Submitted:
20 September 2023
Posted:
25 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
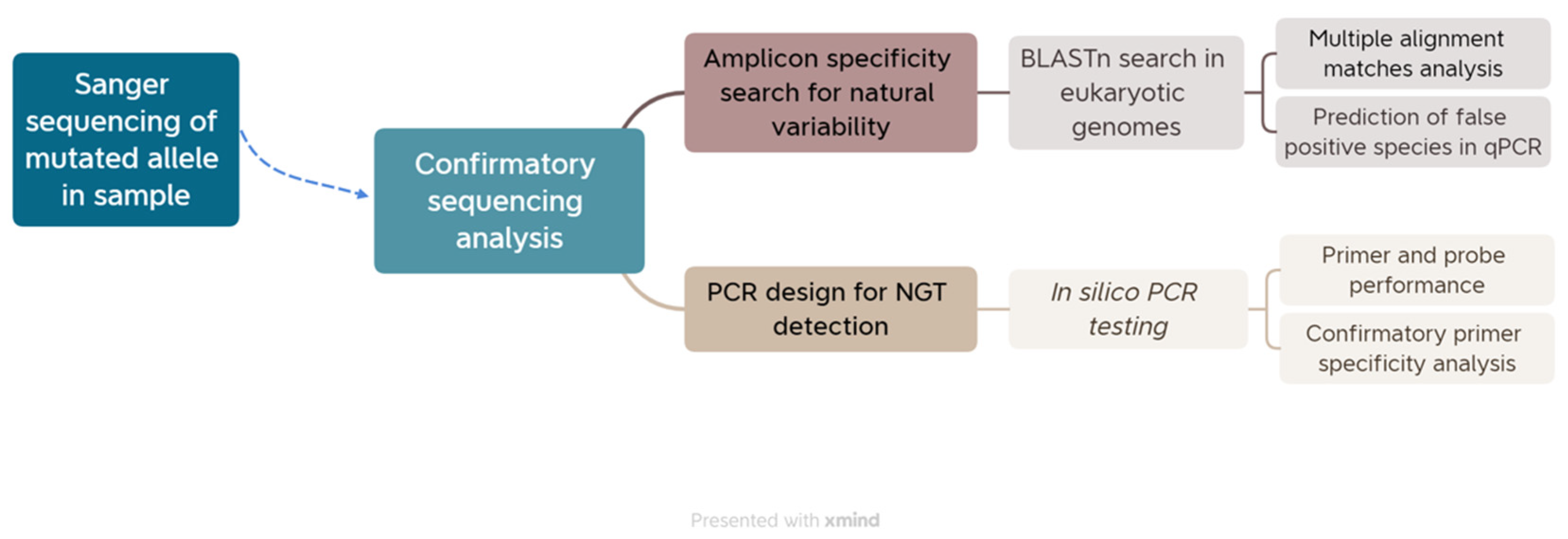
2.1. Overall Description of the Stepwise Approach
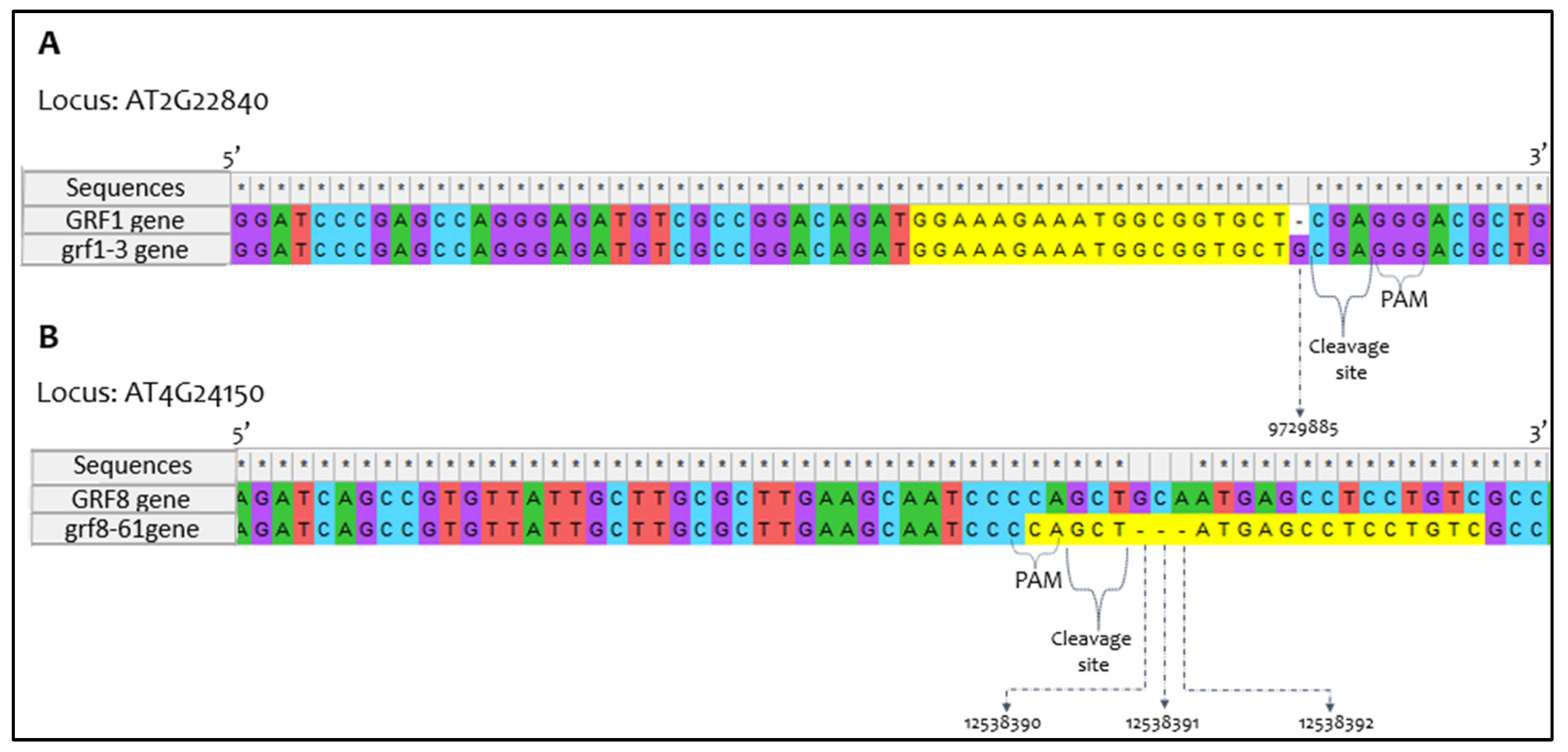
2.2. Confirmatory Sequencing Analysis of CRISPR/Cas9 Mutations
2.3. Amplicon Sequence Search for Natural Variants
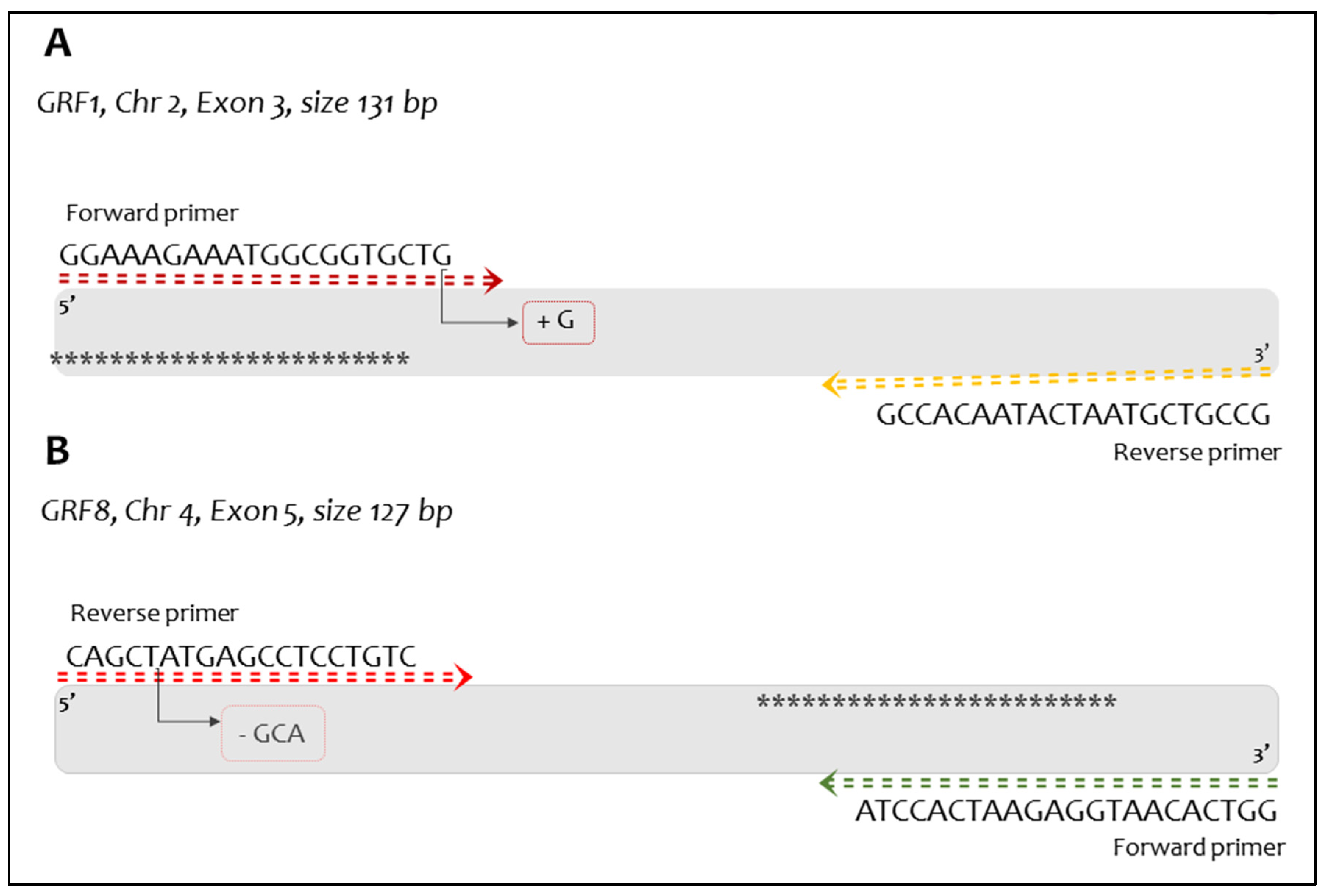
2.4. RT-PCR Primer and Probe Design
2.5. In-Silico PCR Testing
3. Results
3.1. Sequence Confirmation of Mutated Alleles
3.3. In-Silico Specificity Assessment
3.4. In-Silico PCR Performance
| Total of mismatches | Number/hits analyzed | Sequences corresponding perfectly to the primer | Number of Blast hits recovered | Possible discrimination between the grf1-3 genotype and other lines |
|---|---|---|---|---|
