Submitted:
18 September 2023
Posted:
21 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
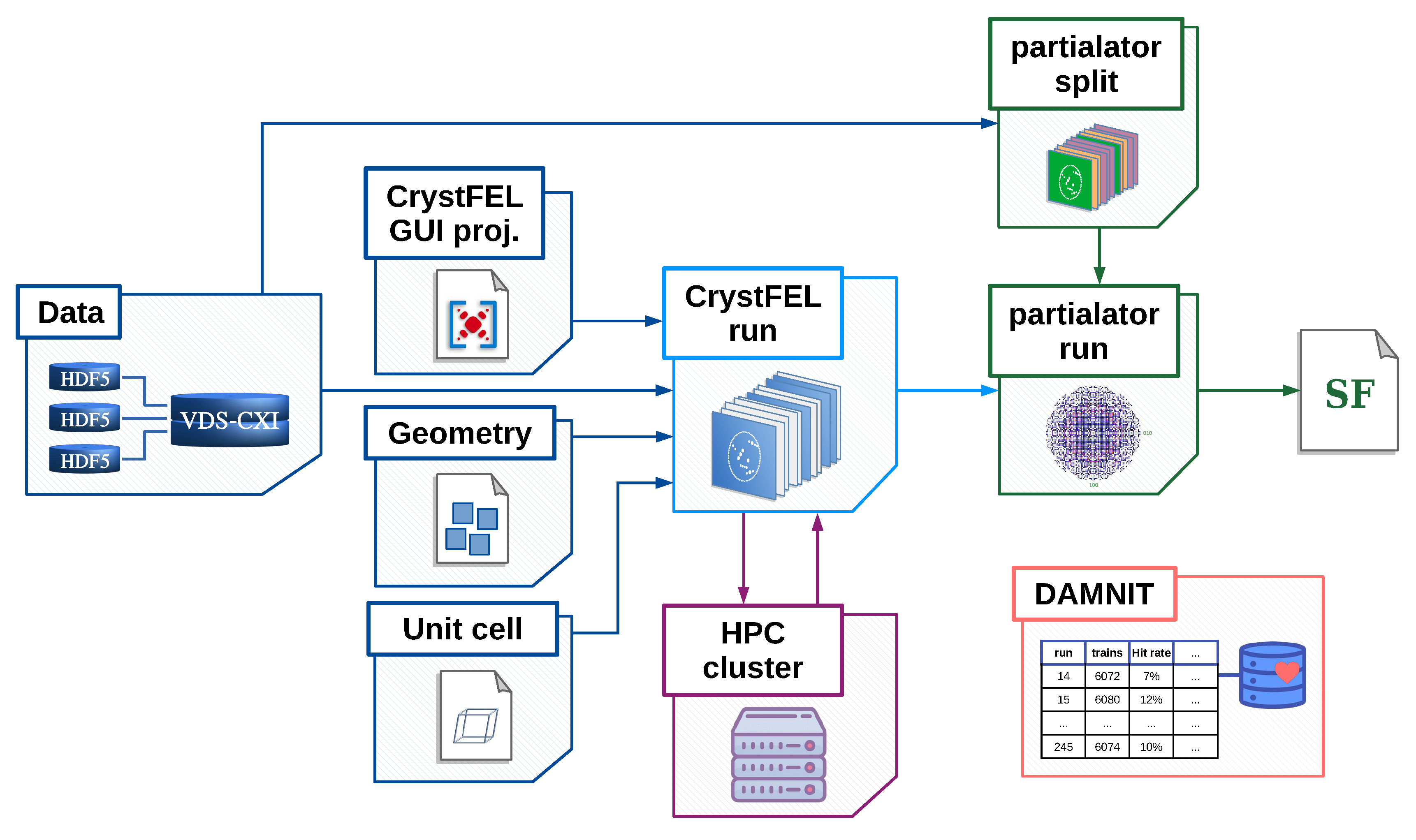
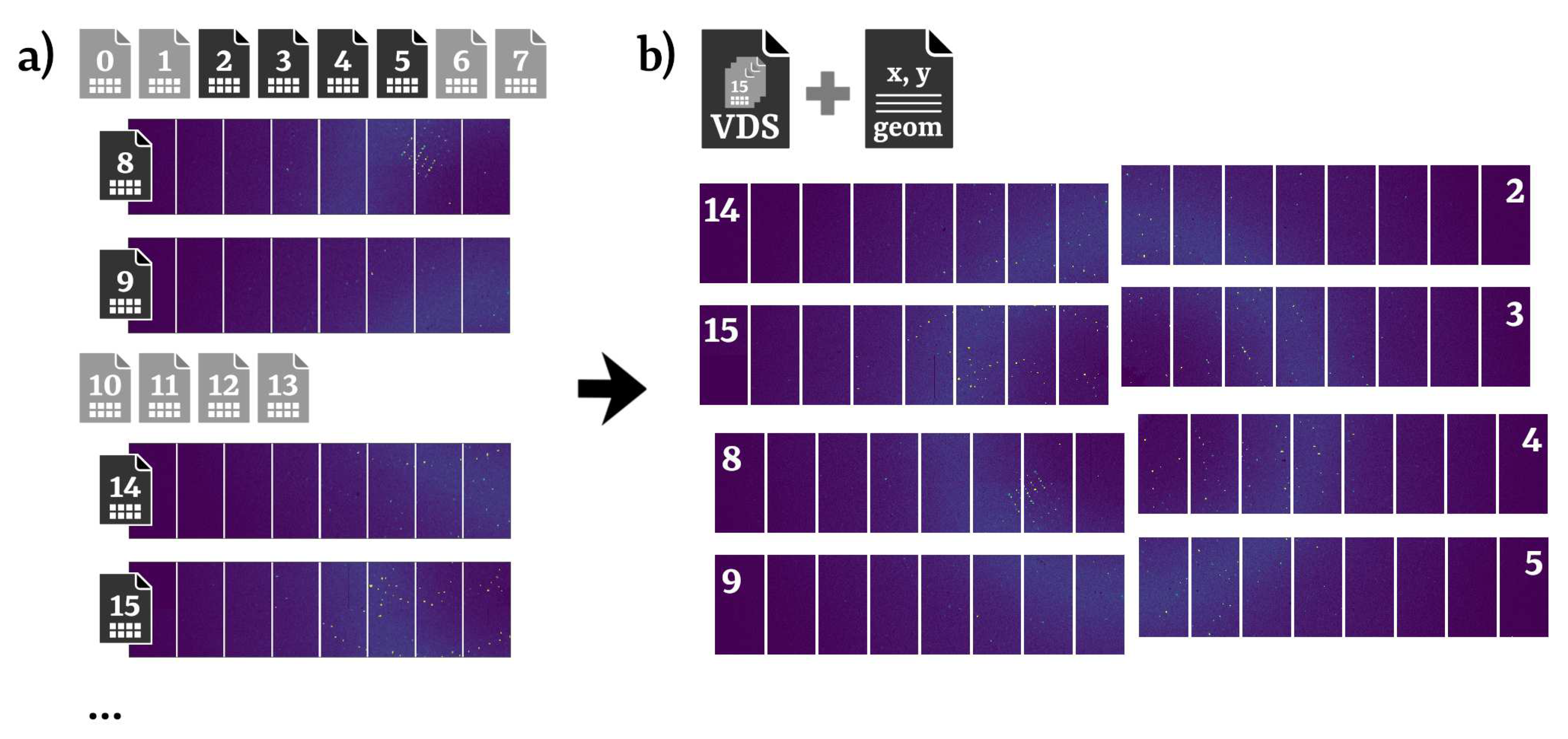
2. Design and implementation of the EXtra-Xwiz pipeline
- indexamajig – main command-line program for indexing and integrating diffraction patterns;
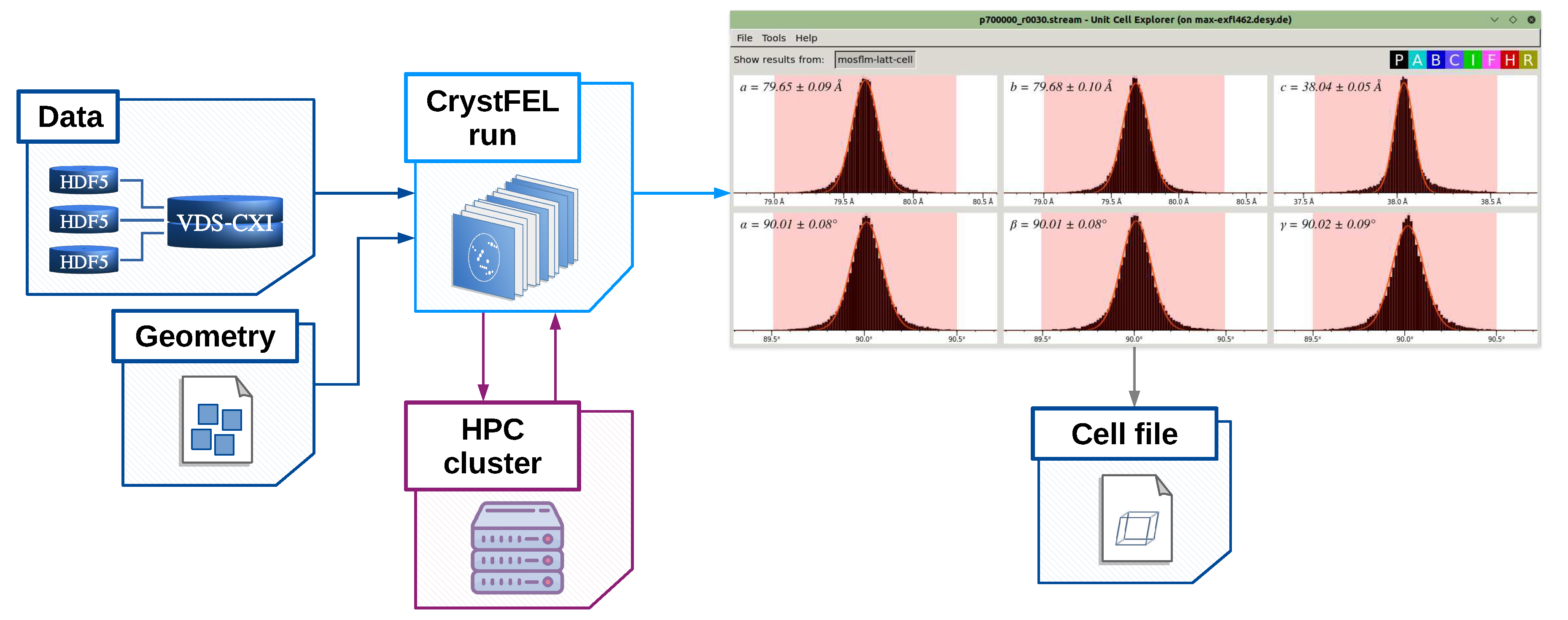
- cell_explorer – a tool for displaying and determining unit cell parameters of the crystalline sample;
- partialator – for scaling, merging, and post-refining reflections data;
- check_hkl – for calculating FOMs based on the full set of merged reflections, such as completeness, average signal strengths, and redundancy;
