (Supplementary information online and
Supplementary Tables S1 and S2) for cardiac and endothelial related genes. Analyzing these data, we observed two interesting and sequential phenomena. At the initial time points following seeding, the mRNA levels of HAND2 were significantly up regulated (~2400 fold, relative to standard tissue culture plate expression levels,
Supplementary Table S3). This upregulation was also observed in repeated experiments at the protein level on day 4 of culture using IHC cross sectional histological staining specifically for HAND2 (
Figure 3). While the mRNA level of HAND2 subsided to lower values at later time points (
Supplementary Table S3), the HAND2 protein level remained persistent at least until day 10 of culture (data not shown). This phenomenon preceded and was concomitant to upregulation of endothelial related genes, including PECAM1 (CD31), for which the hMSCs were originally negative before seeding (
Supplementary Table S3). Intrigued by these results, we further performed protein level evaluation of CD31 expression by the seeded hMSCs revealing a gradual increase in its presence throughout the 30 days of culture as evidenced by both flow cytometry and by immunofluorescent stains (
Figure 3). Collectively, these results suggested that the pcECM induced an MET process in the seeded hMSCs driving their partial commitment towards an endothelial (simple squamous epithelium) phenotype.
Interestingly, HAND2 has only two exons (Chr. 4q34.1, Genecards database
52) but its promoter region and first exon partially overlap an associated antisense long noncoding RNA (lncRNA, HAND2-AS1)—implicated in alleviating numerous human pathologies reviewed in
53,54. We performed bioinformatic analyses of HAND2 and HAND2-AS1 expression data using known human sequencing databases for adult and fetal normal expression as well as pathological expression levels in various cancers. These bioinformatic data correlation and analyses suggested that in normal adult tissues (
Figure 4a,b), during fetal development (
Figure 4a-b insets), and also during cancer progression (
Figure 4c) HAND2 and HAND2-AS1 expression profiles seemed to correlate (
Figure 4d) and obey similar trends. In adults, high HAND2-HAND2-AS1 expression levels appear either in tissues that contain neural crest lineage cells and that appear very early in embryonic organogenesis and development (such as the adrenal glands and the heart) or in tissues that are associated with pregnancy (i.e., the endometrium, ovary, and placenta). Other renewable tissues that had high HAND2-HAND2-AS1 expression levels during development (e.g., stomach, kidney, and intestine) seem to lower their expression to a more moderate level in adulthood. However, the HAND-2 and HAND2-AS1 tissue level expression during homeostasis seems to be highly maintained, as protrusion/imbalance at the tissue level is associated with many different cancer types (
Figure 4c). Among these cancer types shown, are two major categories – those with downregulation of HAND2 and HAND2-AS1 expression relative to healthy tissue controls (most cancers analyzed) and those with HAND2 and HAND2-AS1 upregulation (specific for pancreatic adenocarcinoma, PAAD; and for pheochromocytoma and paraganglioma, PCPG).
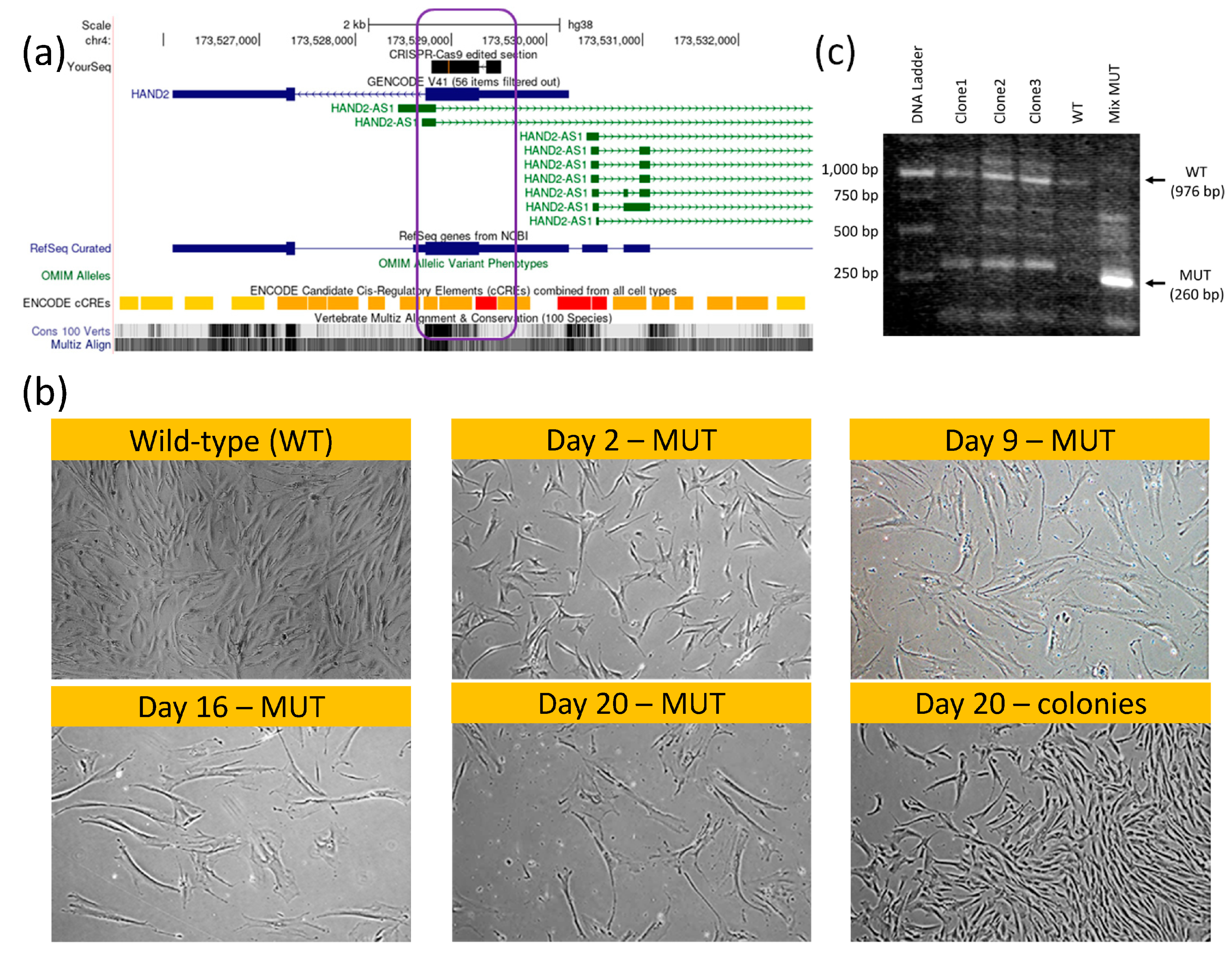
Intrigued by our bioinformatic analyses results, we performed hMSCs knockout (KO) of both HAND2 first exon and HAND2-AS1 regulatory/promoter genomic region (
Figure 5a, indicated by purple rectangle) using CRISPR-Cas9 (Supplementary Fig. S1). Our initial thought was that given the tissue specificity and the small (only two exons) scale of this gene, knocking it down might not be detrimental to hMSCs. Clearly, using such KO hMSCs (HAND2-HAND2-AS1
null) would enable testing the apparent regulation of the HAND2/HAND2-AS1 gene products on pcECM induced hMSC-MET towards endothelial phenotypes.