| 1 | 39 | 0 | 57 | Low |
| 2 | 17 | 0 | ||
| 3 | 1 | 0 |
| Total of mismatches | Number/hits analyzed | Sequences corresponding perfectly to the primers | Number of Blast hits recovered | Possible discrimination between the grf8-3 genotype and other lines |
|---|---|---|---|---|
| 1 | 1 | 1 | 1 | High |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Agapito-Tenfen, S.Z.; Okoli, A.S.; Bernstein, M.J.; Wikmark, O.-G.; Myhr, A.I. Revisiting Risk Governance of GM Plants: The Need to Consider New and Emerging Gene-Editing Techniques. Front. Plant Sci. 2018, 9, 1874. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Saeed, S.; Khan, M.H.U.; Khan, S.U.; Mora-Poblete, F.; Kamran, M.; Faheem, A.; Maqsood, A.; Rauf, M.; Saleem, S.; et al. A Revolution toward Gene-Editing Technology and Its Application to Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef] [PubMed]
- Angulo, J.; et al. Targeted mutagenesis of the Arabidopsis GROWTH-REGULATING FACTOR (GRF) gene family suggests competition of multiplexed sgRNAs for Cas9 apoprotein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ayyadevara, S.; Thaden, J.J.; Reis, R.J.S. Discrimination of Primer 3′-Nucleotide Mismatch by Taq DNA Polymerase during Polymerase Chain Reaction. Anal. Biochem. 2000, 284, 11–18. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth Stage-Based Phenotypic Analysis of Arabidopsis: A Model for High Throughput Functional Genomics in Plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef]
- BRASIL. DECRETO No 4.680, DE 24 DE ABRIL DE 2003. Brasília, 2003.
