1. Introduction
Alcohol-associated liver disease (ALD) is one of the most common liver diseases in the world, and the major medical consequence of excessive drinking. Excessive alcohol consumption can develop alcoholic hepatitis, fibrosis that progresses to cirrhosis in up to 20% of patients[
1]. Furthermore, if the damage is not controlled, approximately 10% of people can progress to hepatocellular carcinoma and high mortality[
2].
The clinical manifestations of the patients with cirrhosis due to ALD widely range from asymptomatic to various decompensation events[
3]. However, the definite diagnosis of ALD is based on a liver biopsy which has drawbacks such as being invasive, inappropriate for monitoring progression and assessment of steatosis somewhat subjective[
4]. The need for novel non-invasive diagnostic biomarkers, including microRNAs[
3], in ALD is necessary.
Apart from diagnostic capabilities, microRNAs also regulate gene expression at the post-transcriptional level by promoting mRNA degradation or inhibiting mRNA translation and exert important biological functions in various pathological and cellular pathways[5-7]. Circulating microRNAs have been proposed as biomarkers in cholangiocarcinoma, hepatocellular carcinoma, non-alcoholic steatohepatitis (NASH), and cirrhosis[8-10]. Therefore, identifying circulating microRNAs could be useful biomarkers to both stratify patients with advanced chronic liver disease and early detection of ALD. This study profiled global circulating microRNAs in heavy alcohol users with an aim to identify microRNA biomarker signatures associated with ALD and study their role in ALD pathogenesis.
2. Methods
2.1. Human Samples
Frozen serum samples (-80
oC) were available through our multinational GenomALC study for an Australian sub-cohort from multiple centres in Sydney recruited between 2011 and 2017. Recruitment and cohort characteristics are detailed previously[11, 12]. Briefly, participants with long-term heavy alcohol use (80g/d (M) and 50g/d (F) for ≥10 years (N=47) were included in the study. Heavy alcohol users without significant liver disease were grouped as controls (n=23) and patients with liver cirrhosis as cases (n=24). Five healthy controls (HC) were volunteers recruited for the study and blood collected in EDTA tubes. Serum was obtained by centrifugation of blood at 1,100 g for 10 minutes at 4°C, aliquoted and immediately stored at -80°C till further use. Cohort characteristics are detailed in
Table 1. The study was approved by the Human Research Ethics Committees (X11-0072, HREC11RPAH88; X17-029, HREC17RPAH331).
2.2. RNA Extraction
Total RNA (including small RNA species) was isolated from 100 µl serum using the RNeasy-HT Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and QiaCube-HT robotic RNA isolation system. Each serum sample was mixed with 500µl TRIzol (ThermoFisher Scientific, USA), and then spiked with 2.5µl of 50nM synthetic control microRNA ath-miR-172a (Sigma Aldrich, Hamburg, Germany). All procedures were performed according to the manufacturer protocol instructions. The quantity and quality of total RNA was measured using the NanoDrop Spectrophotometer, and RNAs with 260/280 ratio greater than 1.6 was considered acceptable.
2.3. OpenArray® Panels - Quantitative Real-time PCR
cDNA was synthesised from total RNA and preamplified for RT-qPCR OpenArray (TaqMan® OpenArray®, ThermoFisher Scientific, USA) using protocols described in detail previously[13, 14].
2.4. Data Analysis
High throughput data generated from TaqMan® OpenArray® RT-qPCR data were uploaded into ThermoFisher iCloud for the global normalisation and detection cut-off threshold determined. microRNAs that were undetectable or with an Amp Score <1.24 and Cq confidence <0.6 were eliminated from further analysis as per the pre-defined QC criteria for this study.[13, 14] The detection threshold cycle (Ct) cut-off was defined at Ct value of 39.
2.5. Statistical Analysis
The relative fold change (ΔΔCt = ΔCt Healthy Control - ΔCt patient sample) between each group. Statistical significance (p-values) calculated by Student t-tests was used to compare two groups while for more than two groups ANOVA test was used in Excel. Regression/correlation analysis (Prism and Excel) was performed using significantly different microRNAs with traditional biomarkers and allele dosage of risk variant information from our genotype data[
15] The statistical program R was used to plot heat maps to visualise differentially expressed microRNAs. Volcano plots were plotted using the GraphPad Prism 9 software (GraphPad, San Diego, CA, USA). The Mann Whitney test was used to compare the differences in microRNA expression between patients and healthy controls. p-values below 0.05 were considered statistically significant for all tests.
2.6. Pathway Analysis of the Dysregulated microRNAs
We used Ingenuity Pathway Analysis (IPA) software (
www.qiagenbioinformatics.com) and microRNA target filter to predict target mRNAs of the differentially expressed microRNAs. For identifying disease and function pathways target mRNAs were overlaid on ‘Toxic’ and ‘Functions’ derived from select microRNAs using ‘Pathway design’ option in IPA. The top ten toxic (Tx) and functional pathway networks, identified for microRNA-mRNA interactions, were plotted. We also used co-analysis function to identify significantly enriched pathways that are associated with the microRNAs.
3. Results
3.1. Cohort Characteristics
Of the 47 heavy alcohol users (n=23 cases, n=24 controls), greater than 99% were European Caucasian. Controls and cases had similar median age (51 vs 53) and proportion of males (66% vs 65%), respectively (
Table 1). Cases with liver cirrhosis had significantly different liver enzymes and liver functions compared to the controls: lower haemoglobin level, platelet count and serum albumin, and higher International Normalised Ratio (INR), mortality end stage liver disease (MELD) score, serum bilirubin and aspartate aminotransferase (AST) level. There was no significant difference in alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT) and serum creatinine. AUDIT score was significantly higher in controls compared to cases (12 vs 5, respectively). The Healthy Control (HC) group comprised of 60% European Caucasian, were slightly older (median age 55 years) and 17% were males (
Table 1).
3.2. Comparison of Global Profiling of the Serum microRNA Expression in Heavy Alcohol Users and Healthy Controls
The following results are from comparisons between different groups defined as: Controls are drinkers with no liver disease; Cases include drinkers with cirrhosis and alcoholic hepatitis; Cirrhosis are drinkers with cirrhosis (no AH); AH are drinkers with alcoholic hepatitis (Supplementary
Table 1).
