Submitted:
30 August 2023
Posted:
31 August 2023
You are already at the latest version
Abstract
Keywords:
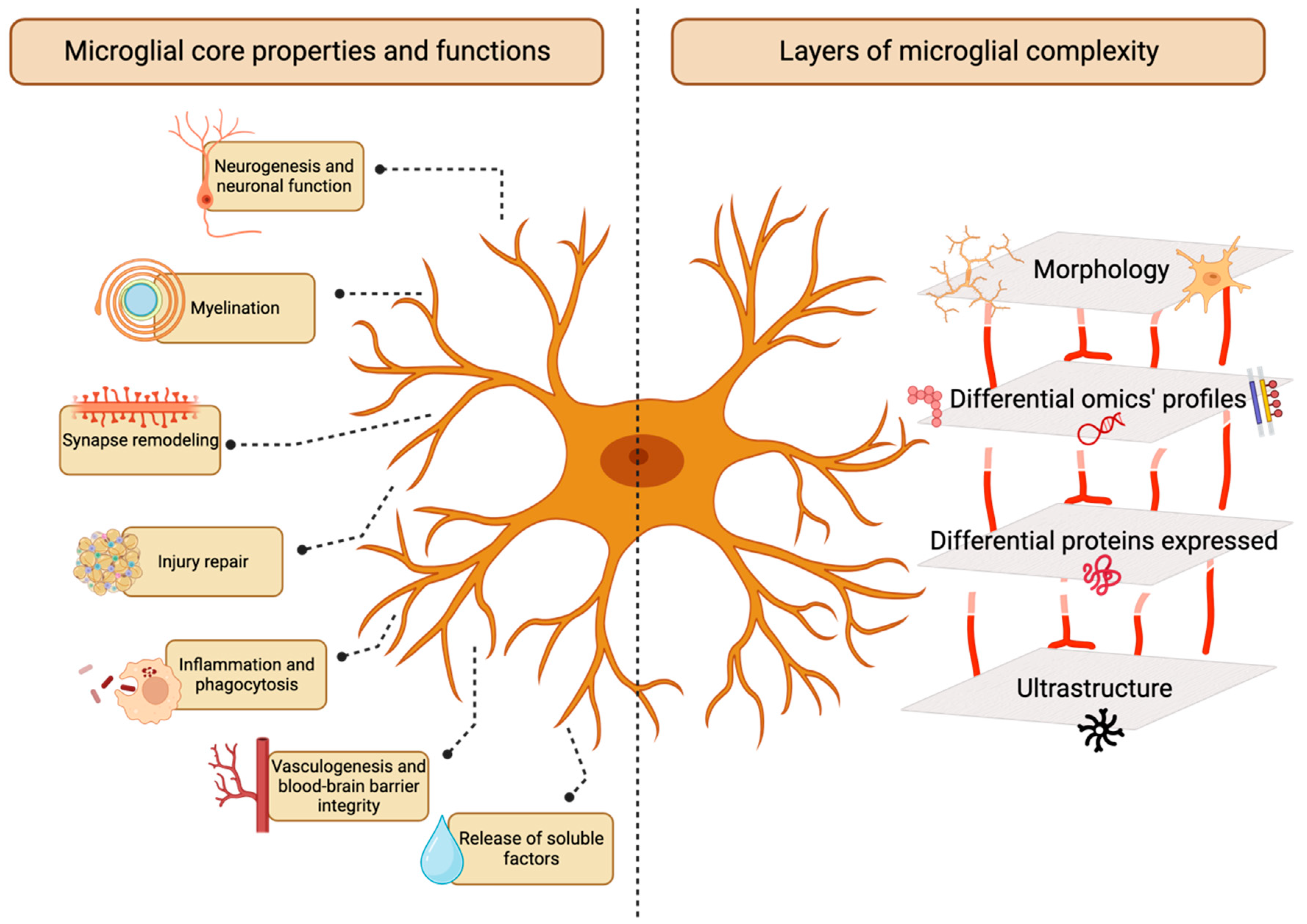
Introduction
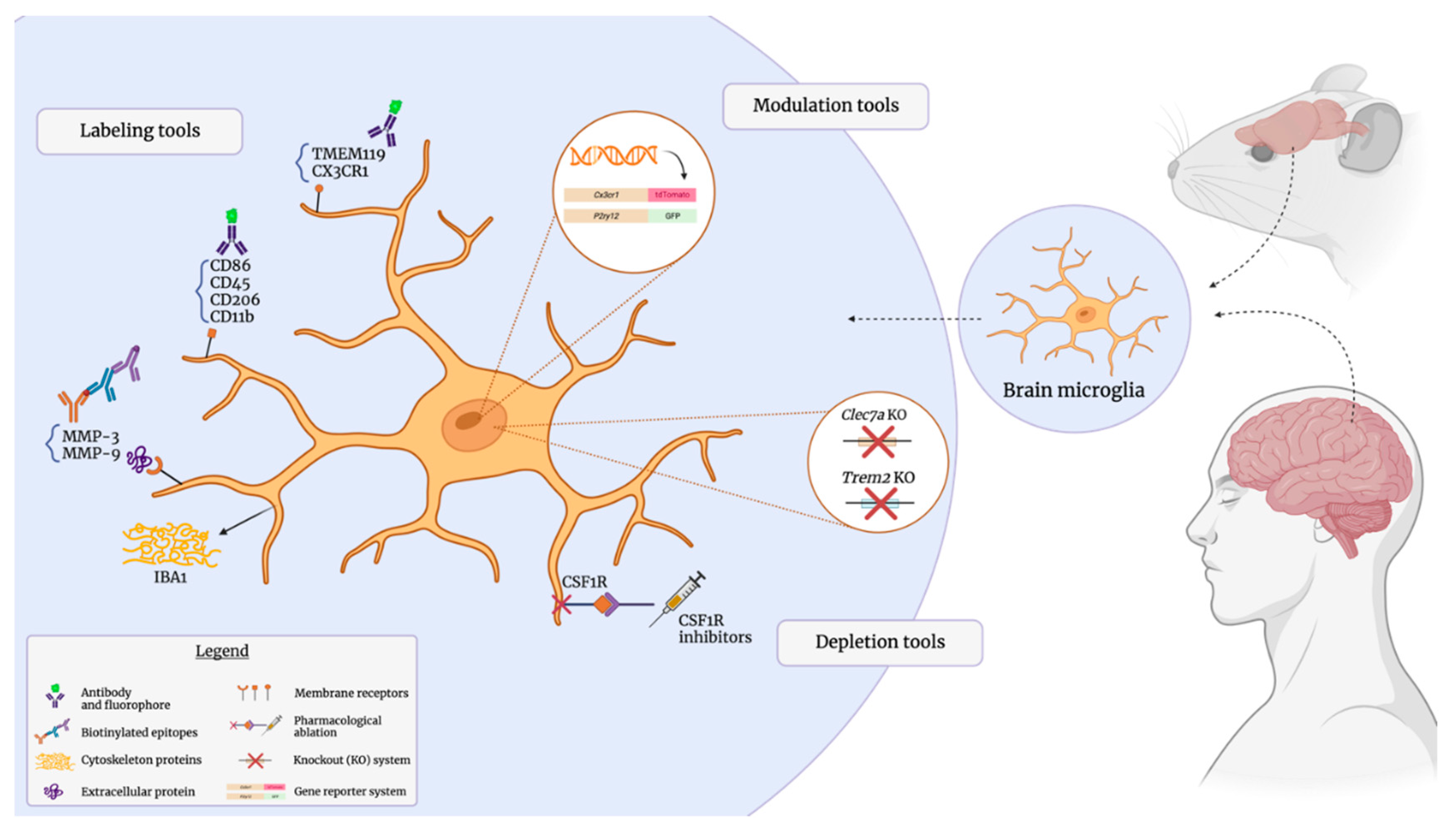
Antibody-Mediated Identification of Microglial Markers
Membrane-Associated Surface Markers
Intracellular Proteins
Extracellular and Secreted Proteins
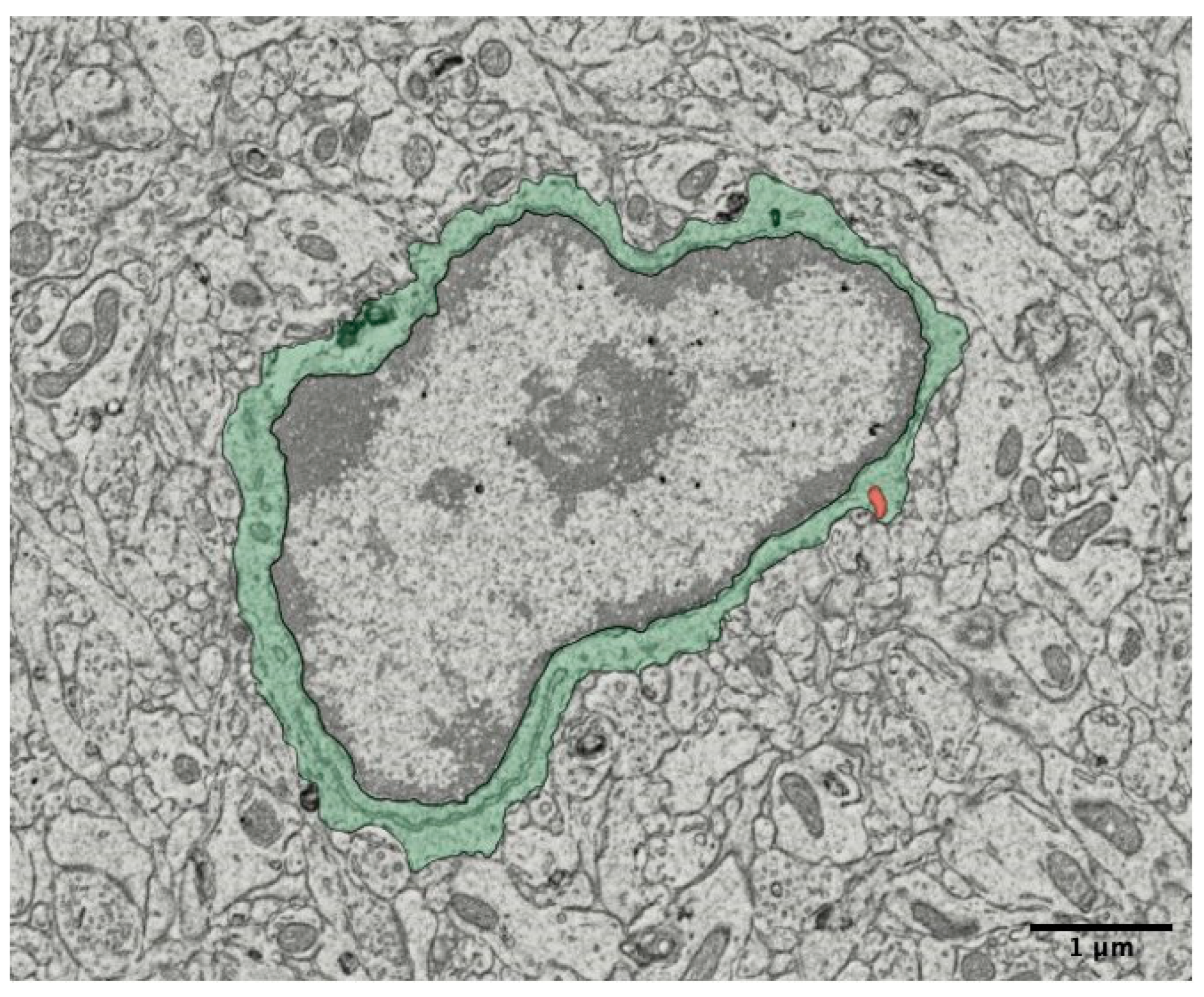
Microglia in Nanoscale: Electron Microscopy Ultrastructure Analysis
Genetically Modulating Microglia with Reporter Models
Constitutive Reporter Models for Microglial Study
Inducible Reporter Models for Microglial Study
Microglial Ablation: Genetic and Pharmacological Approaches
Genetic Depletion of Microglial Subsets
Pharmacological Depletion of Microglia
Fate Mapping: Tracking Microglia through Development, Health, and Disease
Microglial Fate Mapping during Development
Microglial Fate Mapping during Adult Homeostasis
Microglial Fate Mapping during Pathology
A Comparison of Common Microglial Fate Mapping Systems
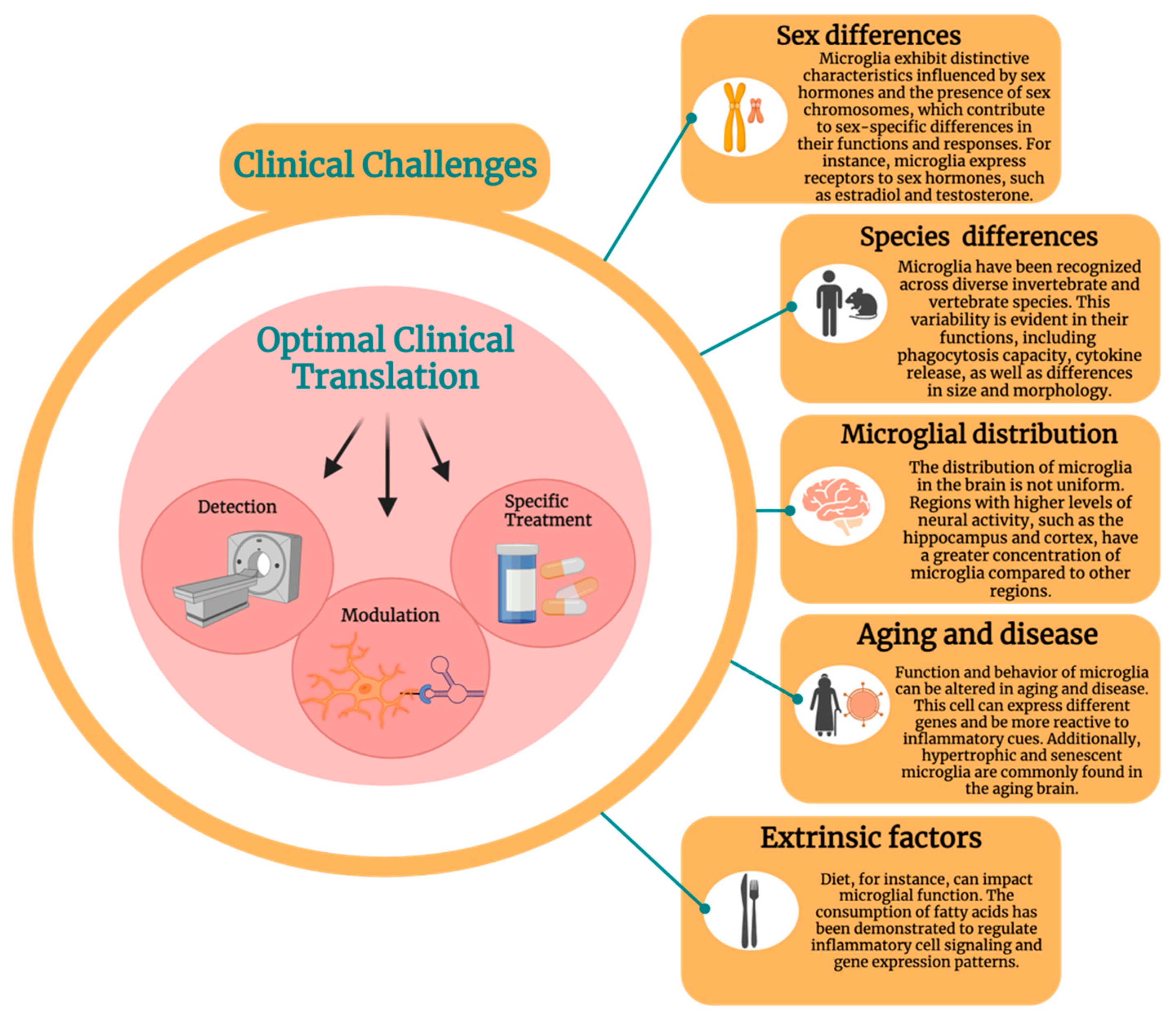
Translational Applications
Discussion
Author Contributions
List of Abbreviations
| AD | Alzheimer disease |
| ATP | adenosine triphosphate |
| BAM | border-associated macrophage |
| CLEC7a, DECTIN-1 | C-type lectin 7a |
| Clod-Lips | clodronate liposomes |
| CNS | central nervous system |
| CSF1R | colony-stimulating factor-1 receptor |
| CX3CL1 | fractalkine |
| CX3CR1 | CX3-motif chemokine receptor 1; fractalkine receptor |
| DAM | disease-associated microglia |
| DW-MRI | diffusion-weighted magnetic resonance imaging |
| DTR | diphtheria toxin receptor |
| ECM | extracellular matrix |
| ELISA | enzyme-linked immunosorbent assay |
| EM | electron microscopy |
| ER | estrogen receptor |
| ERT, ERT2 | tamoxifen-inducible estrogen receptor |
| FACS | fluorescence-activated cell sorting |
| FIRE | fms-intronic regulatory element |
| HEXB | hexosaminidase β-subunit |
| IHC | immunohistochemistry |
| KO | knockout |
| Mac-1-sap | Mac-1-saporin |
| MER | mutated estrogen receptor |
| MGnD | microglia neurodegenerative phenotype |