- Broccanello, C.; Chiodi, C.; Funk, A.; McGrath, J.M.; Panella, L.; Stevanato, P. Comparison of three PCR-based assays for SNP genotyping in plants. Plant Methods 2018, 14, 1–8. [Google Scholar] [CrossRef]
- Capecchi, M.R. The new mouse genetics: Altering the genome by gene targeting. Trends Genet. 1989, 5, 70–76. [Google Scholar] [CrossRef]
- CBD. What is Agricultural Biodiversity? In: 2008, Anais [...]. : Convention on Biological Diversity. 2008. Available online: https://www.cbd.int/agro/whatis.shtml.
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Chhalliyil, P.; Ilves, H.; Kazakov, S.A.; Howard, S.J.; Johnston, B.H.; Fagan, J. A Real-Time Quantitative PCR Method Specific for Detection and Quantification of the First Commercialized Genome-Edited Plant. Foods 2020, 9, 1245. [Google Scholar] [CrossRef]
- Chhalliyil, P.; Ilves, H.; Kazakov, S.A.; Howard, S.J.; Johnston, B.H.; Fagan, J. A Real-Time Quantitative PCR Method Specific for Detection and Quantification of the First Commercialized Genome-Edited Plant. Foods 2020, 9, 1245. [Google Scholar] [CrossRef]
- ENGL. Detection of food and feed plant products obtained by new mutagenesis techniques. [S. l.], n. ENGL. Detection of food and feed plant products obtained by new mutagenesis techniques. [S. l.], n. March, p. 21, 2019. Available online: https://gmo-crl.jrc.ec.europa.eu/doc/JRC116289-GE-report-ENGL.pdf.
- EUROPEAN COMMISSION. EC, Science for Environment Policy Future Brief: Synthetic Biology and Biodiversity. [s.l: S.n.]. Available online: http://ec.europa.eu/science-environment-policy.
- EFSA, G.M.O. Scientific opinion addressing the safety assessment of plants developed using Zinc Finger Nuclease 3 and other Site-Directed Nucleases with similar function. EFSA J. 2012, 10, 2193. [Google Scholar] [CrossRef]
- EUROPEAN LABORATORIES. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. [S. l.], 2015.
- Fraiture, M.-A.; Guiderdoni, E.; Meunier, A.-C.; Papazova, N.; Roosens, N.H. ddPCR strategy to detect a gene-edited plant carrying a single variation point: Technical feasibility and interpretation issues. Food Control. 2022, 137, 108904. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science 2010, 329, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Höijer, I.; Johansson, J.; Gudmundsson, S.; Chin, C.-S.; Bunikis, I.; Häggqvist, S.; Emmanouilidou, A.; Wilbe, M.; den Hoed, M.; Bondeson, M.-L.; et al. Amplification-free long-read sequencing reveals unforeseen CRISPR-Cas9 off-target activity. Genome Biol. 2020, 21, 290. [Google Scholar] [CrossRef] [PubMed]
- Sala, S.; Crenna, E.; Secchi, M.; Pant, R. Global normalisation factors for the Environmental Footprint and Life Cycle Assessment [Internet]. 2017. Available online: https://ec.europa.eu/jrc.