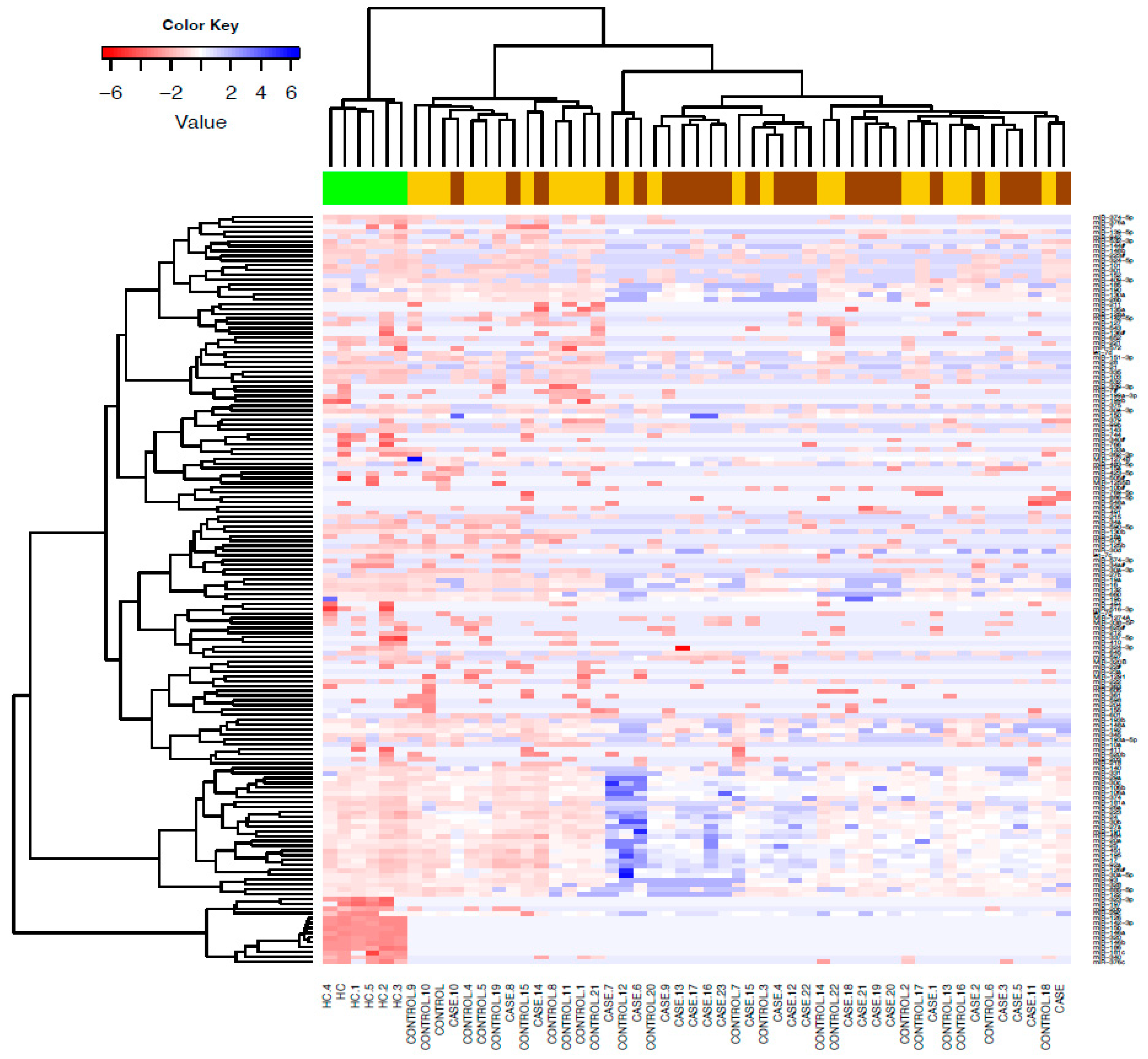
We detected 155 serum microRNAs expressed above threshold in heavy alcohol users (n=47) and HC. Unsupervised hierarchical clustering heat map of expressed microRNAs differentiated patients with liver cirrhosis from HC (
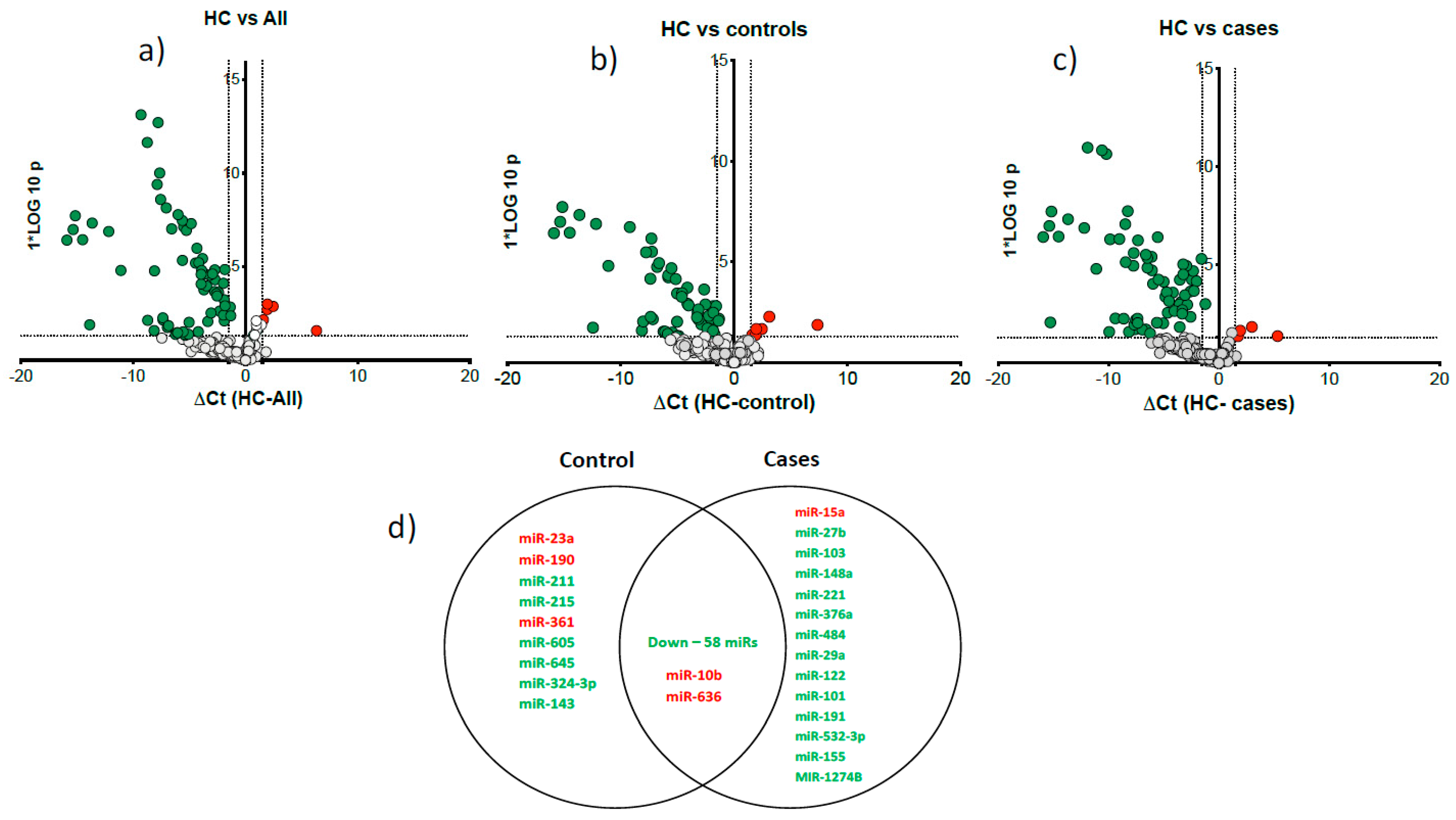
Figure 1). Volcano plots show most microRNAs had lower abundance in all heavy alcohol users (
Figure 2a), in cirrhosis cases (
Figure 2b) and controls (
Figure 2c) compared to HC. Venn diagram (
Figure 2d) show a total of 77 differentially downregulated miRNAs in heavy alcohol users (cases plus controls). Of these, 58 downregulated and two upregulated (miR-10b & miR-636) were common between cases and controls compared to HC, 14 were specific to cirrhosis group and 8 were specific to control group.