Surprisingly, while the gene editing was successful in obtaining double ‘mutant’ KO hMSC cells (Figure. 5b, mix MUT lane,
Supplementary Figure S2), the HAND2/HAND2-AS1 gene products seems to be vital at least at a basal (low) expression level for hMSC proliferation and function. In fact, within the transfected cell populations appearing on the same plate and culture conditions (
Figure 5c) two cell morphologies became apparent following the CRISPR-Cas9 editing. One rare morphology (3 discrete colonies) was identical to wildtype (WT) hMSC. These cells continued to proliferate, and were capable of subculturing, i.e., localized short and low volume trypsinization enabled to selectively lift and propagate them in other tissue culture dishes. Once propagated these colonies’ clones (marked as clone 1-clone 3, lanes 2-4,
Figure 5b) exhibited a heterozygous HAND2 PCR product containing both mutant/KO phenotype of 260bp (short/edited) and a 976 bp (WT) PCR product bands on agarose gel electrophoresis. Most of the cell population however, displayed a senescent-like hypertrophic and cell–cycle arrested morphological phenotype (
Figure 5C, MUT marked morphologies compared to WT/colony standard morphology). These cells did not organize into colonies and remained hypertrophic and quiescent/senescent for at least 3 weeks on plate. The mRNA of these cell types was harvested (labeled mix MUT in
Figure 5b) and their PCR product displayed a homozygous short HAND2 band. This band was further sanger sequenced to determine the actual deletion sequence is correct (as designed,
Supplementary Figure S2). Given the paucity of this cell type and the lack of its proliferation, we were unable to perform additional characterization.
Nevertheless, and taken together with the results of HAND2 involvement in MET induction, these observations further suggest distinct concentration-dependent HAND2/HAND2-AS1 control of cell phenotype. That is, no HAND2/HAND2-AS1 expression (complete absence) causes senescence (or at minimum cell cycle arrest and cellular hypertrophy), basal low–intermediate expression is critical to maintain the mesenchymal phenotype, and high expression levels may induce MET toward epithelial/endothelial phenotypes. To the best of our knowledge, this finding is new, may be of potential interest to the readership of this journal, and merits further elucidation.
4. Discussion
The bioactivity of the pcECM scaffold material in vitro41,44,55,56 and in vivo42,57 (REFs) has been thoroughly described by our group and others including induction of cardiac commitment in human induced pluripotent stem cells (hiPSCs) in the absence of induction media55, stimulation of neo-tissue formation41 (REF), and modulation of host immunity resulting in recruitment of host progenitor cells that assist in cardiac regeneration even after scarring in rat myocardial infarction models42,57. This is in accordance with an established body of evidence on the biological activity, tissue specificity, and induction capacity of decellularized ECM based materials49-51. Thus, in the absence of an immune response (in a completely in vitro setting) the gradual nature of the effect observed may be attributes to dependency on ECM remodelling by the cells, which help release localized high concentration of growth factors retained within the decellularized composite ECM. Such high localized concentrations may be a strong driving force in selectively directing stem cell fate decisions and differentiation states in a tissue specific manner which is dependent on the relative abundance and composition of tissue resident growth factors.
Regardless of the actual activation molecular pathway involved, our findings clearly indicate transient activation of HAND2 in hMSCs seeded on the pcECM which preceded their MET towards endothelial phenotypes. Our bioinformatic analyses further revealed that HAND2 and HAND2-AS1 expression levels are tightly correlated (Pearson coefficient > 0.90) across many different tissues both during embryonic organogenesis (between weeks 10 and 20), as well as in adult normal and pathologic conditions. This tight regulation suggests that HAND2-HAND2-AS1 may be involved in a centrally conserved and important regulatory mechanism, the nature of which is currently unknown. Our findings suggest that this regulatory pathway results in highly active HAND2 expression during organogenesis. This HAND2 expression is subsequently reduced in most adult tissues to basal medium-low levels, except for the endometrium, heart, adrenal and placenta, where HAND2 expression levels remain high even in adulthood. A common denominator to these high HAND2 expressing tissues may be the fact that they are all critical for embryonic support and pregnancy related hormonal regulation. In addition, human protein level analyses58 for HAND2 suggest its constant low-medium level expression, in many tissues, beyond those that were identified by our sequencing data analyses, and at a histologically verified manner.
Taken together, we suggest that this HAND2-HAND2-AS1 regulatory pathway may represent a fine-tuning mechanism regulating plasticity in a spectrum/axis comprising senescence-like phenotype (complete absence of HAND2/HAND2-AS1 expression), maintenance of mesodermal/mesenchyme stemness/proliferation capacity (at basal expression levels), and MET (at high levels). This suggested mechanism is further supported by the similarity of adult hMSCs to neural crest cells, that are considered the fourth germ layer, the literary evidence suggesting HAND2/HAND2-AS1 role as a regulator of many cancer types, and the critical role of MET in cancer progression and metastasis, as outlined below.
At the molecular level, regulation of adult endometrial HAND2 function is the most substantiated. There it is induced by upstream progesterone (P4)-Indian hedgehog (IHH) signaling during menstrual cycles and initiates downstream MET (also termed decidualization59)—a key requirement for blastocyst receptivity during implantation60. In wider adult-tissue pathological contexts (type 2/3 MET), however, HAND2 is associated with endometriosis induction (upregulating proinflammatory IL15 expression61,62), right ventricle protection from pressure overload damage26, familial dilated cardiomyopathy63, onset of mesothelioma malignancies30 and obesity (regulated by glucocorticoid signaling)64. HAND2 is also silenced in colorectal cancer (via hypermethylation correlated with poor prognosis) and has been identified as an epigenetic driver gene and a potential tumor suppressor (via MAPK/ERK signaling)65. Interestingly, HAND2 and its associated lncRNA, HAND2-AS1, were implicated in alleviating numerous human pathologies reviewed in53,54. It is reasonable that HAND-AS1-induced MET can be a shared underlying mechanism initiating atherosclerosis prevention66,67, impairment of fibroblast activation in rheumatoid arthritis68, and inhibition of glioma69, prostate70, ovarian71,72, gastric73, bladder74, cervical75,76, breast77, and pancreatic78 cancer progression with mixed reports in liver cancer79-81.