| MMP | matrix metalloprotease |
| PET | positron emission tomography |
| P2RY12 | purinergic receptor P2Y12 |
| SALL1 | spalt-like transcription factor 1 |
| TMEM119 | transmembrane protein 119 |
| TREM2 | triggering receptor expressed on myeloid cells 2 |
| TSPO | translocator protein |
Funding
Acknowledgments
References
- Acharjee, S., Verbeek, M., Gomez, C. D., Bisht, K., Lee, B., Benoit, L., et al. (2018). Reduced Microglial Activity and Enhanced Glutamate Transmission in the Basolateral Amygdala in Early CNS Autoimmunity. J Neurosci 38, 9019–9033. [CrossRef]
- Acharya, M. M., Green, K. N., Allen, B. D., Najafi, A. R., Syage, A., Minasyan, H., et al. (2016). Elimination of microglia improves cognitive function following cranial irradiation. Sci Rep 6, 31545. [CrossRef]
- Agalave, N. M., Lane, B. T., Mody, P. H., Szabo-Pardi, T. A., and Burton, M. D. (2020). Isolation, culture, and downstream characterization of primary microglia and astrocytes from adult rodent brain and spinal cord. J Neurosci Methods 340, 108742. [CrossRef]
- Ahmed, A., Wang, L.-L., Abdelmaksoud, S., Aboelgheit, A., Saeed, S., and Zhang, C.-L. (2017). Minocycline modulates microglia polarization in ischemia-reperfusion model of retinal degeneration and induces neuroprotection. Sci Rep 7, 14065. [CrossRef]
- Albadri, S., Del Bene, F., and Revenu, C. (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121–122, 77–85. [CrossRef]
- Askew, K., Li, K., Olmos-Alonso, A., Garcia-Moreno, F., Liang, Y., Richardson, P., et al. (2017). Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Rep 18, 391–405. [CrossRef]
- Augusto-Oliveira, M., Arrifano, G. P., Lopes-Araújo, A., Santos-Sacramento, L., Takeda, P. Y., Anthony, D. C., et al. (2019). What Do Microglia Really Do in Healthy Adult Brain? Cells 8, 1293. [CrossRef]
- Avignone, E., Ulmann, L., Levavasseur, F., Rassendren, F., and Audinat, E. (2008). Status Epilepticus Induces a Particular Microglial Activation State Characterized by Enhanced Purinergic Signaling. J. Neurosci. 28, 9133–9144. [CrossRef]