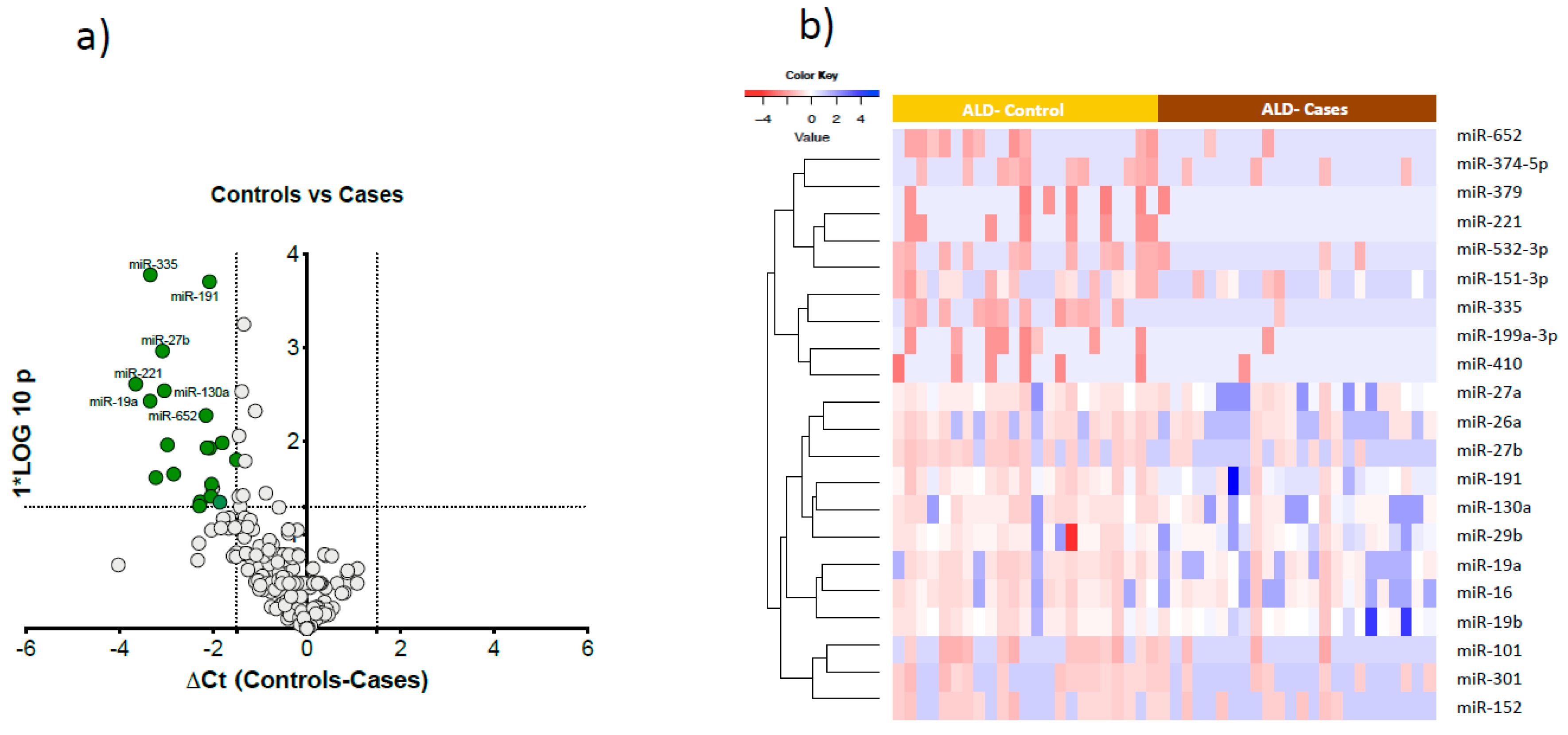
Comparing cases and controls, 21 microRNAs were significantly downregulated in cases compared to controls (
Figure 3a). These 21 microRNAs clearly differentiated cases from controls as visualised in the semi-supervised hierarchical clustering in heavy alcohol users using (
Figure 3b).
Post-hoc analysis of cases and controls by gender, performed to test any gender specific differences in miRNAs expression, showed 24 microRNAs were specific to females and 5 microRNAs were specific to males. (Supplementary
Figure 1). There were four common downregulated microRNAs (miR-27b, miR-221, miR-191, miR-335) in both male and female cases compared to drinking controls.
3.3. Correlation of Downregulated microRNAs with Traditional Biomarkers of Liver Disease
Seventeen microRNAs showed strong correlation with the clinical biomarkers (ALT, AST, Bilirubin, INR, MELD score, Albumin, haemoglobin, platelet count and BMI). Highly significant positive correlation was seen with bilirubin, INR and MELD score and inverse correlation with albumin and platelets count (
Table 2 and Supplementary
Table 1). The seven microRNAs (miR-16, miR-19a, miR-27a, miR-29b, miR-101, miR-130a and miR-191) had highly significant correlations with all traditional biomarkers: bilirubin, INR and MELD score (p<0.001,
Table 2).
3.4. Diagnostic Performance of Conventional Biomarkers and Selected microRNAs to Distinguish Cases from Controls
Conventional markers INR, MELD, bilirubin and platelet count performed better (AUC-ROC 0.97, 0.95, 0.87 and 0.85, respectively) than miRNAs (
Table 3). Of the seven microRNAs above, three microRNAs (miR-27a, miR-130a and miR-191) significantly predicted cases with AUC-ROC 0.8, 0.78 and 0.85, respectively (P<0.020) (
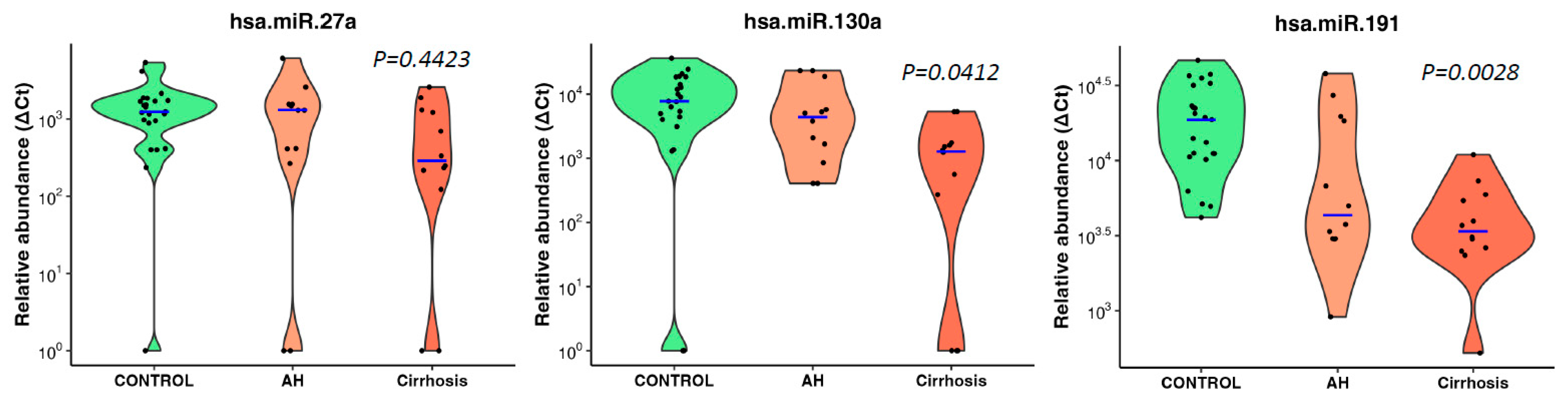
Table 3). The other microRNAs also predicted cases with moderate performance.