- compare_hkl – for calculating FOMs based on the merged reflections split into two sets, such as R-factors and correlation coefficients.
2.1. Structure of the EXtra-Xwiz pipeline
3. Data processing with EXtra-Xwiz by example
$ ssh <user name>@max-exfl-display.desy.de
$ module load exfel EXtra-xwiz/crystals2023
$ xwiz-workflow
[data] proposal = 700000 runs = [30]
frames_range = {start = 0, end = 200000, step = 1}
[crystfel] version = '0.10.2'
[geom] file_path = "agipd_p700000_r0030.geom" [unit_cell] file_path = "hewl.cell"
[indexamajig_run] resolution = 4.0 peak_method = "peakfinder8" peak_threshold = 800 peak_snr = 5 index_method = "mosflm" integration_radii = "2,3,5" ... min_peaks = 10 extra_options = "--no-non-hits-in-stream"
[slurm] partition = "upex" n_nodes_all = 20 duration_all = "10:00:00"
[merging] point_group = "422" scaling_model = "unity" scaling_iterations = 1 max_adu = 100000
[partialator_split]
execute = true
mode = "by_pulse_id"
[partialator_split.manual_datasets]
pump_on = {start=0, end=-1, step=24}
pump_off = [{start=8, step=24}, {start=16, step=24}]
[partialator_split] execute = true mode = "on_off_numbered" xray_signal = ["SPB_LAS_SYS/ADC/UTC1-1:channel_0.output", "data.rawData"] laser_signal = ["SPB_LAS_SYS/ADC/UTC1-1:channel_1.output", "data.rawData"]
$ xwiz-workflow -a
Step # d_lim source N(crystals) N(frames) Indexing rate [%]
1 1.6 indexamajig 46899 639616 7.3
...