- Ayanoğlu, F. B., Elçin, A. E., and Elçin, Y. M. (2020). Bioethical issues in genome editing by CRISPR-Cas9 technology. Turk J Biol 44, 110–120. [CrossRef]
- Baba, Y., Watabe, Y., Sagara, H., and Watanabe, S. (2019). Sall1 plays pivotal roles for lens fiber cell differentiation in mouse. Biochem Biophys Res Commun 512, 927–933. [CrossRef]
- Bai, R., Gao, H., Han, Z., Huang, S., Ge, X., Chen, F., et al. (2017). Flow Cytometric Characterization of T Cell Subsets and Microglia After Repetitive Mild Traumatic Brain Injury in Rats. Neurochem Res 42, 2892–2901. [CrossRef]
- Banerjee, P., Mehta, A. R., Nirujogi, R. S., Cooper, J., James, O. G., Nanda, J., et al. (2023). Cell-autonomous immune dysfunction driven by disrupted autophagy in C9orf72-ALS iPSC-derived microglia contributes to neurodegeneration. Science Advances 9, eabq0651. [CrossRef]
- Barnett, A. M., Crews, F. T., and Coleman, L. G. (2021). Microglial depletion and repopulation: a new era of regenerative medicine? Neural Regen Res 16, 1204–1205. [CrossRef]
- Bassil, R., Shields, K., Granger, K., Zein, I., Ng, S., and Chih, B. (2021). Improved modeling of human AD with an automated culturing platform for iPSC neurons, astrocytes and microglia. Nat Commun 12, 5220. [CrossRef]
- Bemiller, S. M., McCray, T. J., Allan, K., Formica, S. V., Xu, G., Wilson, G., et al. (2017). TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol Neurodegener 12, 74. [CrossRef]
- Bennett, M. L., Bennett, F. C., Liddelow, S. A., Ajami, B., Zamanian, J. L., Fernhoff, N. B., et al. (2016). New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences 113, E1738–E1746. [CrossRef]
- Berdyyeva, T., Xia, C., Taylor, N., He, Y., Chen, G., Huang, C., et al. (2019). PET Imaging of the P2X7 Ion Channel with a Novel Tracer [18F]JNJ-64413739 in a Rat Model of Neuroinflammation. Mol Imaging Biol 21, 871–878. [CrossRef]
- Berger, A. (2003). How does it work? Positron emission tomography. BMJ 326, 1449. [CrossRef]
- Bernier, C. Bernier, C., and Dréno, B. (2001). [Minocycline]. Ann Dermatol Venereol 128, 627–637.