All three microRNAs showed an inverse association trend of decreasing abundance with increasing disease severity (no disease, alcoholic hepatitis, cirrhosis) but only miR-130a (p=0.0412) and miR-191 (p=0.0028) reached significance (Jonckheere-Terpstra nonparametric ordered test,
Figure 4).
3.5. Association of miRNAs with Genetic Variants Linked to Risk of Cirrhosis
We performed correlation analysis of differentially expressed microRNAs in cases vs controls and risk allele dosages from available genotyping data from our previously identified variants associated with alcohol-associated cirrhosis (
PNPLA3:rs2294915,
HSD17B13:rs10433937
, FAF2:rs11134977
, TM6SF2:rs10401969
, SERPINA1:rs28929474,
MBOAT7:rs641738 and
MARC1:rs2642438
). Several microRNAs (miR-19a, miR-26a, miR-101, miR-151-3p, miR-221, and miR-301) showed a significant positive correlation with
HSD17B13 variant (Supplementary
Table 2). miR-27b, miR-130a and miR-335 had a significant positive correlation with single nucleotide polymorphisms in
FAF2:rs11134977,
TM6SF2:rs10401969 and
MARC1:rs2642438, respectively.
3.6. microRNA Signature Distinguished Some Patients with Alcoholic Hepatitis
We further reallocated our ALC patients to alcoholic hepatitis (AH) group using the NIAAA criteria of a bilirubin cut-off of >3.0 mg/dl [
16] in
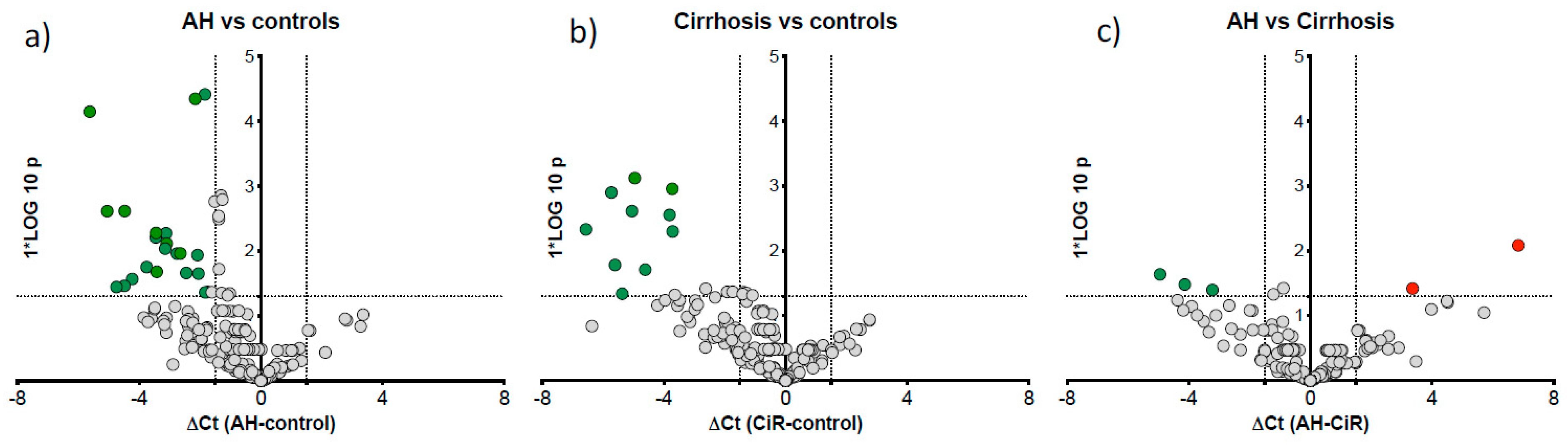
post-hoc analysis to compare microRNA expression specific to AH (n=12) vs cirrhosis (n=12), the remaining ALC patients. Twenty one miRNAs were downregulated in AH vs control (
Figure 5a) and 10 downregulated in ALC vs control (
Figure 5b). Further direct comparison of AH vs cirrhosis cases identified three downregulated (miR-15a, miR-125b-5p and miR-185) and two upregulated (miR-21-5p and miR-28) microRNAs in AH group (
Figure 5c).
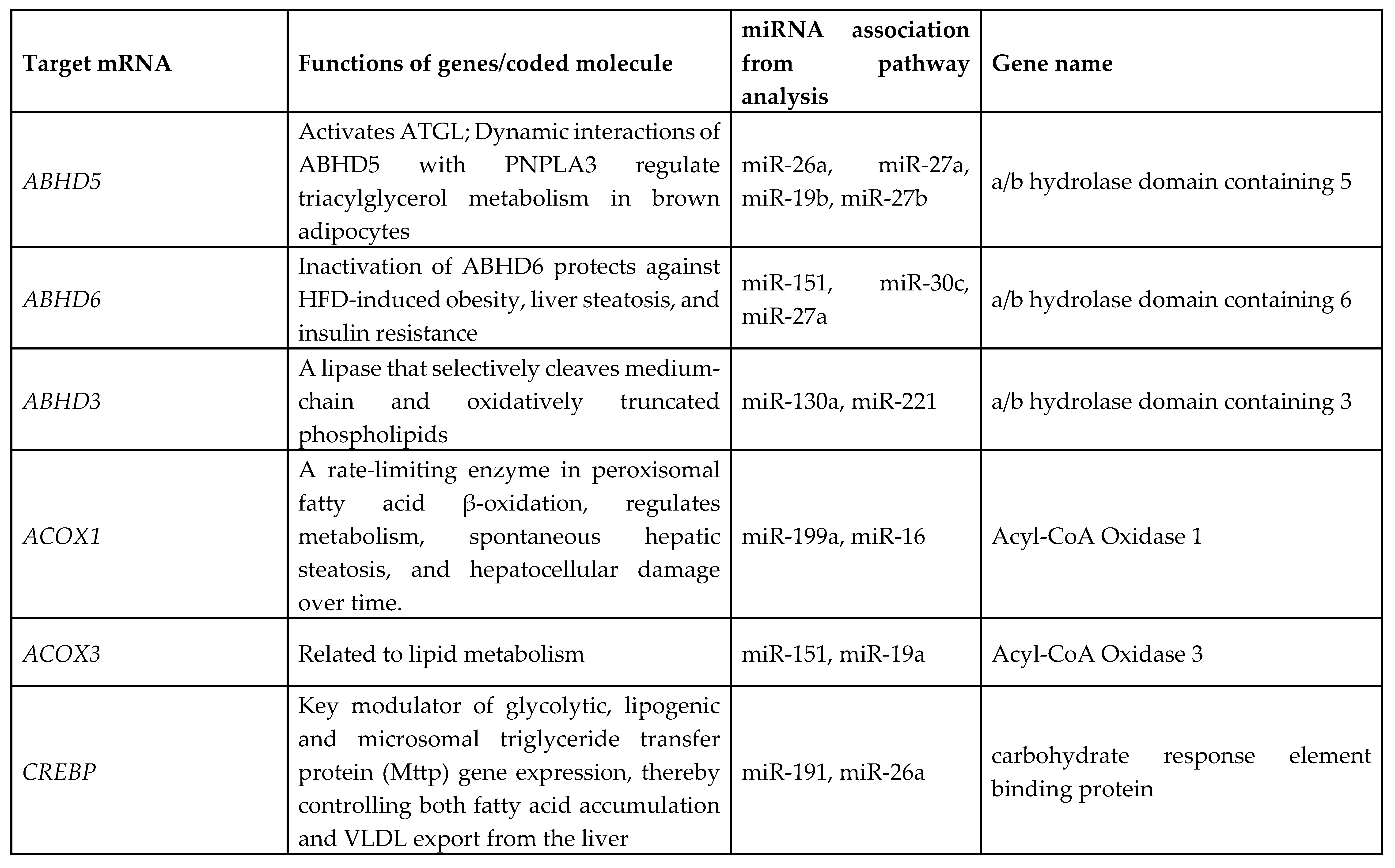
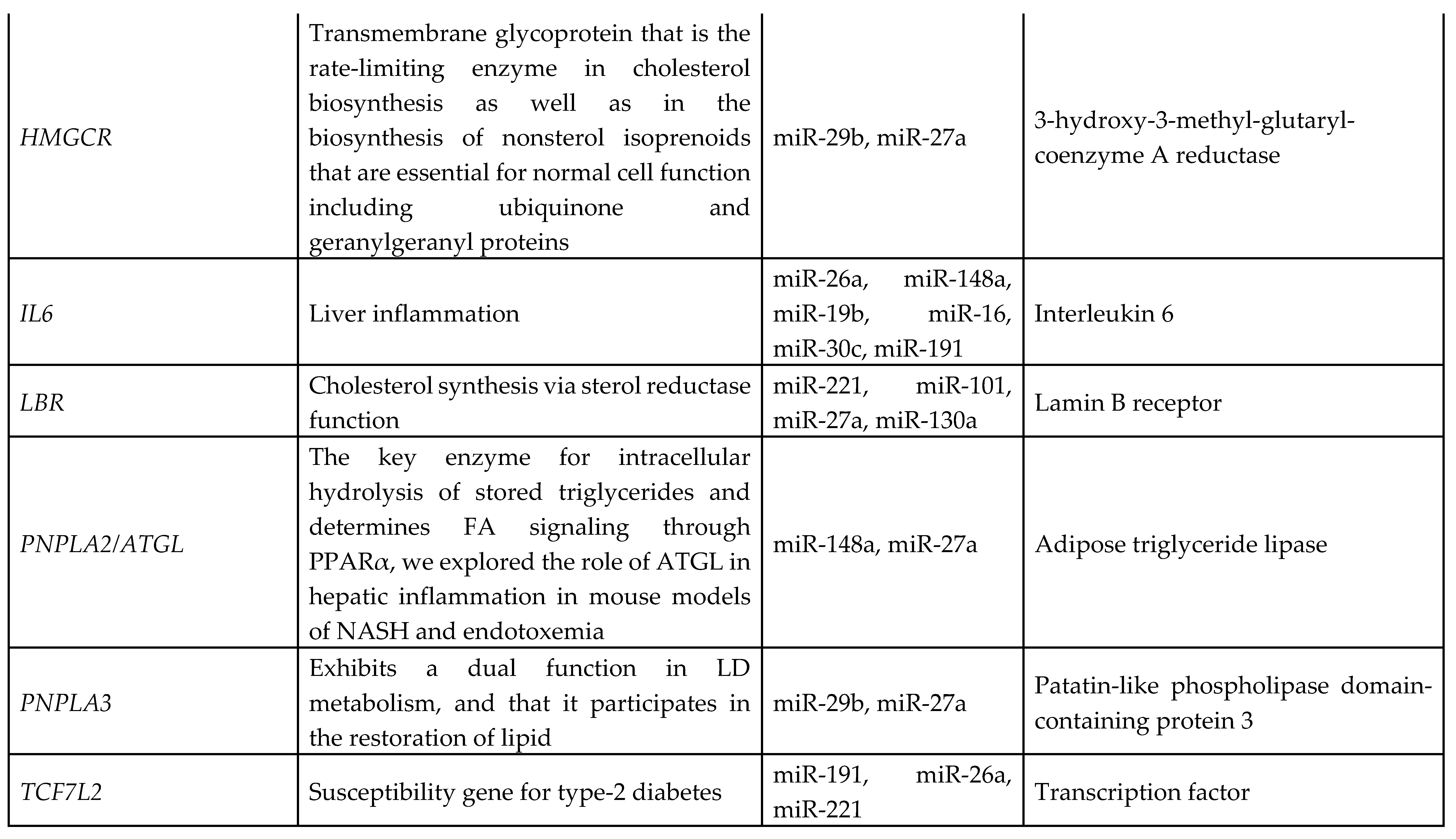
3.7. Identification of Target Genes and Pathway Analysis
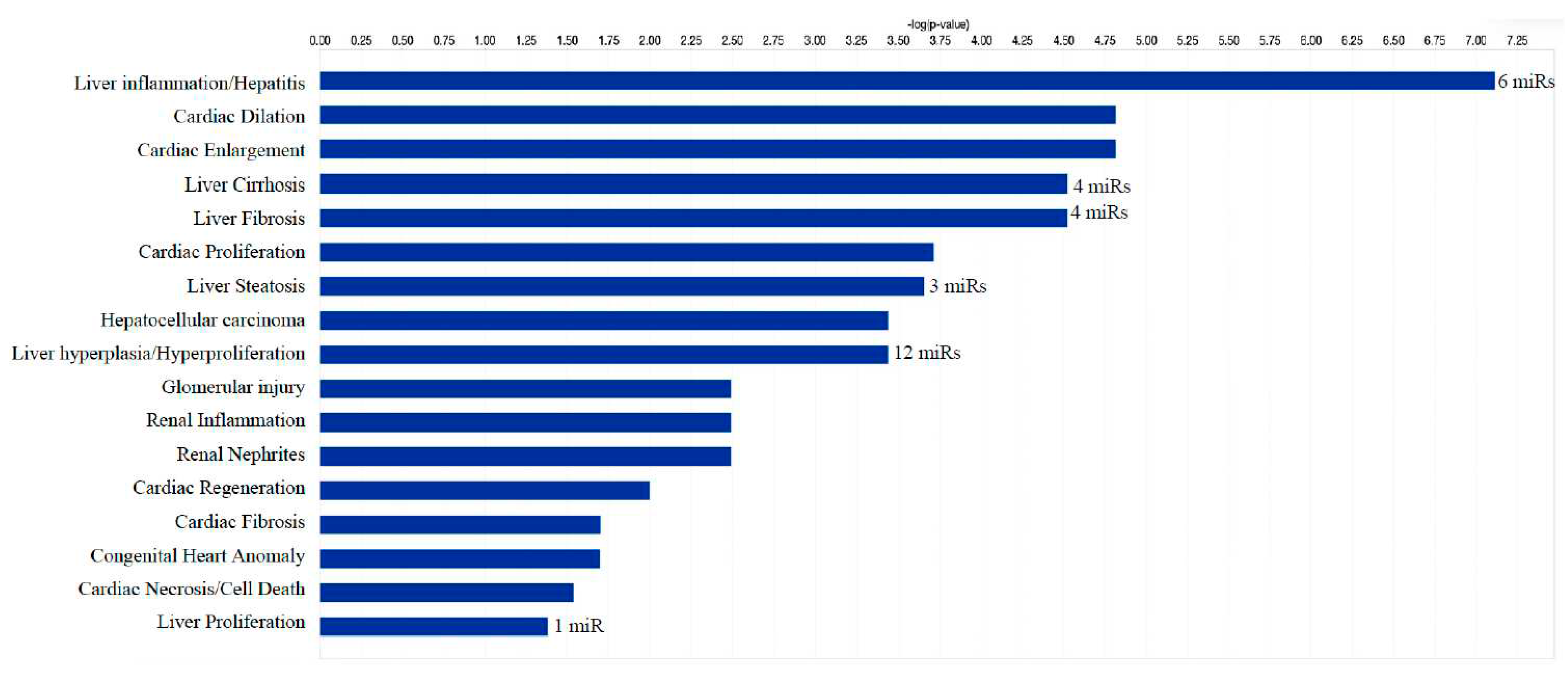
We identified 1212 potential target messenger RNAs (mRNAs) for the 21 differentially expressed microRNAs specific to liver disease/functions using IPA. Co-analysis using significantly dysregulated microRNAs identified pathways for liver pathology (
Figure 6). The liver related pathways were enriched for liver inflammation/hepatitis, liver cirrhosis, liver fibrosis and liver steatosis (