To date, studying HAND2 function in vitro focused on primary human cardiac mesenchymal stromal cells (cMSCs), where its ability to induce MET toward an endothelial (simple squamous epithelium) state in a senescent selective manner (corelated to CD90 pre-expression) was demonstrated18. Other studies using bone marrow human mesenchymal stem/stromal cells (hMSCs) showed that IHH signaling affects cellular senescence through modulation of ROS/mTOR/4EBP1 and p70S6K1/2 signaling—a key molecular pathway in aging82. While HAND2 was not directly explored in the latter study, senescence dependency on IHH signaling may also be relevant to HAND2 activation (as occurring in the endometrium); this merits further elucidation using adequate cell and tissue models.
Collectively, HAND2 and HAND2-AS1 are, therefore, suggested herein as common downstream regulators of MET during embryonic cardiac tissue (and also digit development), in adult heart, endometrium, renewable epithelial and adipose tissues, and within wider tissue contexts of senescence and related pathologies. It is also reasonable that regulation of MET (which results in a wide spectrum of commitment levels) would occur in a manner which is proportional to expression level. Indeed, several different HAND2 activators (cardiac Notch, endometrial progesterone-IHH, and adipose glucocorticoid signaling) along with its lncRNA (HAND2-AS1) epigenetic regulation pinpoint the HAND2-HAND2-AS1 loci as a possible local integrator and driver of MET in tissue development, renewal and senescence. The fascinating MET roles of HAND2 and HAND2-AS1 within such diverse biological contexts remain to be elucidated and to this end necessitate the development of appropriate inflammation-free human 3D cell and tissue models.
Our suggested mechanism has far-reaching implications for both developmental biology, regenerative medicine, tissue engineering, and as a possibly common treatment target for many related pathologies. Nevertheless, further investigations and efforts are required to verify and corroborate our findings and suggested mechanism, better understand this regulatory pathway, identify a more controlled environment that can selectively induce and inhibit this pathway, and ultimately understand the molecular players involved. We also suggest that targeting this pathway may represent a promising new therapeutic approach for a wide range of diseases, including cancer, degenerative disorders, and aging.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, Udi Sarig; Formal analysis, Rachel Vazana-Netzarim, Hod Bruck and Naama Danino; Investigation, Rachel Vazana-Netzarim; Methodology, Rachel Vazana-Netzarim and Shachar Sofer; Supervision, Udi Sarig; Validation, Rachel Vazana-Netzarim and Yishay Elmalem; Writing – original draft, Udi Sarig; Writing – review & editing, Rachel Vazana-Netzarim, Shachar Sofer, Hod Bruck, Naama Danino and Udi Sarig.
Funding
This research was supported by the Ministry of Innovation, Science & Technology, Israel (grant number: 0004628), and by the Singapore National Research Foundation (under the CREATE program).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgement
The authors would like to acknowledge the contribution of Evelyne Bao-Vi Nguyen, Priyadarshini S Mhaisalkar, and Yu Wei Ping for their assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Santos, F.; Moreira, C.; Nóbrega-Pereira, S.; de Jesus, B.B. New Insights into the Role of Epithelial–Mesenchymal Transition during Aging. Int. J. Mol. Sci. 2019, 20, 891. [Google Scholar] [CrossRef]
- Chung, S.; Andrew, D.J. The formation of epithelial tubes. J. Cell Sci. 2008, 121, 3501–3504. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A. Epithelial Plasticity: A Common Theme in Embryonic and Cancer Cells. Science 2013, 342, 1234850. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A. Are You Interested or Afraid of Working on EMT? Methods Mol Biol 2021, 2179, 19–28. [Google Scholar]
- Gilbert, S.F.; Barresi MJ, F. Developmental biology, 12nd ed.; Oxford University Press, 2020. [Google Scholar]
- Amack, J.D. Cellular dynamics of EMT: lessons from live in vivo imaging of embryonic development. Cell Commun. Signal. 2021, 19, 1–16. [Google Scholar] [CrossRef]
- Pei, D.; Shu, X.; Gassama-Diagne, A.; Thiery, J.P. Mesenchymal–epithelial transition in development and reprogramming. Nature 2019, 21, 44–53. [Google Scholar] [CrossRef]
- Gouignard, N.; Andrieu, C.; Theveneau, E. Neural crest delamination and migration: Looking forward to the next 150 years. genesis 2018, 56, e23107. [Google Scholar] [CrossRef]
- Kalcheim, C. Neural Crest Cells: Methods and Protocols (eds Quenten Schwarz & Sophie Wiszniak) 1-19; Springer: New York, 2019. [Google Scholar]
- Escot, S.; Blavet, C.; Faure, E.; Zaffran, S.; Duband, J.-L.; Fournier-Thibault, C. Disruption of CXCR4 signaling in pharyngeal neural crest cells causes DiGeorge syndrome-like malformations. Development 2016, 143, 582–588. [Google Scholar] [CrossRef]
- Cortes, C.; Francou, A.; De Bono, C.; Kelly, R.G. Epithelial Properties of the Second Heart Field. Circ. Res. 2018, 122, 142–154. [Google Scholar] [CrossRef]
- Kim, J.-H.; Choi, M.H. Embryonic Development and Adult Regeneration of the Adrenal Gland. Endocrinol. Metab. 2020, 35, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Marconi, G.D.; Fonticoli, L.; Rajan, T.S.; Pierdomenico, S.D.; Trubiani, O.; Pizzicannella, J.; Diomede, F. Epithelial-Mesenchymal Transition (EMT): The Type-2 EMT in Wound Healing, Tissue Regeneration and Organ Fibrosis. Cells 2021, 10, 1587. [Google Scholar] [CrossRef]
- Jia, D.; Park, J.H.; Kaur, H.; Jung, K.H.; Yang, S.; Tripathi, S.; Galbraith, M.; Deng, Y.; Jolly, M.K.; Kaipparettu, B.A.; et al. Towards decoding the coupled decision-making of metabolism and epithelial-to-mesenchymal transition in cancer. Br. J. Cancer 2021, 124, 1902–1911. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S.; Bayliss, P.E.; Hanai, J.-I. A prospective epigenetic paradigm between cellular senescence and epithelial-mesenchymal transition in organismal development and aging. Transl. Res. 2014, 165, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Martini, H.; Iacovoni, J.S.; Maggiorani, D.; Dutaur, M.; Marsal, D.J.; Roncalli, J.; Itier, R.; Dambrin, C.; Pizzinat, N.; Mialet-Perez, J.; et al. Aging induces cardiac mesenchymal stromal cell senescence and promotes endothelial cell fate of the CD90 + subset. Aging Cell 2019, 18, e13015. [Google Scholar] [CrossRef]