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- HUANG, M.M.; ARNHEIM, N.; GOODMAN, M.F. Extension of base mispairs by Taq DNA polymerase: Implications for single. Nucleic.Acids.Res. 1992, 20, 4567–4573. [Google Scholar] [CrossRef]
- ICSWGSB, International Civil Society Working Group On Synthetic Biology. Synthetic Biology and the CBD Five key decisions for COP 13 & COP-MOP 8. [S. l.], p. 8pp, 2016. Available online: www.boell.de/en/2016/11/02/synthetic-biology-and-cbd-five-keydecisions- cop-13-cop-mop-8.
- Isaac, R.S.; Jiang, F.; A Doudna, J.; A Lim, W.; Narlikar, G.J.; Almeida, R. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife 2016, 5. [Google Scholar] [CrossRef]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Jones, H.D. Regulatory uncertainty over genome editing. Nat. Plants 2015, 1, 14011. [Google Scholar] [CrossRef]
- Karginov, F.V.; Hannon, G.J. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell 2010, 37, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.; Lee, J.; Kim, J.-S. Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol. Cells 2015, 38, 475–481. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-T.; Shaw, P.-C. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [CrossRef]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef]
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312. [Google Scholar] [CrossRef]
- Mishra, R.; Zhao, K. Genome editing technologies and their applications in crop improvement. Plant Biotechnol. Rep. 2018, 12, 57–68. [Google Scholar] [CrossRef]
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Wilhelm, R. What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map. Environ. Évid. 2019, 8, 1–33. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.-D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K. Genome Editing with Engineered Nucleases in Plants. Plant Cell Physiol. 2014, 56, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Pröbsting, M.; Schenke, D.; Hossain, R.; Häder, C.; Thurau, T.; Wighardt, L.; Schuster, A.; Zhou, Z.; Ye, W.; Rietz, S.; et al. Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 2020, 18, 2328–2344. [Google Scholar] [CrossRef] [PubMed]
- Ribarits, A.; Eckerstorfer, M.; Simon, S.; Stepanek, W. Genome-Edited Plants: Opportunities and Challenges for an Anticipatory Detection and Identification Framework. Foods 2021, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Salsman, J.; Dellaire, G. Precision genome editing in the CRISPR era. Biochem. Cell Biol. 2017, 95, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Shelake, R.M.; Pramanik, D.; Kim, J.-Y. Evolution of plant mutagenesis tools: A shifting paradigm from random to targeted genome editing. Plant Biotechnol. Rep. 2019, 13, 423–445. [Google Scholar] [CrossRef]
- Shillito, R.D.; Whitt, S.; Ross, M.; Ghavami, F.; De Vleesschauwer, D.; D’halluin, K.; Van Hoecke, A.; Meulewaeter, F. Detection of genome edits in plants—From editing to seed. Vitr. Cell. Dev. Biol. - Plant 2021, 57, 595–608. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2014, 33, 187–197. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2014, 33, 187–197. [Google Scholar] [CrossRef]
- Verkuijl, S.A.; Rots, M.G. The influence of eukaryotic chromatin state on CRISPR–Cas9 editing efficiencies. Curr. Opin. Biotechnol. 2018, 55, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Wahler, D.; Schauser, L.; Bendiek, J.; Grohmann, L. Next-Generation Sequencing as a Tool for Detailed Molecular Characterisation of Genomic Insertions and Flanking Regions in Genetically Modified Plants: A Pilot Study Using a Rice Event Unauthorised in the EU. Food Anal. Methods 2013, 6, 1718–1727. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Weidner, C.; Edelmann, S.; Moor, D.; Lieske, K.; Savini, C.; Jacchia, S.; Sacco, M.G.; Mazzara, M.; Lämke, J.; Eckermann, K.N.; et al. Assessment of the Real-Time PCR Method Claiming to be Specific for Detection and Quantification of the First Commercialised Genome-Edited Plant. Food Anal. Methods 2022, 15, 2107–2125. [Google Scholar] [CrossRef]
- WENDY, Cannon; PEDERSON, David. Mechanisms and conseqeunces of double-strand break formation in chromatin. Physiology & behavior, [S. l.], v. 176, n. 5, p. 139–148, 2017. [CrossRef]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.O.W.; O’brien, A.R.; Bauer, D.C. The Current State and Future of CRISPR-Cas9 gRNA Design Tools. Front. Pharmacol. 2018, 9, 749. [Google Scholar] [CrossRef]