- Betlazar, C., Harrison-Brown, M., Middleton, R. J., Banati, R., and Liu, G.-J. (2018). Cellular Sources and Regional Variations in the Expression of the Neuroinflammatory Marker Translocator Protein (TSPO) in the Normal Brain. Int J Mol Sci 19, 2707. [CrossRef]
- Bijata, M., Bączyńska, E., Müller, F. E., Bijata, K., Masternak, J., Krzystyniak, A., et al. (2022). Activation of the 5-HT7 receptor and MMP-9 signaling module in the hippocampal CA1 region is necessary for the development of depressive-like behavior. Cell Reports 38. [CrossRef]
- Bijata, M., Labus, J., Guseva, D., Stawarski, M., Butzlaff, M., Dzwonek, J., et al. (2017). Synaptic Remodeling Depends on Signaling between Serotonin Receptors and the Extracellular Matrix. Cell Reports 19, 1767–1782. [CrossRef]
- Bisht, K., Okojie, K. A., Sharma, K., Lentferink, D. H., Sun, Y.-Y., Chen, H.-R., et al. (2021). Capillary-associated microglia regulate vascular structure and function through PANX1-P2RY12 coupling in mice. Nat Commun 12, 5289. [CrossRef]
- Bisht, K., Sharma, K. P., Lecours, C., Gabriela Sánchez, M., El Hajj, H., Milior, G., et al. (2016). Dark microglia: A new phenotype predominantly associated with pathological states. Glia 64, 826–839. [CrossRef]
- Blecharz-Lang, K. G., Patsouris, V., Nieminen-Kelhä, M., Seiffert, S., Schneider, U. C., and Vajkoczy, P. (2022). Minocycline Attenuates Microglia/Macrophage Phagocytic Activity and Inhibits SAH-Induced Neuronal Cell Death and Inflammation. Neurocrit Care 37, 410–423. [CrossRef]
- Bohnert, S., Seiffert, A., Trella, S., Bohnert, M., Distel, L., Ondruschka, B., et al. (2020). TMEM119 as a specific marker of microglia reaction in traumatic brain injury in postmortem examination. Int J Legal Med 134, 2167–2176. [CrossRef]
- Bollinger, J. L., Collins, K. E., Patel, R., and Wellman, C. L. (2017). Behavioral stress alters corticolimbic microglia in a sex- and brain region-specific manner. PLOS ONE 12, e0187631. [CrossRef]
- Bollinger, J. L., Dadosky, D. T., Flurer, J. K., Rainer, I. L., Woodburn, S. C., and Wohleb, E. S. (2023). Microglial P2Y12 mediates chronic stress-induced synapse loss in the prefrontal cortex and associated behavioral consequences. Neuropsychopharmacol. 48, 1347–1357. [CrossRef]
- Bordeleau, M., Fernández de Cossío, L., Lacabanne, C., Savage, J. C., Vernoux, N., Chakravarty, M., et al. (2021). Maternal high-fat diet modifies myelin organization, microglial interactions, and results in social memory and sensorimotor gating deficits in adolescent mouse offspring. Brain Behav Immun Health 15, 100281. [CrossRef]
- Brown, G. D., Taylor, P. R., Reid, D. M., Willment, J. A., Williams, D. L., Martinez-Pomares, L., et al. (2002). Dectin-1 Is A Major β-Glucan Receptor On Macrophages. Journal of Experimental Medicine 196, 407–412. [CrossRef]
- Bruzelius, A., Hidalgo, I., Boza-Serrano, A., Hjelmér, A.-G., Tison, A., Deierborg, T., et al. (2021). The human bone marrow harbors a CD45- CD11B+ cell progenitor permitting rapid microglia-like cell derivative approaches. Stem Cells Transl Med 10, 582–597. [CrossRef]
- Butler, C. A., Popescu, A. S., Kitchener, E. J. A., Allendorf, D. H., Puigdellívol, M., and Brown, G. C. (2021). Microglial phagocytosis of neurons in neurodegeneration, and its regulation. J Neurochem 158, 621–639. [CrossRef]
- Butovsky, O., and Weiner, H. L. (2018a). Microglial signatures and their role in health and disease. Nat Rev Neurosci 19, 622–635. [CrossRef]
- Butovsky, O., and Weiner, H. L. (2018b). Microglial signatures and their role in health and disease. Nat Rev Neurosci 19, 622–635. [CrossRef]
- Buttgereit, A., Lelios, I., Yu, X., Vrohlings, M., Krakoski, N. R., Gautier, E. L., et al. (2016). Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol 17, 1397–1406. [CrossRef]
- Cao, Z., Harvey, S. S., Chiang, T., Foltz, A. G., Lee, A. G., Cheng, M. Y., et al. (2021). Unique Subtype of Microglia in Degenerative Thalamus After Cortical Stroke. Stroke 52, 687–698. [CrossRef]
- Carrier, M., Dolhan, K., Bobotis, B. C., Desjardins, M., and Tremblay, M.-È. (2022). The implication of a diversity of non-neuronal cells in disorders affecting brain networks. Front Cell Neurosci 16, 1015556. [CrossRef]
- Carrier, M., Robert, M.-È., González Ibáñez, F., Desjardins, M., and Tremblay, M.-È. (2020). Imaging the Neuroimmune Dynamics Across Space and Time. Front Neurosci 14, 903. [CrossRef]
- Carrier, M., Šimončičová, E., St-Pierre, M.-K., McKee, C., and Tremblay, M.-È. (2021). Psychological Stress as a Risk Factor for Accelerated Cellular Aging and Cognitive Decline: The Involvement of Microglia-Neuron Crosstalk. Front Mol Neurosci 14, 749737. [CrossRef]
- Casali, B. T., and Reed-Geaghan, E. G. (2021). Microglial Function and Regulation during Development, Homeostasis and Alzheimer’s Disease. Cells 10, 957. [CrossRef]
- In Chaney, A; TREM1 as a novel PET imaging biomarker of peripheral infiltrating myeloid cells and potential therapeutic target in multiple sclerosis: , Cropper, H., Johnson, E., Stevens, M., and James, M. (2019). Imaging the invaders; Chaney, A., Cropper, H., Johnson, E., Stevens, M., and James, M. (2019). Imaging the invaders: TREM1 as a novel PET imaging biomarker of peripheral infiltrating myeloid cells and potential therapeutic target in multiple sclerosis. Journal of Nuclear Medicine 60, 1506b–1506b.