Crystallographic FOMs:
overall outer shell
Completeness 100.0 100.0
Signal-over-noise 4.224 0.99
CC_1/2 0.8974 0.03244
CC* 0.9726 0.2507
R_split 27.28 80.6
3.1. Automatic scan over EXtra-Xwiz configuration parameters
$ xwiz-scan-parameters
[scan.SNR]
'indexamajig_run.peak_snr' = {start = 3, end = 7, step = 2}
'indexamajig_run.peak_threshold' = [1000, 800, 700]
index_rate(%) ... cc_half cc_star r_split
peak_snr peak_threshold
3 1000 0.080 ... 0.051 0.313 105.40
5 800 7.332 ... 0.894 0.972 27.44
7 700 7.219 ... 0.919 0.979 25.81
3.2. Running EXtra-Xwiz tutorial at VISA
- navigate to https://visa.xfel.eu;
- click "Create a new instance";
- click "Search for experiments" and select the proposal "p700000 - SFX on Hen egg-white lysozyme, AGIPD detector";
- click on "EXtra-Xwiz_Crystals2023" environment;
- choose the virtual hardware;
- create the instance.
[slurm] partition = "local" [indexamajig_run] n_cores = 1 ...
[data] ... frames_list_file = "indexed_p700000_r0030.lst"
4. Discussion and outlook
5. Conclusions
Acknowledgments
Abbreviations
| SFX | Serial femtosecond crystallography |
| XFEL | X-Ray Free-Electron Laser |
| EuXFEL | European XFEL facility |
| EXDF | EuXFEL Data Format |
| AGIPD | Adaptive-Gain Integrating Pixel Detector |
| LPD | Large Pixel Detector |
| HPC | High-Performance Computing |
| HDF5 | Hierarchical Data Format v.5 |
| CBF | Crystallographic Binary Format |
| CXIDB | Coherent X-ray Imaging Data Bank |
| GUI | Graphical User Interface |
| VDS | Virtual Dataset File |
| FOM | Figure Of Merit |
| SNR | Signal-to-Noise Ratio |
| HEWL | Hen Egg-White Lysozyme |
| VISA | Virtual Infrastructure for Scientific Analysis |
References
- Smyth, M.S.; Martin, J.H.J. x Ray crystallography. Molecular Pathology 2000, 53, 8–14, [https://mp.bmj.com/content/53/1/8.full.pdf]. [CrossRef]
- Shi, Y. A Glimpse of Structural Biology through X-Ray Crystallography. Cell 2014, 159, 995–1014. [CrossRef]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25. [CrossRef]
- The Protein Data Bank: Statistics. https://www.rcsb.org/stats. Accessed: 2023-08-15.
- Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Research 2000, 28, 235–242, [https://academic.oup.com/nar/article-pdf/28/1/235/9895144/280235.pdf]. [CrossRef]
- Als-Nielsen, J.; McMorrow, D. Elements of Modern X-ray Physics, 2nd ed.; Wiley: New Jersey, United States, 2011.
- Perutz, M.F. et al. Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature 1960, 185, 416–422. [CrossRef]
- Ramakrishnan, V. et al. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [CrossRef]
- Garman, E.F. Radiation damage in macromolecular crystallography: what is it and why should we care? Acta Crystallographica Section D 2010, 66, 339–351. [CrossRef]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallographica Section D 2006, 62, 32–47. [CrossRef]
- Standfuss, J.; Spence, J. Serial crystallography at synchrotrons and X-ray lasers. IUCrJ 2017, 4, 100–101. [CrossRef]
- Neutze, R. et al. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757.