Figure 6). The most common ‘tox’ analysis pathways targeted by these microRNAs include fatty acid metabolism, liver necrosis/cell death, hepatic fibrosis, hepatic stellate cell activation, mitochondrial dysfunction, liver proliferation, increased liver damage, liver steatosis and liver hepatitis (
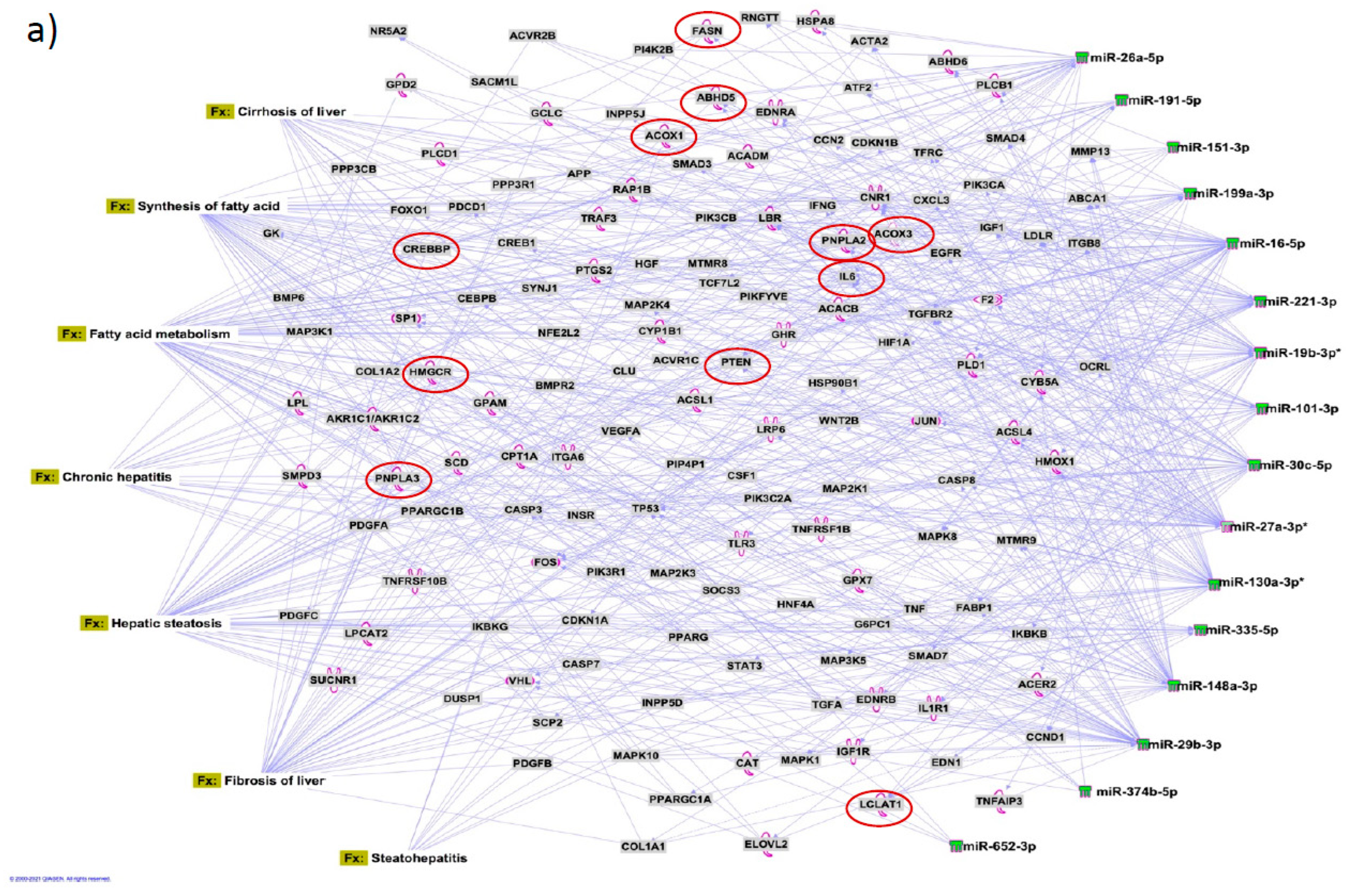
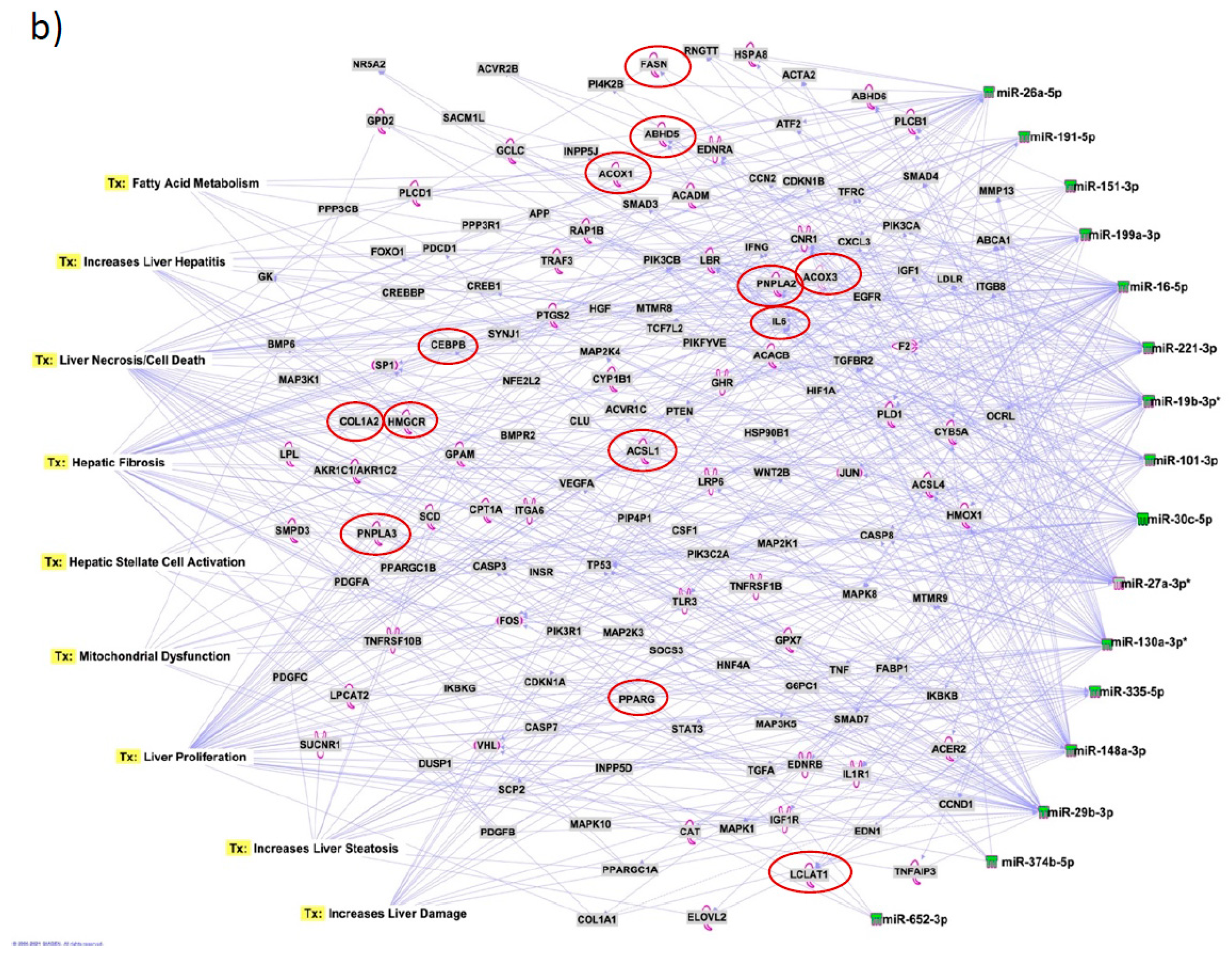
Figure 7a). Several target mRNAs in disease/function pathways that regulate cirrhosis of liver, chronic hepatitis, hepatic steatosis, fibrosis of liver, synthesis of fatty acids, fatty acid metabolism and steatohepatitis were identified (
Figure 7b). We also plotted the network diagram with seven microRNAs and the target mRNAs associated these microRNAs. One of the microRNA interaction networks had several mRNAs related to carbohydrate and lipid metabolism, for example, carbohydrate response element binding protein (
CREBP), patatin-like phospholipase domain-containing protein 3 (
PNPLA3) and both adipose triglyceride lipase (
ATGL) and its activator a/b hydrolase domain containing 5 (
ABHD5) (
Figure 7a, 7b,
Table 4). Similar pathway analysis for target mRNAs associated with AH specific microRNAs revealed mRNAs related to inflammation, fibrosis, signaling and alcohol metabolism (Supplementary
Figure 2).
4. Discussion
One of the important findings of our study is the identification of a signature panel of microRNAs that specifically distinguished alcohol-associated liver cirrhosis from heavy alcohol users without liver disease. Unlike previous reports that compared ALD with healthy controls, our comparison avoided the chance of identifying miRNAs in response to alcohol exposure alone. Second, the abundance of a subset of microRNAs inversely correlating with progressive diseases stages (no disease, AH, cirrhosis) demonstrates their potential as indicators of disease severity. Third, among the downregulated microRNAs, seven (miR-16, miR-19a, miR-27a, miR-29b, miR-101, miR-130a, and miR-191) exhibiting a highly significant correlation with liver disease biomarkers, indicate their potential as robust biomarkers for liver cirrhosis in heavy drinkers. Interestingly, three microRNAs (miR-27a, miR-130a, and miR-191) showed promising predictive