- Chaney, A. Chaney, A., Wilson, E., Jain, P., Cropper, H., Swarovski, M., Lucot, K., et al. (2020). TREM1-PET imaging of pro-inflammatory myeloid cells distinguishes active disease from remission in Multiple Sclerosis. Journal of Nuclear Medicine 61, 199–199.
- Chappell-Maor, L., Kolesnikov, M., Kim, J.-S., Shemer, A., Haimon, Z., Grozovski, J., et al. (2020). Comparative analysis of CreER transgenic mice for the study of brain macrophages: A case study. Eur J Immunol 50, 353–362. [CrossRef]
- Chen, M., Ona, V. O., Li, M., Ferrante, R. J., Fink, K. B., Zhu, S., et al. (2000). Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med 6, 797–801. [CrossRef]
- Chistiakov, D. A., Killingsworth, M. C., Myasoedova, V. A., Orekhov, A. N., and Bobryshev, Y. V. (2017). CD68/macrosialin: not just a histochemical marker. Lab Invest 97, 4–13. [CrossRef]
- Choi, Y., Kim, H.-S., Shin, K. Y., Kim, E.-M., Kim, M., Kim, H.-S., et al. (2007). Minocycline attenuates neuronal cell death and improves cognitive impairment in Alzheimer’s disease models. Neuropsychopharmacology 32, 2393–2404. [CrossRef]
- Colognato, H., and Tzvetanova, I. D. (2011). Glia unglued: how signals from the extracellular matrix regulate the development of myelinating glia. Dev Neurobiol 71, 924–955. [CrossRef]
- Conen, S., Gregory, C. J., Hinz, R., Smallman, R., Corsi-Zuelli, F., Deakin, B., et al. (2021). Neuroinflammation as measured by positron emission tomography in patients with recent onset and established schizophrenia: implications for immune pathogenesis. Mol Psychiatry 26, 5398–5406. [CrossRef]
- Cornell, J., Salinas, S., Huang, H.-Y., and Zhou, M. (2022). Microglia regulation of synaptic plasticity and learning and memory. Neural Regen Res 17, 705–716. [CrossRef]
- Coughlin, J. M., Du, Y., Lesniak, W. G., Harrington, C. K., Brosnan, M. K., O’Toole, R., et al. (2022). First-in-human use of 11C-CPPC with positron emission tomography for imaging the macrophage colony-stimulating factor 1 receptor. EJNMMI Research 12, 64. [CrossRef]
- Cuní-López, C., Stewart, R., Quek, H., and White, A. R. (2022). Recent Advances in Microglia Modelling to Address Translational Outcomes in Neurodegenerative Diseases. Cells 11, 1662. [CrossRef]
- Daley, D., Mani, V. R., Mohan, N., Akkad, N., Ochi, A., Heindel, D. W., et al. (2017). Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med 23, 556–567. [CrossRef]
- Damasceno, J. D., Reis-Cunha, J., Crouch, K., Beraldi, D., Lapsley, C., Tosi, L. R. O., et al. (2020). Conditional knockout of RAD51-related genes in Leishmania major reveals a critical role for homologous recombination during genome replication. PLoS Genet 16, e1008828. [CrossRef]
- Daugherty, D. J., Selvaraj, V., Chechneva, O. V., Liu, X.-B., Pleasure, D. E., and Deng, W. (2013). A TSPO ligand is protective in a mouse model of multiple sclerosis. EMBO Mol Med 5, 891–903. [CrossRef]
- Davalos, D., Grutzendler, J., Yang, G., Kim, J. V., Zuo, Y., Jung, S., et al. (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8, 752–758. [CrossRef]
- Deczkowska, A., Keren-Shaul, H., Weiner, A., Colonna, M., Schwartz, M., and Amit, I. (2018a). Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081. [CrossRef]
- Deczkowska, A., Keren-Shaul, H., Weiner, A., Colonna, M., Schwartz, M., and Amit, I. (2018b). Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081. [CrossRef]
- Deerhake, M. E., Danzaki, K., Inoue, M., Cardakli, E. D., Nonaka, T., Aggarwal, N., et al. (2021). Dectin-1 limits autoimmune neuroinflammation and promotes myeloid cell-astrocyte crosstalk via Card9-independent expression of Oncostatin M. Immunity 54, 484-498.e8. [CrossRef]
- Di Biase, M. A., Zalesky, A., O’keefe, G., Laskaris, L., Baune, B. T., Weickert, C. S., et al. (2017). PET imaging of putative microglial activation in individuals at ultra-high risk for psychosis, recently diagnosed and chronically ill with schizophrenia. Transl Psychiatry 7, e1225. [CrossRef]
- Dolan, M.-J., Therrien, M., Jereb, S., Kamath, T., Gazestani, V., Atkeson, T., et al. (2023). Exposure of iPSC-derived human microglia to brain substrates enables the generation and manipulation of diverse transcriptional states in vitro. Nat Immunol 24, 1382–1390. [CrossRef]
- Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [CrossRef]
- Dräger, N. M., Sattler, S. M., Huang, C. T.-L., Teter, O. M., Leng, K., Hashemi, S. H., et al. (2022). A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat Neurosci 25, 1149–1162. [CrossRef]
- Drissen, R., Thongjuea, S., Theilgaard-Mönch, K., and Nerlov, C. (2019). Identification of two distinct pathways of human myelopoiesis. Sci Immunol 4, eaau7148. [CrossRef]
- Du, Y., Ma, Z., Lin, S., Dodel, R. C., Gao, F., Bales, K. R., et al. (2001). Minocycline prevents nigrostriatal dopaminergic neurodegeneration in the MPTP model of Parkinson’s disease. Proc Natl Acad Sci U S A 98, 14669–14674. [CrossRef]
- Dubbelaar, M. L., Kracht, L., Eggen, B. J. L., and Boddeke, E. W. G. M. (2018). The Kaleidoscope of Microglial Phenotypes. Front Immunol 9, 1753. [CrossRef]
- El Hajj, H., Savage, J. C., Bisht, K., Parent, M., Vallières, L., Rivest, S., et al. (2019). Ultrastructural evidence of microglial heterogeneity in Alzheimer’s disease amyloid pathology. J Neuroinflammation 16, 87. [CrossRef]
- Elmore, M. R. P., Najafi, A. R., Koike, M. A., Dagher, N. N., Spangenberg, E. E., Rice, R. A., et al. (2014). Colony-Stimulating Factor 1 Receptor Signaling Is Necessary for Microglia Viability, Unmasking a Microglia Progenitor Cell in the Adult Brain. Neuron 82, 380–397. [CrossRef]