- Yan, J.; Xue, D.; Chuai, G.; Gao, Y.; Zhang, G.; Liu, Q. Benchmarking and integrating genome-wide CRISPR off-target detection and prediction. Nucleic Acids Res. 2020, 48, 11370–11379. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Zhao, S.; Yan, X.; Si, N.; Gao, H.; Li, Y.; Zhai, S.; Xiao, F.; Wu, G.; et al. An Editing-Site-Specific PCR Method for Detection and Quantification of CAO1-Edited Rice. Foods 2021, 10, 1209. [Google Scholar] [CrossRef]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef]
| Hits | Accession | Blast against the query grf1-3 | (MSA) Mismatches (bp) against each accession | |||
|---|---|---|---|---|---|---|
| Quey cover | Per. Ident | Forward | Probe | Reverse | ||
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR782543.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699746.2 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699771.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699766.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699761.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699756.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR699751.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR215053.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana growth-regulating factor 1 (GRF1), mRNA | NM_127849.4 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana chromosome 2 | CP116281.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | OX298798.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | OX298803.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 1254 chromosome 2 sequence | CP086755.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 5856 chromosome 2 sequence | CP086750.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 6021 chromosome 2 sequence | CP086745.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 6024 chromosome 2 sequence | CP086740.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 9412 chromosome 2 sequence | CP086735.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana ecotype 9470 chromosome 2 sequence | CP086730.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana chromosome 2 | CP087127.2 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana isolate t2t_salk_col chromosome 2 | CP096025.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | OW119597.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR881467.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR797808.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR797803.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR797798.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR797793.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana genome assembly, chromosome: 2 | LR797788.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana chromosome 2 | CP002685.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana At2g22840 mRNA for hypothetical protein, partial cds, clone: RAAt2g22840 | AB493560.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana isolate CS906 GRL1 (GRL1) gene, partial cds | EU550462.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana isolate CS902 GRL1 (GRL1) gene, partial cds | EU550456.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana isolate CS6799 GRL1 (GRL1) gene, partial cds | EU550455.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana isolate CS901 GRL1 (GRL1) gene, partial cds | EU550445.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana transcription activator (GRF1) mRNA, complete cds | AY102634.1 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana chromosome 2 clone T20K9 map CIC06C07, complete sequence | AC004786.3 | 100% | 99.24% | 1 | 0 | 0 |
| Arabidopsis thaliana Full-length cDNA Complete sequence from clone GSLTPGH12ZD08 of Hormone Treated Callus of strain col-0 of Arabidopsis thaliana (thale cress) | BX820248.1 | 100% | 99.24% | 1 | 0 | 0 |
| PREDICTED: Arabidopsis xampl subsp. xampl growth-regulating factor 1 (LOC9316532), mRNA | XM_002878592.2 | 100% | 98.47% | 1 | 0 | 0 |
| PREDICTED: Camelina sativa growth-regulating factor 1-like (LOC104713726), mRNA | XM_010430916.2 | 100% | 98.47% | 1 | 0 | 0 |
| PREDICTED: Camelina sativa growth-regulating factor 1 (LOC104751923), mRNA | XM_010473979.2 | 100% | 98.47% | 1 | 0 | 0 |
| PREDICTED: Camelina sativa growth-regulating factor 1-like (LOC104704976), mRNA | XM_010420970.1 | 100% | 98.47% | 1 | 0 | 0 |
| Camelina hispida cultivar hispida voucher DAO 902780 chromosome 2 | CP094632.1 | 100% | 97.71% | 1 | 0 | 1 |
| Arabidopsis arenosa genome assembly, chromosome: 4 | LR999454.1 | 100% | 97.71% | 1 | 0 | 1 |
| Raphanus sativus genome assembly, chromosome: 6 | LR778315.1 | 98% | 96.12% | 1 | 0 | 2 (gap) |