- Wang, M.; Wu, Y.; He, Y.; Liu, J.; Chen, Y.; Huang, J.; Qi, G.; Li, P. SIRT1 upregulation promotes epithelial-mesenchymal transition by inducing senescence escape in endometriosis. Sci. Rep. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Aach, J.; Lunshof, J.; Iyer, E.; Church, G.M. Addressing the ethical issues raised by synthetic human entities with embryo-like features. eLife 2017, 6. [Google Scholar] [CrossRef]
- Hahn, S.; Nam, M.-O.; Noh, J.H.; Lee, D.H.; Han, H.W.; Kim, D.H.; Hahm, K.B.; Hong, S.P.; Yoo, J.-H.; Yoo, J. Organoid-based epithelial to mesenchymal transition (OEMT) model: from an intestinal fibrosis perspective. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Campbell, K.; Theveneau, E.e. The Epithelial-to Mesenchymal Transition: Methods and Protocols; Springer: US, 2021. [Google Scholar]
- Firulli, B.A.; Hadzic, D.B.; McDaid, J.R.; Firulli, A.B. The Basic Helix-Loop-Helix Transcription factorsdHAND and eHAND Exhibit Dimerization Characteristics That Suggest Complex Regulation of Function. J. Biol. Chem. 2000, 275, 33567–33573. [Google Scholar] [CrossRef]
- Srivastava, D.; Thomas, T.; Lin, Q.; Kirby, M.L.; Brown, D.; Olson, E.N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997, 16, 154–160. [Google Scholar] [CrossRef]
- Videira, R.F.; Koop, A.M.C.; Ottaviani, L.; Poels, E.M.; Kocken, J.M.M.; Dos Remedios, C.; Mendes-Ferreira, P.; Van De Kolk, K.W.; Sarvaas, G.J.D.M.; Lourenço, A.; et al. The adult heart requires baseline expression of the transcription factor Hand2 to withstand right ventricular pressure overload. Cardiovasc. Res. 2021, 118, 2688–2702. [Google Scholar] [CrossRef] [PubMed]
- VanDusen, N.J.; Casanovas, J.; Vincentz, J.W.; Firulli, B.A.; Osterwalder, M.; Lopez-Rios, J.; Zeller, R.; Zhou, B.; Grego-Bessa, J.; De La Pompa, J.L.; et al. Hand2 Is an Essential Regulator for Two Notch-Dependent Functions within the Embryonic Endocardium. Cell Rep. 2014, 9, 2071–2083. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.M.; Firulli, B.A.; VanDusen, N.J.; Morikawa, Y.; Conway, S.J.; Cserjesi, P.; Vincentz, J.W.; Firulli, A.B.; N, D.; G, B.; et al. Hand2 Loss-of-Function in Hand1 -Expressing Cells Reveals Distinct Roles in Epicardial and Coronary Vessel Development. Circ. Res. 2011, 108, 940–949. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.G.; McAnally, J.; Richardson, J.A.; Charité, J.; Olson, E.N. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 2002, 129, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Prummel, K.D.; Crowell, H.L.; Nieuwenhuize, S.; Brombacher, E.C.; Daetwyler, S.; Soneson, C.; Kresoja-Rakic, J.; Kocere, A.; Ronner, M.; Ernst, A.; et al. Hand2 delineates mesothelium progenitors and is reactivated in mesothelioma. Nat. Commun. 2022, 13, 1–21. [Google Scholar] [CrossRef]
- de Martin, X.; Sodaei, R.; Santpere, G. Mechanisms of Binding Specificity among bHLH Transcription Factors. Int. J. Mol. Sci. 2021, 22, 9150. [Google Scholar] [CrossRef]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef]
- Lenas, P.; Ikonomou, L. Developmental engineering: design of clinically efficacious bioartificial tissues through developmental and systems biology. Sci. China Life Sci. 2018, 61, 978–981. [Google Scholar] [CrossRef]
- Lenas, P.; Moos, M.; Luyten, F.P.; Ph. D.; M.D.; Fisher, J.N.; Peretti, G.M.; Scotti, C.; Dennis, S.C.; Berkland, C.J.; et al. Developmental Engineering: A New Paradigm for the Design and Manufacturing of Cell-Based Products. Part II. From Genes to Networks: Tissue Engineering from the Viewpoint of Systems Biology and Network Science. Tissue Eng. Part B: Rev. 2009, 15, 395–422. [Google Scholar] [CrossRef] [PubMed]
- Lenas, P.; Moos, M.; Luyten, F.P.; Ph. D.; M.D.; Gerges, I.; Tamplenizza, M.; Lopa, S.; Recordati, C.; Martello, F.; et al. Developmental Engineering: A New Paradigm for the Design and Manufacturing of Cell-Based Products. Part I: From Three-Dimensional Cell Growth to Biomimetics ofIn VivoDevelopment. Tissue Eng. Part B: Rev. 2009, 15, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Zarkesh, I.; Ashtiani, M.K.; Shiri, Z.; Aran, S.; Braun, T.; Baharvand, H. Synthetic developmental biology: Engineering approaches to guide multicellular organization. Stem Cell Rep. 2022, 17, 715–733. [Google Scholar] [CrossRef]
- Zohorsky, K.; Mequanint, K. Designing Biomaterials to Modulate Notch Signaling in Tissue Engineering and Regenerative Medicine. Tissue Eng. Part B: Rev. 2021, 27, 383–410. [Google Scholar] [CrossRef]
- Ho, C.; Morsut, L. Novel synthetic biology approaches for developmental systems. Stem Cell Rep. 2021, 16, 1051–1064. [Google Scholar] [CrossRef] [PubMed]
- Eitan, Y.; Sarig, U.; Dahan, N.; Machluf, M. Acellular Cardiac Extracellular Matrix as a Scaffold for Tissue Engineering:In VitroCell Support, Remodeling, and Biocompatibility. Tissue Eng. Part C: Methods 2010, 16, 671–683. [Google Scholar] [CrossRef]
- Sarig, U.; Au-Yeung, G.C.; Wang, Y.; Bronshtein, T.; Dahan, N.; Boey, F.Y.; Venkatraman, S.S.; Machluf, M. Thick Acellular Heart Extracellular Matrix with Inherent Vasculature: A Potential Platform for Myocardial Tissue Regeneration. Tissue Eng. Part A 2012, 18, 2125–2137. [Google Scholar] [CrossRef]
- Sarig, U.; Sarig, H.; Gora, A.; Krishnamoorthi, M.K.; Au-Yeung, G.C.T.; De-Berardinis, E.; Chaw, S.Y.; Mhaisalkar, P.; Bogireddi, H.; Ramakrishna, S.; et al. Biological and mechanical interplay at the Macro- and Microscales Modulates the Cell-Niche Fate. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Sarig, U.; Sarig, H.; De-Berardinis, E.; Chaw, S.-Y.; Nguyen, E.B.; Ramanujam, V.S.; Thang, V.D.; Al-Haddawi, M.; Liao, S.; Seliktar, D.; et al. Natural myocardial ECM patch drives cardiac progenitor based restoration even after scarring. Acta Biomater. 2016, 44, 209–220. [Google Scholar] [CrossRef]
- Lonza. hMSC - Human Mesenchymal Stem Cells, (2023). https://bioscience.lonza.com/lonza_bs/CH/en/Primary-and-Stem-Cells/p/000000000000186706/hMSC---Human-Mesenchymal-Stem-Cells.