- Erblich, B., Zhu, L., Etgen, A. M., Dobrenis, K., and Pollard, J. W. (2011). Absence of Colony Stimulation Factor-1 Receptor Results in Loss of Microglia, Disrupted Brain Development and Olfactory Deficits. PLOS ONE 6, e26317. [CrossRef]
- Fattorelli, N., Martinez-Muriana, A., Wolfs, L., Geric, I., De Strooper, B., and Mancuso, R. (2021). Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat Protoc 16, 1013–1033. [CrossRef]
- Faust, T. E., Feinberg, P. A., Oâ Connor, C., Kawaguchi, R., Chan, A., Strasburger, H., et al. (2023). A comparative analysis of microglial inducible Cre lines. bioRxiv, 2023.01.09.523268. [CrossRef]
- Feil, R., Wagner, J., Metzger, D., and Chambon, P. (1997). Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237, 752–757. [CrossRef]
- Filipello, F., Morini, R., Corradini, I., Zerbi, V., Canzi, A., Michalski, B., et al. (2018). The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 48, 979-991.e8. [CrossRef]
- Fischer, H.-G., and Reichmann, G. (2001). Brain Dendritic Cells and Macrophages/Microglia in Central Nervous System Inflammation1. The Journal of Immunology 166, 2717–2726. [CrossRef]
- Friedrich, G., and Soriano, P. (1991). Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 5, 1513–1523. [CrossRef]
- Fu, H., Zhao, Y., Hu, D., Wang, S., Yu, T., and Zhang, L. (2020). Depletion of microglia exacerbates injury and impairs function recovery after spinal cord injury in mice. Cell Death Dis 11, 528. [CrossRef]
- Fujiwara, T., Yakoub, M. A., Chandler, A., Christ, A. B., Yang, G., Ouerfelli, O., et al. (2021). CSF1/CSF1R Signaling Inhibitor Pexidartinib (PLX3397) Reprograms Tumor-Associated Macrophages and Stimulates T-cell Infiltration in the Sarcoma Microenvironment. Mol Cancer Ther 20, 1388–1399. [CrossRef]
- Gao, H., Di, J., Clausen, B. H., Wang, N., Zhu, X., Zhao, T., et al. (2023). Distinct myeloid population phenotypes dependent on TREM2 expression levels shape the pathology of traumatic versus demyelinating CNS disorders. Cell Reports 42. [CrossRef]
- Garcia, J. A., Cardona, S. M., and Cardona, A. E. (2013). Analyses of microglia effector function using CX3CR1-GFP knock-in mice. Methods Mol Biol 1041, 307–317. [CrossRef]
- Garcia-Hernandez, R., Cerdán Cerdá, A., Trouve Carpena, A., Drakesmith, M., Koller, K., Jones, D. K., et al. (2022). Mapping microglia and astrocyte activation in vivo using diffusion MRI. Science Advances 8, eabq2923. [CrossRef]
- Garrido-Mesa, N., Zarzuelo, A., and Gálvez, J. (2013). Minocycline: far beyond an antibiotic. Br J Pharmacol 169, 337–352. [CrossRef]
- Gerhard, A., Banati, R. B., Goerres, G. B., Cagnin, A., Myers, R., Gunn, R. N., et al. (2003). [11C](R)-PK11195 PET imaging of microglial activation in multiple system atrophy. Neurology 61, 686–689. [CrossRef]
- Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [CrossRef]
- Goldman, R. D. (2000). Antibodies: indispensable tools for biomedical research. Trends Biochem Sci 25, 593–595. [CrossRef]
- Goldmann, T., Wieghofer, P., Jordão, M. J. C., Prutek, F., Hagemeyer, N., Frenzel, K., et al. (2016). Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol 17, 797–805. [CrossRef]
- Goldmann, T., Wieghofer, P., Müller, P. F., Wolf, Y., Varol, D., Yona, S., et al. (2013). A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16, 1618–1626. [CrossRef]
- Gómez Morillas, A., Besson, V. C., and Lerouet, D. (2021). Microglia and Neuroinflammation: What Place for P2RY12? Int J Mol Sci 22, 1636. [CrossRef]
- Gomez Perdiguero, E., Klapproth, K., Schulz, C., Busch, K., Azzoni, E., Crozet, L., et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. [CrossRef]
- Gomez-Nicola, D., and Perry, V. H. (2015). Microglial dynamics and role in the healthy and diseased brain: a paradigm of functional plasticity. Neuroscientist 21, 169–184. [CrossRef]
- Gong, J., Szego, É. M., Leonov, A., Benito, E., Becker, S., Fischer, A., et al. (2019). Translocator Protein Ligand Protects against Neurodegeneration in the MPTP Mouse Model of Parkinsonism. J Neurosci 39, 3752–3769. [CrossRef]
- González-Prieto, M., Gutiérrez, I. L., García-Bueno, B., Caso, J. R., Leza, J. C., Ortega-Hernández, A., et al. (2021). Microglial CX3CR1 production increases in Alzheimer’s disease and is regulated by noradrenaline. Glia 69, 73–90. [CrossRef]
- Gratuze, M., Leyns, C. E. G., and Holtzman, D. M. (2018). New insights into the role of TREM2 in Alzheimer’s disease. Molecular Neurodegeneration 13, 66. [CrossRef]
- Green, K. N., Crapser, J. D., and Hohsfield, L. A. (2020a). To Kill a Microglia: A Case for CSF1R Inhibitors. Trends Immunol 41, 771–784. [CrossRef]
- Green, K. N., Crapser, J. D., and Hohsfield, L. A. (2020b). To Kill Microglia: A Case for CSF1R Inhibitors. Trends Immunol 41, 771–784. [CrossRef]
- Greter, M., Lelios, I., and Croxford, A. L. (2015). Microglia Versus Myeloid Cell Nomenclature during Brain Inflammation. Front Immunol 6, 249. [CrossRef]
- Guerreiro, R., Wojtas, A., Bras, J., Carrasquillo, M., Rogaeva, E., Majounie, E., et al. (2013). TREM2 Variants in Alzheimer’s Disease. N Engl J Med 368, 117–127. [CrossRef]
- Guillot-Sestier, M.-V., Araiz, A. R., Mela, V., Gaban, A. S., O’Neill, E., Joshi, L., et al. (2021). Microglial metabolism is a pivotal factor in sexual dimorphism in Alzheimer’s disease. Commun Biol 4, 1–13. [CrossRef]
- Haenseler, W., Sansom, S. N., Buchrieser, J., Newey, S. E., Moore, C. S., Nicholls, F. J., et al. (2017). A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports 8, 1727–1742. [CrossRef]
- Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253-271.e6. [CrossRef]