| PREDICTED: Raphanus sativus growth-regulating factor 1 (LOC108836427), mRNA | XM_018609585.1 | 98% | 96.12% | 1 | 0 | 2 (gap) |
| PREDICTED: Brassica rapa growth-regulating factor 1 (LOC103858395), mRNA | XM_009135745.3 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica oleracea HDEM genome, scaffold: C3 | LR031872.1 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica rapa genome, scaffold: A03 | LR031572.1 | 100% | 95.42% | 1 | 0 | 1 |
| PREDICTED: Capsella rubella growth-regulating factor 1 (LOC17887921), mRNA | XM_006293922.2 | 100% | 95.42% | 1 | 1 | 1 |
| PREDICTED: Brassica napus growth-regulating factor 1-like (LOC125584397), mRNA | XM_048752816.1 | 100% | 95.42% | 1 | 0 | 1 |
| PREDICTED: Brassica napus growth-regulating factor 1 (LOC106389497), mRNA | XM_013829762.3 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica rapa genome assembly, chromosome: A03 | LS974619.2 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica napus genome assembly, chromosome: C03 | HG994367.1 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica napus genome assembly, chromosome: A03 | HG994357.1 | 100% | 95.42% | 1 | 0 | 1 |
| Brassica rapa subsp. Pekinensis growth-regulating xamp 1 mRNA, partial cds | JN698986.1 | 100% | 95.42% | 1 | 0 | 1 |
| PREDICTED: Brassica oleracea var. oleracea growth-regulating factor 1 (LOC106328366), mRNA | XM_013766798.1 | 100% | 94.66% | 1 | 0 | 1 |
| PREDICTED: Eutrema salsugineum growth-regulating factor 1 (LOC18021800), mRNA | XM_006404687.2 | 100% | 93.89% | 1 | 2 | 2 |
| Arabis alpina genome assembly, chromosome: 6 | LT669793.1 | 93% | 95.90% | 1 | 2 | 9 (gap) |
| Description | ccession | Blast against the query grf8-61 | (MSA) Mismatches (bp) against each accession | |||
|---|---|---|---|---|---|---|
| Query Cover | Per. Ident | Forward | Probe | Reverse | ||
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR782545.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699748.2 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699773.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699768.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699758.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699753.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR215055.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana chromosome 4 | CP116283.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | OX298800.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | OX298805.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana ecotype 1254 chromosome 4 sequence | CP086757.1 | 98% | 99.20% | 0 | 0 | 4 (gaps) 1 mismach |
| Arabidopsis thaliana ecotype 5856 chromosome 4 sequence | CP086752.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana ecotype 6021 chromosome 4 sequence | CP086747.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana ecotype 6024 chromosome 4 sequence | CP086742.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana ecotype 9412 chromosome 4 sequence | CP086737.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana ecotype 9470 chromosome 4 sequence | CP086732.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana chromosome 4 | CP087129.2 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana isolate t2t_salk_col chromosome 4 | CP096027.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | OW119599.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR881469.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR797810.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR797805.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR797800.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR797795.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana chromosome 4 | CP002687.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana DNA chromosome 4, contig xample No. 61 | AL161561.2 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana DNA chromosome 4, BAC clone T19F6, partial sequence (ESSA xample) | AL109619.1 | 96% | 100.00% | 0 | 0 | 5 (gaps) |
| Arabidopsis thaliana chromosome IV BAC T19F6 genomic sequence, complete sequence | AC002343.1 | 98% | 99.20% | 0 | 0 | 4 (gaps) 1 mismach |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR699763.1 | 96% | 97.54% | 0 | 2 | 5 (gaps) |
| Arabidopsis thaliana genome assembly, chromosome: 4 | LR797790.1 | 96% | 97.54% | 0 | 2 | 5 (gaps) |
| Arabidopsis thaliana growth-regulating factor 8 (GRF8), partial mRNA | NM_118547.2 | 83% | 100.00% | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