- Chapman, H.N. X-Ray Free-Electron Lasers for the Structure and Dynamics of Macromolecules. Annual Review of Biochemistry 2019, 88, 35–58, PMID: 30601681. [CrossRef]
- Chapman, H. et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [CrossRef]
- Fromme, P.; Spence, J.C. Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Current Opinion in Structural Biology 2011, 21, 509–516. [CrossRef]
- Schlichting, I. Serial femtosecond crystallography: the first five years. IUCrJ 2015, 2, 246–255. [CrossRef]
- Barends, T. R. M. et al. Serial femtosecond crystallography. Nat. Rev. Methods Primers 2022, 2, 59. [CrossRef]
- Wiedorn, M. O. et al. Megahertz serial crystallography. Nat. Communications 2018, 9, 4025. [CrossRef]
- de Wijn, R.; Melo, D.V.M.; Koua, F.H.M.; Mancuso, A.P. Potential of Time-Resolved Serial Femtosecond Crystallography Usin High Repetition Rate XFEL Sources. Appl. Sci. 2022, 12, 2551. [CrossRef]
- Pandey, S.; Poudyal, I.; Malla, T.N. Pump-Probe Time-Resolved Serial Femtosecond Crystallography at X-Ray Free Electron Lasers. Crystals 2020, 10, 628. [CrossRef]
- Aquila, A. et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Optics Express 2012, 20, 2706. [CrossRef]
- Kupitz, C. et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 2014, 513, 261–265. [CrossRef]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Current Opinion in Structural Biology 2020, 65, 193–208. Catalysis and Regulation; Protein Nucleic Acid Interaction. [CrossRef]
- Powell, H.R. X-ray data processing. Bioscience Reports 2017, 37, BSR20170227, [https://portlandpress.com/bioscirep/article-pdf/37/5/BSR20170227/430355/bsr-2017-0227c.pdf]. [CrossRef]
- Kabsch, W. XDS. Acta Cryst. D 2010, 66, 125–132. [CrossRef]
- Battye, T.G.G. et al. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica Section D 2011, 67, 271–281. [CrossRef]
- Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Cryst. 2018, D74, 85–97. [CrossRef]
- Brewster, A.S. et al. Improving signal strength in serial crystallography with DIALS geometry refinement. Acta Crystallographica Section D 2018, 74, 877–894. [CrossRef]
- White, T.A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Cryst. 2012, 45, 335–341. [CrossRef]
- Lamzin, V.S.; Perrakis, A. Current state of automated crystallographic data analysis. Nature Structural Biology 2000, 7, 978–981. [CrossRef]
- Winter, G.; McAuley, K.E. Automated data collection for macromolecular crystallography. Methods 2011, 55, 81–93. Methods in Structural Proteomics. [CrossRef]
- Perrakis, A.; Morris, R.; Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nature Structural Biology 1999, 6, 458–463. [CrossRef]
- Alharbi, E. et al. Comparison of automated crystallographic model-building pipelines. Acta Crystallographica Section D Structural Biology 2019, 75, 1119–1128. [CrossRef]
- Hamilton, W.C. The Revolution in Crystallography. Science 1970, 169, 133–141. [CrossRef]
- Abola, E. et al. Automation of X-ray crystallography. Nature Structural Biology 2000, 7, 973–977. [CrossRef]
- Sauter, N.K. XFEL diffraction: developing processing methods to optimize data quality. Journal of Synchrotron Radiation 2015, 22, 239–248. [CrossRef]
- Decking, W. et al. A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 2020, 14, 391–397. [CrossRef]
- Tschentscher, Th.. Investigating ultrafast structural dynamics using high repetition rate x-ray FEL radiation at European XFEL. Eur. Phys. J. Plus 2023, 138, 274. [CrossRef]
- Allahgholi, A. et al. The adaptive gain integrating pixel detector. JINST 2016, 11. [CrossRef]
- Hart, M. et al. Development of the LPD, a high dynamic range pixel detector for the European XFEL. 2012 IEEE Nuclear Science Symposium and Medical Imaging Conference Record (NSS/MIC, 2012, pp. 534–537. [CrossRef]
- Mozzanica, A. et al. The JUNGFRAU Detector for Applications at Synchrotron Light Sources and XFELs. Synchr. Rad. News 2018, 31, 16–20. [CrossRef]
- .