- Han, J., Fan, Y., Zhou, K., Blomgren, K., and Harris, R. A. (2021). Uncovering sex differences of rodent microglia. Journal of Neuroinflammation 18, 74. [CrossRef]
- Han, J., Harris, R. A., and Zhang, X.-M. (2017). An updated assessment of microglia depletion: current concepts and future directions. Mol Brain 10, 25. [CrossRef]
- Han, X., Li, Q., Lan, X., El-Mufti, L., Ren, H., and Wang, J. (2019). Microglial Depletion with Clodronate Liposomes Increases Proinflammatory Cytokine Levels, Induces Astrocyte Activation, and Damages Blood Vessel Integrity. Mol Neurobiol 56, 6184–6196. [CrossRef]
- Hanisch, U.-K., and Kettenmann, H. (2007). Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10, 1387–1394. [CrossRef]
- Hayashi, S., and McMahon, A. P. (2002). Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 244, 305–318. [CrossRef]
- Haynes, S. E., Hollopeter, G., Yang, G., Kurpius, D., Dailey, M. E., Gan, W.-B., et al. (2006). The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9, 1512–1519. [CrossRef]
- Hellström Erkenstam, N., Smith, P. L. P., Fleiss, B., Nair, S., Svedin, P., Wang, W., et al. (2016). Temporal Characterization of Microglia/Macrophage Phenotypes in a Mouse Model of Neonatal Hypoxic-Ischemic Brain Injury. Front Cell Neurosci 10, 286. [CrossRef]
- Hendrickx, D. A. E., van Eden, C. G., Schuurman, K. G., Hamann, J., and Huitinga, I. (2017). Staining of HLA-DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. Journal of Neuroimmunology 309, 12–22. [CrossRef]
- Heuss, N. D., Pierson, M. J., Roehrich, H., McPherson, S. W., Gram, A. L., Li, L., et al. (2018). Optic nerve as a source of activated retinal microglia post-injury. Acta Neuropathol Commun 6, 66. [CrossRef]
- Hickman, S. E., Allison, E. K., Coleman, U., Kingery-Gallagher, N. D., and El Khoury, J. (2019). Heterozygous CX3CR1 Deficiency in Microglia Restores Neuronal β-Amyloid Clearance Pathways and Slows Progression of Alzheimer’s Like-Disease in PS1-APP Mice. Frontiers in Immunology 10. Available at:.
- Hickman, S. E., Kingery, N. D., Ohsumi, T. K., Borowsky, M. L., Wang, L., Means, T. K., et al. (2013). The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 16, 1896–1905. [CrossRef]
- Hirrlinger, J., Scheller, A., Hirrlinger, P. G., Kellert, B., Tang, W., Wehr, M. C., et al. (2009). Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One 4, e4286. [CrossRef]
- Hoeffel, G., Chen, J., Lavin, Y., Low, D., Almeida, F. F., See, P., et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678. [CrossRef]
- Holtman, I. R., Skola, D., and Glass, C. K. (2017). Transcriptional control of microglia phenotypes in health and disease. J Clin Invest 127, 3220–3229. [CrossRef]
- Honarpisheh, P., Lee, J., Banerjee, A., Blasco-Conesa, M. P., Honarpisheh, P., d’Aigle, J., et al. (2020). Potential caveats of putative microglia-specific markers for assessment of age-related cerebrovascular neuroinflammation. J Neuroinflammation 17, 366. [CrossRef]
- Hoogland, I. C. M., Houbolt, C., van Westerloo, D. J., van Gool, W. A., and van de Beek, D. (2015). Systemic inflammation and microglial activation: systematic review of animal experiments. Journal of Neuroinflammation 12, 114. [CrossRef]
- Hopperton, K. E., Mohammad, D., Trépanier, M. O., Giuliano, V., and Bazinet, R. P. (2018). Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: a systematic review. Mol Psychiatry 23, 177–198. [CrossRef]
- Hu, W., Pan, D., Wang, Y., Bao, W., Zuo, C., Guan, Y., et al. (2020). PET Imaging for Dynamically Monitoring Neuroinflammation in APP/PS1 Mouse Model Using [18F]DPA714. Front Neurosci 14, 810. [CrossRef]
- Huang, Y., Xu, Z., Xiong, S., Sun, F., Qin, G., Hu, G., et al. (2018). Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci 21, 530–540. [CrossRef]
- Hui, C. W., Vecchiarelli, H. A., Gervais, É., Luo, X., Michaud, F., Scheefhals, L., et al. (2020). Sex Differences of Microglia and Synapses in the Hippocampal Dentate Gyrus of Adult Mouse Offspring Exposed to Maternal Immune Activation. Front Cell Neurosci 14, 558181. [CrossRef]
- Hupp, S., and Iliev, A. I. (2020). CSF-1 receptor inhibition as a highly effective tool for depletion of microglia in mixed glial cultures. J Neurosci Methods 332, 108537. [CrossRef]
- Ibanez, F. G., Picard, K., Bordelau, M., Sharma, K., Bisht, K., and Tremblay, M.-E. (2019). Immunofluorescence Staining Using IBA1 and TMEM119 for Microglial Density, Morphology and Peripheral Myeloid Cell Infiltration Analysis in Mouse Brain. J. Vis. Exp., e60510. [CrossRef]
- Imai, Y., Ibata, I., Ito, D., Ohsawa, K., and Kohsaka, S. (1996). A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun 224, 855–862. [CrossRef]
- Indra, A. K., Warot, X., Brocard, J., Bornert, J. M., Xiao, J. H., Chambon, P., et al. (1999). Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res 27, 4324–4327. [CrossRef]
- Janssen, B., Vugts, D. J., Windhorst, A. D., and Mach, R. H. (2018). PET Imaging of Microglial Activation-Beyond Targeting TSPO. Molecules 23, 607. [CrossRef]
- Jefferson, R. A., Burgess, S. M., and Hirsh, D. (1986). beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A 83, 8447–8451. [CrossRef]
- Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6, 3901–3907. [CrossRef]
- Jia, M., Zhang, W., Zhu, J., Huang, C., Zhou, J., Lian, J., et al. (2021). Microglia-Specific Expression of HEXA and HEXB Leads to Poor Prognosis in Glioblastoma Patients. Front Oncol 11, 685893. [CrossRef]
- Jiang, T., Xing, B., and Rao, J. (2008). Recent developments of biological reporter technology for detecting gene expression. Biotechnol Genet Eng Rev 25, 41–75. [CrossRef]