- Sarig, U.; Nguyen, E.B.-V.; Wang, Y.; Ting, S.; Bronshtein, T.; Sarig, H.; Dahan, N.; Gvirtz, M.; Reuveny, S.; Oh, S.K.; et al. Pushing the Envelope in Tissue Engineering: Ex Vivo Production of Thick Vascularized Cardiac Extracellular Matrix Constructs. Tissue Eng. Part A 2015, 21, 1507–1519. [Google Scholar] [CrossRef]
- Duff, M.O.; Olson, S.; Wei, X.; Garrett, S.C.; Osman, A.; Bolisetty, M.; Plocik, A.; Celniker, S.E.; Graveley, B.R. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 2015, 521, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.A.; Morey, R.; Palpant, N.J.; Wang, P.L.; Afari, N.; Jiang, C.; Parast, M.M.; Murry, C.E.; Laurent, L.C.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 1–26. [Google Scholar] [CrossRef] [PubMed]
- dbGAP. Common Fund (CF) Genotype-Tissue Expression Project (GTEx) (dbGaP Study Accession: phs000424.v9.p2), https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v9.p2 (2023).
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Badylak, S.F. The extracellular matrix as a biologic scaffold material. Biomaterials 2007, 28, 3587–3593. [Google Scholar] [CrossRef] [PubMed]
- Mewhort, H.E.; Turnbull, J.D.; Satriano, A.; Chow, K.; Flewitt, J.A.; Andrei, A.-C.; Guzzardi, D.G.; Svystonyuk, D.A.; White, J.A.; Fedak, P.W. Epicardial infarct repair with bioinductive extracellular matrix promotes vasculogenesis and myocardial recovery. J. Hear. Lung Transplant. 2016, 35, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Saldin, L.T.; Cramer, M.C.; Velankar, S.S.; White, L.J.; Badylak, S.F. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater. 2017, 49, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Safran, M. et al. in Practical Guide to Life Science Databases Ch. Chapter 2, 27-56 (2021).
- Gu, X.; Zheng, Q.; Chu, Q.; Zhu, H. HAND2-AS1: A functional cancer-related long non-coding RNA. Biomed. Pharmacother. 2021, 137, 111317. [Google Scholar] [CrossRef]
- Da, C.-M.; Cheng, Z.-Y.; Gong, C.-Y.; Nan, W.; Zhou, K.-S.; Zhao, G.-H.; Zhang, H.-H. Role of HAND2-AS1 in human tumors. Clin. Chim. Acta 2020, 511, 189–197. [Google Scholar] [CrossRef]
- Krishnamoorthi, M.K.; Sarig, U.; Baruch, L.; Ting, S.; Reuveny, S.; Oh, S.K.-W.; Goldfracht, I.; Gepstein, L.; Venkatraman, S.S.; Tan, L.P.; et al. Robust Fabrication of Composite 3D Scaffolds with Tissue-Specific Bioactivity: A Proof-of-Concept Study. ACS Appl. Bio Mater. 2020, 3, 4974–4986. [Google Scholar] [CrossRef]
- Au-Yeung, G.C.T.; Sarig, U.; Sarig, H.; Bogireddi, H.; Bronshtein, T.; Baruch, L.; Spizzichino, A.; Bortman, J.; Freddy, B.Y.C.; Machluf, M.; et al. Restoring the biophysical properties of decellularized patches through recellularization. Biomater. Sci. 2017, 5, 1183–1194. [Google Scholar] [CrossRef]
- Efraim, Y.; Sarig, H.; Cohen Anavy, N.; Sarig, U.; de Berardinis, E.; Chaw, S.-Y.; Krishnamoorthi, M.; Kalifa, J.; Bogireddi, H.; Duc, T.V.; et al. Biohybrid cardiac ECM-based hydrogels improve long term cardiac function post myocardial infarction. Acta Biomater. 2017, 50, 220–233. [Google Scholar] [CrossRef]
- The-Human-Protein-Atlas. The tissue section - Tissue-based map of the human proteome, https://www.proteinatlas.org/ENSG00000164107-HAND2/tissue (2023).
- Zhang, X.-H.; Liang, X.; Liang, X.-H.; Wang, T.-S.; Qi, Q.-R.; Deng, W.-B.; Sha, A.-G.; Yang, Z.-M. The Mesenchymal—Epithelial Transition During In Vitro Decidualization. Reprod. Sci. 2013, 20, 354–360. [Google Scholar] [CrossRef] [PubMed]
- DeMayo, F.J.; Lydon, J.P. 90 YEARS OF PROGESTERONE: New insights into progesterone receptor signaling in the endometrium required for embryo implantation. J. Mol. Endocrinol. 2020, 65, T1–T14. [Google Scholar] [CrossRef]
- Yu, J.-J.; Sun, H.-T.; Zhang, Z.-F.; Shi, R.-X.; Liu, L.-B.; Shang, W.-Q.; Wei, C.-Y.; Chang, K.-K.; Shao, J.; Wang, M.-Y.; et al. IL15 promotes growth and invasion of endometrial stromal cells and inhibits killing activity of NK cells in endometriosis. Reproduction 2016, 152, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Bellelis, P.; Barbeiro, D.F.; Gueuvoghlanian-Silva, B.Y.; Kalil, J.; Abrão, M.S.; Podgaec, S. Interleukin-15 and Interleukin-7 are the Major Cytokines to Maintain Endometriosis. Gynecol. Obstet. Investig. 2019, 84, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.-J.; Li, R.-G.; Wang, Z.-S.; Zhang, M.; Qu, X.-K.; Qiao, Q.; Li, X.-M.; Di, R.-M.; Qiu, X.-B.; et al. HAND2 loss-of-function mutation causes familial dilated cardiomyopathy. Eur. J. Med Genet. 2018, 62, 103540. [Google Scholar] [CrossRef]
- Giroud, M.; Tsokanos, F.-F.; Caratti, G.; Kotschi, S.; Khani, S.; Jouffe, C.; Vogl, E.S.; Irmler, M.; Glantschnig, C.; Gil-Lozano, M.; et al. HAND2 is a novel obesity-linked adipogenic transcription factor regulated by glucocorticoid signalling. Diabetologia 2021, 64, 1850–1865. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Yu, X.; Chen, W.; Chen, D.; Cai, J.; Jiang, Y.; Liu, X.; Wu, Z.; Wang, L.; Grady, W.M.; et al. Epigenetic silencing and tumor suppressor gene of HAND2 by targeting ERK signaling in colorectal cancer. Cell Commun. Signal. 2022, 20, 1–11. [Google Scholar] [CrossRef]
- Zeng, G.-G.; Li, H.; Tang, C.-K. HAND2-AS1: A novel therapeutic target for atherosclerosis. Int. J. Cardiol. 2022, 360, 76–76. [Google Scholar] [CrossRef]
- Ma, L.; He, S.; Li, H.; Zhang, S.; Yin, Y. HAND2-AS1 targeting miR-1208/SIRT1 axis alleviates foam cell formation in atherosclerosis. Int. J. Cardiol. 2022, 346, 53–61. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Y.; Ma, C.; Guan, C.; Ma, X.; Meng, S. Mesenchymal stem cell-originated exosomal lncRNA HAND2-AS1 impairs rheumatoid arthritis fibroblast-like synoviocyte activation through miR-143-3p/TNFAIP3/NF-κB pathway. J. Orthop. Surg. Res. 2021, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Y. LncRNA HAND2-AS1 attenuates glioma cell proliferation, invasion and migration by targeting CDK6. Neurol. Res. 2022, 44, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Yang, J.; Zhang, D.; Cui, M.; Li, L. lncRNA HAND2-AS1 Regulates Prostate Cancer Cell Growth Through Targeting the miR-106a-5p/RBM24 Axis [Retraction]. OncoTargets Ther. 2022, ume 15, 133–134. [Google Scholar] [CrossRef]