- Turkot, O., Dall’Antonia, F. et al. Towards automated analysis of serial crystallography data at the European XFEL. X-Ray Free-Electron Lasers: Advances in Source Development and Instrumentation VI; Tschentscher, T. et al., Ed. International Society for Optics and Photonics, SPIE, 2023, Vol. 12581, p. 125810M. [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking crystallographic model and data quality. Science 2012, 336, 1030–1033. [CrossRef]
- White, T.A. et al. Recent developments in CrystFEL. Journal of Applied Crystallography 2016, 49, 680–689. [CrossRef]
- White, T. CrystFEL: data processing for FEL crystallography. https://www.desy.de/~twhite/crystfel. Accessed: 2023-09-15.
- Dall’Antonia, F., Turkot, O. et al. Code repository of EXtra-Xwiz: pipeline for SFX data analysis at European XFEL. https://github.com/European-XFEL/EXtra-Xwiz/tree/crystals2023. Accessed: 2023-09-17.
- White, T. Processing serial crystallography data with CrystFEL: a step-by-step guide. Acta Cryst. 2019, D75, 1–15. [CrossRef]
- Könnecke, M. et al. The NeXus data format. Journal of Applied Crystallography 2015, 48, 301–305. [CrossRef]
- Bernstein, H.J. et al. Gold Standard for macromolecular crystallography diffraction data. IUCrJ 2020, 7, 784–792. [CrossRef]
- Maia, F.R.N.C. The Coherent X-ray Imaging Data Bank. Nat. Methods 2012, 9, 854–855. [CrossRef]
- Kluyver, T. et al. EXtra-data: library for accessing data at European XFEL. https://extra-data.readthedocs.io. Accessed: 2023-09-15.
- Yoo, A.B.; Jette, M.A.; Grondona, M. SLURM: Simple Linux Utility for Resource Management. Job Scheduling Strategies for Parallel Processing. Springer, 2003, pp. 44–60.
- White, T.A. Post-refinement method for snapshot serial crystallography. Philosophical Transactions of the Royal Society B: Biological Sciences 2014, 369, 20130330. [CrossRef]
- Wrigley, J. et al. DAMNIT: tool for interactive data and metadata inspection at European XFEL. https://rtd.xfel.eu/docs/damnit/en/latest/. Accessed: 2023-05-07.
- Götz, A.; Konrad, U.; Le Gall, E.; Ounsy, M.; Servan, S. VISA sustainability sheet, 2023. [CrossRef]
- Gelisio, L. et al. EuXFEL Data Analysis User Documentation. https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/. Accessed: 2023-09-15.
- Preston-Werner, T. TOML: Tom’s Obvious Minimal Language. https://toml.io/en/. Accessed: 2023-09-15.
- Dall’Antonia, F., Turkot, O. et al. Documentation on EXtra-Xwiz: pipeline for SFX data analysis at European XFEL. https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/software/extra-xwiz/. Accessed: 2023-09-15.
- Kluyver, T. et al. EXtra-geom: library for describing physical layout of multi-module detectors at European XFEL. https://extra-geom.readthedocs.io. Accessed: 2023-05-15.
- White, T. Symmetry Classification for Serial Crystallography Experiments. https://www.desy.de/~twhite/crystfel/twin-calculator.pdf. Accessed: 2023-09-15.
- Le Gall, E. et al. Documentation on VISA: Virtual Infrastructure for Scientific Analysis. https://visa.readthedocs.io/en/latest/index.html. Accessed: 2023-09-15.
- Ferreira de Lima, D.E. et al. Automatic online data analysis optimization: application to serial femtosecond crystallography. International Union of Crystallography 2024. In preparation.
- Yefanov, O. et al. Accurate determination of segmented X-ray detector geometry. Optics Express 2015, 23, 28459. [CrossRef]
| 1 | For an extensive description the reader is referred to, e.g., [6] |
| 2 | For a comprehensive introduction to structure determination using XFELs the reader is referred to [13] |
| 3 | For a detailed explanation, the reader is referred to, e.g., [30] |
| 4 | Hierarchical Data Format v.5 [42] |
| 5 | Detailed description of all available configuration options is available at the EXtra-Xwiz documentation [59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