accuracy in distinguishing cirrhosis cases from controls, suggesting their capacity for clinical utility as diagnostic biomarkers for alcohol-associated liver cirrhosis. However, given that the conventional biomarkers were initially used to differentiate the cases from controls, these biomarkers with greater AUC-ROCs outperformed miRNAs as expected.
As liver biopsy, the current gold standard for diagnosis, is invasive and carries certain limitations[
17] less-invasive diagnostic biomarkers are preferred for identifying and monitoring liver cirrhosis[
18]. Circulating microRNAs have emerged as promising biomarkers in various liver diseases, including hepatocellular carcinoma (HCC) and non-alcoholic fatty liver disease and steatohepatitis (NAFLD/NASH)[19-22]. Our protocol offers a convenient diagnostic approach using minimal volume of serum without the need of additional sample collection beyond the clinical requirement for treatment. The identification of our microRNAs distinguishing heavy drinkers with cirrhosis and their inverse association with disease severity offers a potential approach for diagnosing and monitoring the progression of this condition. Measuring the specific panel of downregulated microRNAs in serum samples may be developed as a reliable semi-quantitative and accessible means for detection of disease stages.
MicroRNAs have been previously implicated in various pathological and cellular pathways related to liver diseases[23-25]. Our pathway analysis revealed that the dysregulated microRNAs were associated with critical pathways involved in cell death (Caspases, FASN), fibrosis (TGF, Col1, SMADs, MMPs) inflammation (IL6, TNF, PPARG), steatosis and fatty acid metabolism (ABHD family, ACOX1, ACOX3, HMGCR, LCLAT1), specifically regulating triglyceride metabolism in the hepatic lipid droplets (PNPLA2 (aka ATGL), ABHD5, PNPLA3).
Several target mRNAs identified through disease-function and tox pathway networks are associated with triglyceride metabolism, particularly PNPLA3 (miR-29b, miR-27a), PNPLA2/ATGL: miR-148a, miR-27a)) and ABHD5 (miR-26a, miR-27a, miR-19b, miR-27b). Interestingly, PNPLA3 is the strongest and the most replicated genetic risk for both alcohol- and non-alcohol-associated liver cirrhosis as reported in genome-wide association