- Li, L.; Li, L.; Hu, L.; Li, T.; Xie, D.; Liu, X. Long non-coding RNA HAND2-AS1/miR-106a/PTEN axis re-sensitizes cisplatin-resistant ovarian cells to cisplatin treatment. Mol. Med. Rep. 2021, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA HAND2-AS1 Acts as a Tumor Suppressor in High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 4059. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-J.; Song, Z.-W.; Fei, J.-G.; Chen, F. Long non-coding RNA HAND2-AS1: A potential therapeutic target in the treatment of gastric cancer? Dig. Liver Dis. 2021, 54, 148–148. [Google Scholar] [CrossRef]
- Shan, L.; Liu, W.; Zhan, Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on bladder cancer via restoration of RARB as a sponge of microRNA-146. Cancer Cell Int. 2021, 21, 1–14. [Google Scholar] [CrossRef]
- Gao, Y.; Zou, T.; Liang, W.; Zhang, Z.; Qie, M. Long non-coding RNA HAND2-AS1 delays cervical cancer progression via its regulation on the microRNA-21-5p/TIMP3/VEGFA axis. Cancer Gene Ther. 2020, 28, 619–633. [Google Scholar] [CrossRef]
- Gong, J.; Fan, H.; Deng, J.; Zhang, Q. LncRNA HAND2-AS1 represses cervical cancer progression by interaction with transcription factor E2F4 at the promoter of C16orf74. J. Cell. Mol. Med. 2020, 24, 6015–6027. [Google Scholar] [CrossRef]
- Xing, L.; Tang, X.; Wu, K.; Huang, X.; Yi, Y.; Huan, J. LncRNA HAND2-AS1 suppressed the growth of triple negative breast cancer via reducing secretion of MSCs derived exosomal miR-106a-5p. Aging 2020, 13, 424–436. [Google Scholar] [CrossRef]
- Ding, J.; Li, Y.; Zhang, Y.; Fan, B.; Li, Q.; Zhang, J.; Zhang, J. Identification of key lncRNAs in the tumorigenesis of intraductal pancreatic mucinous neoplasm by coexpression network analysis. Cancer Med. 2020, 9, 3840–3851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, P.; Luo, J.; Wang, J.; Liu, Z.; Wu, W.; Du, Y.; Ye, B.; Wang, D.; He, L.; et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019, 38, e101110. [Google Scholar] [CrossRef] [PubMed]
- Jing, G.; Zheng, X.; Ji, X. lncRNA HAND2-AS1 overexpression inhibits cancer cell proliferation in hepatocellular carcinoma by downregulating RUNX2 expression. J. Clin. Lab. Anal. 2021, 35, e23717. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Jin, F.; Lin, Y. lncRNA HAND2-AS1 Inhibits Liver Cancer Cell Proliferation and Migration by Upregulating SOCS5 to Inactivate the JAK-STAT Pathway. Cancer Biotherapy Radiopharm. 2020, 35, 143–152. [Google Scholar] [CrossRef]
- Al-Azab, M.; Wang, B.; Elkhider, A.; Walana, W.; Li, W.; Yuan, B.; Ye, Y.; Tang, Y.; Almoiliqy, M.; Adlat, S.; et al. Indian Hedgehog regulates senescence in bone marrow-derived mesenchymal stem cell through modulation of ROS/mTOR/4EBP1, p70S6K1/2 pathway. Aging 2020, 12, 5693–5715. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Decellularized porcine cardiac extracellular matrix (pcECM) is acellular and preserves cardio-vascular tissue specific ultrastructure and bioactive growth factors. Decellularized pcECM was paraffin cross sectioned and compared to native porcine left ventricular tissue using hematoxylin and eosin (H&E) and Masson’s tri-chrome (MTC) stains, as indicated. Sequential paraffin cross sections of the pcECM were also immunohistochemically (IHC) stained, as indicated, for two representative cardio-vascular related growth factors: basic fibroblast growth factor (bFGF) and vascular endothelial growth factor A (VEGF), respectively. IHC staining for epidermal growth factor (EGF) is also presented as control, along with the isotype staining for each case. Scale bars: 100μm. Microscope magnifications: 200x. Representative images are presented out of at least n=3 biological samples and n=2 IHC / histological section stains tested for each group.
Figure 1.
Decellularized porcine cardiac extracellular matrix (pcECM) is acellular and preserves cardio-vascular tissue specific ultrastructure and bioactive growth factors. Decellularized pcECM was paraffin cross sectioned and compared to native porcine left ventricular tissue using hematoxylin and eosin (H&E) and Masson’s tri-chrome (MTC) stains, as indicated. Sequential paraffin cross sections of the pcECM were also immunohistochemically (IHC) stained, as indicated, for two representative cardio-vascular related growth factors: basic fibroblast growth factor (bFGF) and vascular endothelial growth factor A (VEGF), respectively. IHC staining for epidermal growth factor (EGF) is also presented as control, along with the isotype staining for each case. Scale bars: 100μm. Microscope magnifications: 200x. Representative images are presented out of at least n=3 biological samples and n=2 IHC / histological section stains tested for each group.
Figure 2.
Human mesenchymal stem cells (hMSCs) characterization. Negative (CD31-, isotype control) and positive (CD90+CD105+CD73+, as indicated) marker stains for bone marrow hMSCs (Lonza, Switzerland) show consensus marker expression. Cellular morphology even at passage 5 remained characteristic of hMSCs as indicated by brightfield phase contrast microscopy. Furthermore, the cells differentiation potential appeared to be conserved at that passage as well as shown for hMSC derived differentiated adipocytes- (oil red), chondrocytes- (Alcian blue) and osteocytes- (Alizarin red S) like cells, as indicated. Brightfield image shows untreated hMSC control morphology. Magnifications: Brightfield–100x; Fluorescent images–200x; Differentiation images: 40x.