studies[15, 26, 27]. PNPLA3 exerts its effect by binding to a/b hydrolase domain 5 (ABHD5) dissociating it from adipose triglyceride lipase (ATGL), inactivating the major triglyceride lipase and resulting in triglyceride accumulation. However, we did not find significant correlation between any microRNA and the specific PNPLA3:rs2294915 risk variant. But miR130a was significantly correlated with another lipid droplet protein FAF2:rs11134977, that binds to ATGL and also inhibits triglyceride metabolism. Furthermore, six microRNAs (miR-19a, miR-26a, miR-101, miR-151-3p, miR-221, miR-301) had significant positive correlation with HSD17B13:rs10433937 risk variant, another lipid droplet associated protein. The association of dysregulated miRNAs with target mRNAs that are involved in steatosis (lipid pathways, SREMP, ACOX3, HMGCR, LCLAT1), particularly triglyceride metabolism (PNPLA3, ATGL ABDH, FAF2) supports the new understanding that lipids play a key role in this disease pathogenesis and miRNAs may regulate this pathway contributing to the underlying molecular mechanisms of lipid genetics in alcohol-associated cirrhosis. Future studies are needed to test this. Other target mRNAs involved in inflammation (TNF, IFN, IL6, CXCL3) and fibrosis (TGFs, SMAD) were also identified.
Surprisingly, in this study we did not identify the previous most commonly reported miRNAs associated with alcohol-associated cirrhosis (miR21, miR122, miR155) summarised in the two recent comprehensive reviews[28, 29]. This may be because earlier studies were performed either in in vivo cross species models or compared ALD/ALC with healthy controls. Our main comparison was within drinkers, with cirrhosis vs without liver disease, and a relatively small cohort of HC. With reference to the above reviews, we did identify some common miRNAs previously identified in ALD (miR19, miR21-5p, miR26, miR199), NAFLD (miR27b, miR30c) and viral hepatitis (miR27a, miR125b, 130a) suggesting common regulation in liver diseases. Our findings identifying several dysregulated miRNAs in cases (miR21-5p, miR29b, miR101, miR199a, miR221, miR335, miR652) also reported in HCC, begs the speculation of a precancerous profile on pathway to HCC as cirrhosis predominantly underlies development of hepatocellular carcinoma.
5. Strengths and Limitations
A major strength of this study is the use of clinically and genetically well characterized patients from the GenomALC-1 cohort. In addition, choosing drinkers as controls to compare liver cirrhosis patients is designed to minimise the specific effect of alcohol-associated microRNAs. However, we cannot rule out the possibility of bias towards alcohol-associated microRNAs in current drinkers, especially in the control group. Global microRNA profiling of all three groups on the same array platform provides greater strength to our study for direct comparisons between groups and reliability of results.
One of our study limitations is that the sample size in our study was relatively small, and the results should be validated in larger cohorts and diverse populations. Furthermore, due to the volunteer nature of participation for healthy subjects we were only able to recruit controls who were younger likely introducing age bias Additionally, the functional roles of the identified microRNAs in the pathogenesis of cirrhosis need to be elucidated through further experimental studied. Despite these limitations, our study has revealed interesting information on the involvement of microRNAs and their target mRNAs in identifying several pathways relevant in liver diseases. Most interestingly, our data identifying several lipid related target genes support the recent genetic studies and school of thought regarding lipid biology as a key pathway in regulating liver diseases.
In conclusion, our findings highlight that microRNAs can have clinical relevance in their potential as diagnostic and/or therapeutic targets for alcohol-associated liver disease/cirrhosis. The study also underscores the role of triglyceride and lipid metabolism in this disease by identifying new miRNA and mRNA as potential therapeutic targets. Further research in this area may lead to the development of targeted therapeutic strategies for the management of liver diseases.
Author Contributions
Conceived, designed, supervised the study, provided samples (DS); Performed experimental work (FS, MVJ); Collected and analysed microRNA data (FS, MVJ, AAH); provided genotype data (S-AT); Drafted first manuscript (FS). All Authors critically reviewed and approved the manuscript:.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Human Research Ethics Committees (HREC) of Sydney Local Health District (X11-0072, HREC11RPAH88; X17-029, HREC17RPAH331, 2011).” for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy issues.
Acknowledgments
This project was funded by the National Institutes of Health (NIH), National Institute of Alcohol Abuse & Alcoholism (NIAAA) (UO1 AA018389) for sample collection. We thank Hardikar Laboratory (Islet Biology and Diabetes Group) at NHMRC Clinical Trials Centre, University of Sydney for the infrastructure support to conduct laboratory experiments. AAH was supported through JDRF Australia Clinical Research Network CDA funding.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Mendez-Sanchez, N., P. Almeda-Valdes, and M. Uribe, Alcoholic liver disease. An update. Ann Hepatol, 2005. 4(1): p. 32-42.
- Lin, C.W. , et al., Heavy alcohol consumption increases the incidence of hepatocellular carcinoma in hepatitis B virus-related cirrhosis. J Hepatol, 2013. 58(4): p. 730-5.
- Liu, S.Y., I. T. Tsai, and Y.C. Hsu, Alcohol-Related Liver Disease: Basic Mechanisms and Clinical Perspectives. Int J Mol Sci, 2021. 22(10).
- Cequera, A. and M.C. Garcia de Leon Mendez, [Biomarkers for liver fibrosis: advances, advantages and disadvantages]. Rev Gastroenterol Mex, 2014. 79(3): p. 187-99.
- Pu, M. , et al., Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci, 2019. 76(3): p. 441-451.
- O'Brien, J. , et al., Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne), 2018. 9: p. 402.
- Gulyaeva, L.F. and N.E. Kushlinskiy, Regulatory mechanisms of microRNA expression. J Transl Med, 2016. 14(1): p. 143.
- Wang, X. , et al., MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut, 2021. 70(4): p. 784-795.
- Musaddaq, G. , et al., Circulating liver-specific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomarkers, 2019. 24(2): p. 103-109.
- Bala, S. , et al., Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology, 2012. 56(5): p. 1946-57.
- Whitfield, J.B. , et al., Brief report: genetics of alcoholic cirrhosis-GenomALC multinational study. Alcohol Clin Exp Res, 2015. 39(5): p. 836-42.
- Whitfield, J.B. , et al., Obesity, Diabetes, Coffee, Tea, and Cannabis Use Alter Risk for Alcohol-Related Cirrhosis in 2 Large Cohorts of High-Risk Drinkers. Am J Gastroenterol, 2021. 116(1): p. 106-115.
- Shihana, F. , et al., Circulating human microRNA biomarkers of oxalic acid-induced acute kidney injury. Arch Toxicol, 2020.
- Shihana, F. , et al., Urinary microRNAs as non-invasive biomarkers for toxic acute kidney injury in humans. Sci Rep, 2021. 11(1): p. 9165.
- Schwantes-An, T.H. , et al., Genome-wide Association Study and Meta-analysis on Alcohol-Associated Liver Cirrhosis Identifies Genetic Risk Factors. Hepatology, 2021. 73(5): p. 1920-1931.
- Yang, Z. , et al., Transcriptomic Analysis Reveals the MicroRNAs Responsible for Liver Regeneration Associated With Mortality in Alcohol-Associated Hepatitis. Hepatology, 2021. 74(5): p. 2436-2451.
- Sumida, Y., A. Nakajima, and Y. Itoh, Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol, 2014. 20(2): p. 475-85.
- Soresi, M. , et al., Non invasive tools for the diagnosis of liver cirrhosis. World J Gastroenterol, 2014. 20(48): p. 18131-50.
- Rana, M. , et al., Circulating Micrornas: Diagnostic Value As Biomarkers In the Detection Of Non-Alcoholic Fatty Liver Diseases and Hepatocellular Carcinoma. Microrna, 2023.
- Pant, K. and S.K. Venugopal, Circulating microRNAs: Possible role as non-invasive diagnostic biomarkers in liver disease. Clin Res Hepatol Gastroenterol, 2017. 41(4): p. 370-377.
- Cai, C., Y. Lin, and C. Yu, Circulating miRNAs as Novel Diagnostic Biomarkers in Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Can J Gastroenterol Hepatol, 2019. 2019: p. 2096161.
- Enache, L.S. , et al., Circulating RNA molecules as biomarkers in liver disease. Int J Mol Sci, 2014. 15(10): p. 17644-66.
- Bakshi, S. , et al., Altered expressions of circulating microRNAs 122 and 192 during antitubercular drug induced liver injury indicating their role as potential biomarkers. Hum Exp Toxicol, 2021. 40(9): p. 1474-1484.
- Sanjay, S. and C. Girish, Role of miRNA and its potential as a novel diagnostic biomarker in drug-induced liver injury. Eur J Clin Pharmacol, 2017. 73(4): p. 399-407.
- Schueller, F. , et al., The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int J Mol Sci, 2018. 19(1).
- Romeo, S. , et al., Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics, 2008. 40(12): p. 1461-1465.
- Buch, S. , et al., A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet, 2015. 47: p. 1443-1448.
- Abdel Halim, A.S. , et al., MicroRNAs: Small molecules with big impacts in liver injury. J Cell Physiol, 2023. 238(1): p. 32-69.
- Doghish, A.S. , et al., The role of miRNAs in liver diseases: Potential therapeutic and clinical applications. Pathol Res Pract, 2023. 243: p. 154375.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).