Figure 2.
Human mesenchymal stem cells (hMSCs) characterization. Negative (CD31-, isotype control) and positive (CD90+CD105+CD73+, as indicated) marker stains for bone marrow hMSCs (Lonza, Switzerland) show consensus marker expression. Cellular morphology even at passage 5 remained characteristic of hMSCs as indicated by brightfield phase contrast microscopy. Furthermore, the cells differentiation potential appeared to be conserved at that passage as well as shown for hMSC derived differentiated adipocytes- (oil red), chondrocytes- (Alcian blue) and osteocytes- (Alizarin red S) like cells, as indicated. Brightfield image shows untreated hMSC control morphology. Magnifications: Brightfield–100x; Fluorescent images–200x; Differentiation images: 40x.
Figure 3.
pcECM induced early HAND2 expression followed by gradual MET towards an endothelial phenotype. Three to four days after seeding, hMSCs on pcECM expressed high levels of HAND2 at the mRNA (Supplementary Table T3) and protein levels (HAND2 – Day 4 nuclei localization, IHC bottom left image as indicated). This was accompanied by hMSCs exhibiting an unusual squamous monolayer epithelial-like morphology (arrows in HAND2 IHC image) and was followed by a gradual increase in the expression of endothelial cell markers through long-term (up to 30 days) cultures (Supplementary Table T3). Protein level expression of endothelial markers is demonstrated by CD31/PECAM+ presence in both trypsinized hMSCs (using fluorescent activated cell sorting, FACS, at two and four weeks after seeding, top row) and in situ immunofluorescence imaging (CD31 – Day 30 as indicated). Scale bars: IHC HAND2 stain: 500µm. CD31 IF stain: 100µm.
Figure 3.
pcECM induced early HAND2 expression followed by gradual MET towards an endothelial phenotype. Three to four days after seeding, hMSCs on pcECM expressed high levels of HAND2 at the mRNA (Supplementary Table T3) and protein levels (HAND2 – Day 4 nuclei localization, IHC bottom left image as indicated). This was accompanied by hMSCs exhibiting an unusual squamous monolayer epithelial-like morphology (arrows in HAND2 IHC image) and was followed by a gradual increase in the expression of endothelial cell markers through long-term (up to 30 days) cultures (Supplementary Table T3). Protein level expression of endothelial markers is demonstrated by CD31/PECAM+ presence in both trypsinized hMSCs (using fluorescent activated cell sorting, FACS, at two and four weeks after seeding, top row) and in situ immunofluorescence imaging (CD31 – Day 30 as indicated). Scale bars: IHC HAND2 stain: 500µm. CD31 IF stain: 100µm.
Figure 4.
Bioinformatic analyses of HAND2 and its associated lncRNA HAND2-AS1 expression patterns in adult, embryonic and pathological tissues Bioinformatic analyses of HAND2 (NCBI Gene accession: 9464, a) and HAND2-AS1 (NCBI Gene accession: 79804, b) expression in adult human tissues (reads per kilobase per million reads, RPKM) based on the national center for biotechnology information (NCBI) bioproject #PRJNA280600 (“RNA sequencing of total RNA from 20 human tissues”, 45). Insets in (a) and (b) show corresponding HAND2 and HAND2-AS1 embryonic expression, respectively, at sampled tissues between weeks 10 to 20 of human gestation as sourced from the NCBI bioproject #PRJNA270632 (“Tissue-specific circular RNA induction during human fetal development”, 46); Expression levels of HAND2 and HAND2-AS1 (RPKM, as indicated) in a collection of cancer samples (red dots), relative to neighboring normal tissue samples (green dots) are shown side by side for various cancer types abbreviated as follows: BLCA– Bladder Urothelial Carcinoma; BRCA– Breast invasive carcinoma; CESC– Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL– Cholangiocarcinoma; COAD– Colon adenocarcinoma; ESCA– Esophageal carcinoma; HNSC– Head and Neck squamous cell carcinoma; LIHC– Liver hepatocellular carcinoma; LUAD– Lung adenocarcinoma; LUSC– Lung squamous cell carcinoma; PAAD– Pancreatic adenocarcinoma; PCPG– Pheochromocytoma and Paraganglioma; PRAD– Prostate adenocarcinoma; READ– Rectum adenocarcinoma; SARC– Sarcoma; SKCM– Skin Cutaneous Melanoma; STAD– Stomach adenocarcinoma; THCA– Thyroid carcinoma; THYM– Thymoma; UCEC– Uterine Corpus Endometrial Carcinoma. Cancer data was obtained from the national institute of health (NIH) and national cancer institute (NCI) supported cancer genome atlas program (TCGA); and from the Genotype-Tissue Expression (GTEx) project databases (data sources appear in the materials and methods section). Correlation between HAND2 and HAND2-AS1 expression levels in normal tissues (d, left), tumor tissues (d, middle), and combined normal and tumor tissues (d, right), combining both the TCGA and GTEx data sets.
Figure 4.
Bioinformatic analyses of HAND2 and its associated lncRNA HAND2-AS1 expression patterns in adult, embryonic and pathological tissues Bioinformatic analyses of HAND2 (NCBI Gene accession: 9464, a) and HAND2-AS1 (NCBI Gene accession: 79804, b) expression in adult human tissues (reads per kilobase per million reads, RPKM) based on the national center for biotechnology information (NCBI) bioproject #PRJNA280600 (“RNA sequencing of total RNA from 20 human tissues”, 45). Insets in (a) and (b) show corresponding HAND2 and HAND2-AS1 embryonic expression, respectively, at sampled tissues between weeks 10 to 20 of human gestation as sourced from the NCBI bioproject #PRJNA270632 (“Tissue-specific circular RNA induction during human fetal development”, 46); Expression levels of HAND2 and HAND2-AS1 (RPKM, as indicated) in a collection of cancer samples (red dots), relative to neighboring normal tissue samples (green dots) are shown side by side for various cancer types abbreviated as follows: BLCA– Bladder Urothelial Carcinoma; BRCA– Breast invasive carcinoma; CESC– Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL– Cholangiocarcinoma; COAD– Colon adenocarcinoma; ESCA– Esophageal carcinoma; HNSC– Head and Neck squamous cell carcinoma; LIHC– Liver hepatocellular carcinoma; LUAD– Lung adenocarcinoma; LUSC– Lung squamous cell carcinoma; PAAD– Pancreatic adenocarcinoma; PCPG– Pheochromocytoma and Paraganglioma; PRAD– Prostate adenocarcinoma; READ– Rectum adenocarcinoma; SARC– Sarcoma; SKCM– Skin Cutaneous Melanoma; STAD– Stomach adenocarcinoma; THCA– Thyroid carcinoma; THYM– Thymoma; UCEC– Uterine Corpus Endometrial Carcinoma. Cancer data was obtained from the national institute of health (NIH) and national cancer institute (NCI) supported cancer genome atlas program (TCGA); and from the Genotype-Tissue Expression (GTEx) project databases (data sources appear in the materials and methods section). Correlation between HAND2 and HAND2-AS1 expression levels in normal tissues (d, left), tumor tissues (d, middle), and combined normal and tumor tissues (d, right), combining both the TCGA and GTEx data sets.
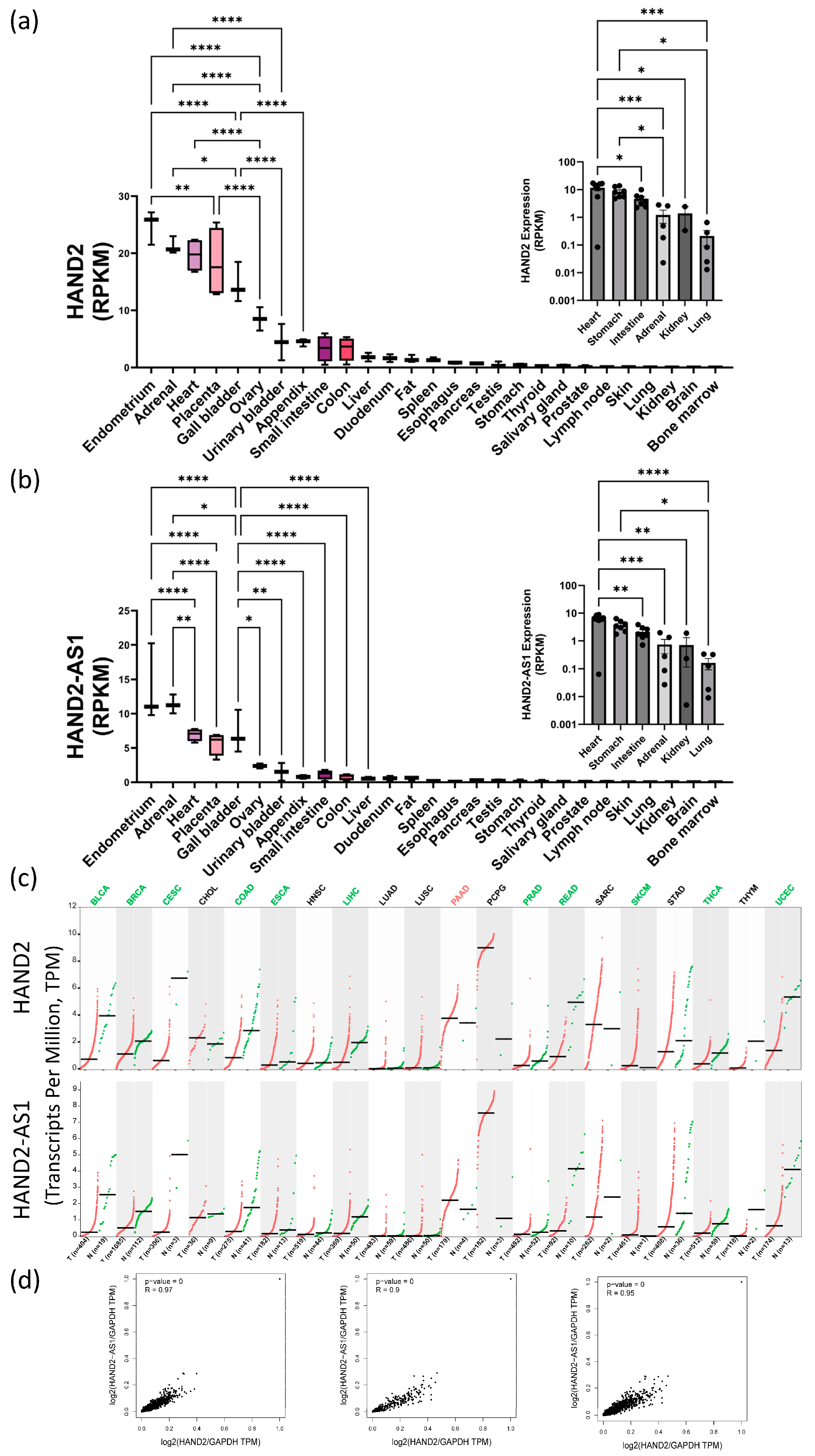
Figure 5.
HAND2/HAND2-AS1 locus knockout (KO) hMSCs using CRISPR-Cas9 genomic editing. Genome view of HAND2 (sense strand, blue) and HAND2-AS1 isoforms (antisense strand, green) sharing a common promoter region (red ENCODE region, a), in which permanent KO (b, CRISPR‒Cas9 black sequence top lane; KO region is graphically displayed by purple rectangle; KO vector map is provided in
Supplementary Figure S1) led to a hMSC senescent morphology (b, MUT-marked images show time progression of proliferation-arrest and hypertrophy within the same wells, without passaging) compared to wild-type (WT) and to heterozygous cells that managed to form characteristic hMSC colonies (b, as indicated). Given the scarcity of full MUT cells (non-dividing) we compared HAND2 PCR products using gel electrophoresis (c) for the original (WT, untreated) and total gene-edited plates (mixed MUT) comprising mostly HAND2
-/- senescent cells. Colony clones (clone 1–clone 3) displayed a mixed WT-MUT band signature suggesting heterozygosity to the CRISPR-Cas9 editing. Mixed MUT band comprising mostly HAND2
-/- senescent cells displayed a shorter bright mutant 260 bp band. Sanger sequencing of that mutant (MUT) band confirmed deletions compared to the original sequence as per vector design (
Supplementary Figure S2).
Figure 5.
HAND2/HAND2-AS1 locus knockout (KO) hMSCs using CRISPR-Cas9 genomic editing. Genome view of HAND2 (sense strand, blue) and HAND2-AS1 isoforms (antisense strand, green) sharing a common promoter region (red ENCODE region, a), in which permanent KO (b, CRISPR‒Cas9 black sequence top lane; KO region is graphically displayed by purple rectangle; KO vector map is provided in
Supplementary Figure S1) led to a hMSC senescent morphology (b, MUT-marked images show time progression of proliferation-arrest and hypertrophy within the same wells, without passaging) compared to wild-type (WT) and to heterozygous cells that managed to form characteristic hMSC colonies (b, as indicated). Given the scarcity of full MUT cells (non-dividing) we compared HAND2 PCR products using gel electrophoresis (c) for the original (WT, untreated) and total gene-edited plates (mixed MUT) comprising mostly HAND2
-/- senescent cells. Colony clones (clone 1–clone 3) displayed a mixed WT-MUT band signature suggesting heterozygosity to the CRISPR-Cas9 editing. Mixed MUT band comprising mostly HAND2
-/- senescent cells displayed a shorter bright mutant 260 bp band. Sanger sequencing of that mutant (MUT) band confirmed deletions compared to the original sequence as per vector design (
Supplementary Figure S2).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).