- Jin, X., Ishii, H., Bai, Z., Itokazu, T., and Yamashita, T. (2012). Temporal Changes in Cell Marker Expression and Cellular Infiltration in a Controlled Cortical Impact Model in Adult Male C57BL/6 Mice. PLOS ONE 7, e41892. [CrossRef]
- Johnson, N. R., Yuan, P., Castillo, E., Lopez, T. P., Yue, W., Bond, A., et al. (2023). CSF1R inhibitors induce a sex-specific resilient microglial phenotype and functional rescue in a tauopathy mouse model. Nat Commun 14, 118. [CrossRef]
- Jonsson, T., Stefansson, H., Steinberg, S., Jonsdottir, I., Jonsson, P. V., Snaedal, J., et al. (2013). Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. N Engl J Med 368, 107–116. [CrossRef]
- Jordão, M. J. C., Sankowski, R., Brendecke, S. M., Sagar, null, Locatelli, G., Tai, Y.-H., et al. (2019). Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554. [CrossRef]
- Jullien, N., Sampieri, F., Enjalbert, A., and Herman, J. (2003). Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Research 31, e131. [CrossRef]
- Jung, S., Aliberti, J., Graemmel, P., Sunshine, M. J., Kreutzberg, G. W., Sher, A., et al. (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114. [CrossRef]
- Jurga, A. M., Paleczna, M., and Kuter, K. Z. (2020). Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front Cell Neurosci 14, 198. [CrossRef]
- Kaiser, T., and Feng, G. (2019). Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia. eNeuro 6, ENEURO.0448-18.2019. [CrossRef]
- Keren-Shaul, H., Spinrad, A., Weiner, A., Matcovitch-Natan, O., Dvir-Szternfeld, R., Ulland, T. K., et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276-1290.e17. [CrossRef]
- Kiani Shabestari, S., Morabito, S., Danhash, E. P., McQuade, A., Sanchez, J. R., Miyoshi, E., et al. (2022). Absence of microglia promotes diverse pathologies and early lethality in Alzheimer’s disease mice. Cell Reports 39, 110961. [CrossRef]
- Kim, J., Kim, J.-H., Do, J. Y., Lee, J. Y., Yanai, R., Lee, I., et al. (2021a). Key Role of Microglial Matrix Metalloproteinases in Choroidal Neovascularization. Frontiers in Cellular Neuroscience 15. Available at:.
- Kim, J.-S., Kolesnikov, M., Peled-Hajaj, S., Scheyltjens, I., Xia, Y., Trzebanski, S., et al. (2021b). A Binary Cre Transgenic Approach Dissects Microglia and CNS Border-Associated Macrophages. Immunity 54, 176-190.e7. [CrossRef]
- Kim, S., and Son, Y. (2021). Astrocytes Stimulate Microglial Proliferation and M2 Polarization In Vitro through Crosstalk between Astrocytes and Microglia. Int J Mol Sci 22, 8800. [CrossRef]
- Kim, Y. S., Kim, S. S., Cho, J. J., Choi, D. H., Hwang, O., Shin, D. H., et al. (2005). Matrix Metalloproteinase-3: A Novel Signaling Proteinase from Apoptotic Neuronal Cells That Activates Microglia. J. Neurosci. 25, 3701–3711. [CrossRef]
- Kingham, P. J., and Pocock, J. M. (2001). Microglial secreted cathepsin B induces neuronal apoptosis. Journal of Neurochemistry 76, 1475–1484. [CrossRef]
- Kishimoto, I., Okano, T., Nishimura, K., Motohashi, T., and Omori, K. (2019). Early Development of Resident Macrophages in the Mouse Cochlea Depends on Yolk Sac Hematopoiesis. Frontiers in Neurology 10. Available at:.
- Konishi, H., and Kiyama, H. (2018). Microglial TREM2/DAP12 Signaling: A Double-Edged Sword in Neural Diseases. Front Cell Neurosci 12, 206. [CrossRef]
- Könnecke, H., and Bechmann, I. (2013). The role of microglia and matrix metalloproteinases involvement in neuroinflammation and gliomas. Clin Dev Immunol 2013, 914104. [CrossRef]
- Krais, J. J., and Johnson, N. (2020). BRCA1 Mutations in Cancer: Coordinating Deficiencies in Homologous Recombination with Tumorigenesis. Cancer Res 80, 4601–4609. [CrossRef]
- Krasemann, S., Madore, C., Cialic, R., Baufeld, C., Calcagno, N., Fatimy, R. E., et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566-581.e9. [CrossRef]
- Kuil, L. E., López Martí, A., Carreras Mascaro, A., van den Bosch, J. C., van den Berg, P., van der Linde, H. C., et al. (2019). Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development. Glia 67, 1705–1718. [CrossRef]
- Kwon, H. S., and Koh, S.-H. (2020). Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener 9, 42. [CrossRef]
- Lam, D., Enright, H. A., Cadena, J., Peters, S. K. G., Sales, A. P., Osburn, J. J., et al. (2019). Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci Rep 9, 4159. [CrossRef]
- Lauro, C., Chece, G., Monaco, L., Antonangeli, F., Peruzzi, G., Rinaldo, S., et al. (2019). Fractalkine Modulates Microglia Metabolism in Brain Ischemia. Front. Cell. Neurosci. 13, 414. [CrossRef]
- Lavisse, S., Goutal, S., Wimberley, C., Tonietto, M., Bottlaender, M., Gervais, P., et al. (2021). Increased microglial activation in patients with Parkinson disease using [18F]-DPA714 TSPO PET imaging. Parkinsonism Relat Disord 82, 29–36. [CrossRef]
- Lawson, L. J., Perry, V. H., and Gordon, S. (1992). Turnover of resident microglia in the normal adult mouse brain. Neuroscience 48, 405–415. [CrossRef]
- Lecours, C., St-Pierre, M.-K., Picard, K., Bordeleau, M., Bourque, M., Awogbindin, I. O., et al. (2020). Levodopa partially rescues microglial numerical, morphological, and phagolysosomal alterations in a monkey model of Parkinson’s disease. Brain Behav Immun 90, 81–96. [CrossRef]
- Lee, E.-J., Moon, P.-G., Baek, M.-C., and Kim, H.-S. (2014). Comparison of the Effects of Matrix Metalloproteinase Inhibitors on TNF-α Release from Activated Microglia and TNF-α Converting Enzyme Activity. Biomol Ther (Seoul) 22, 414–419. [CrossRef]
- Lee, E.-J., Woo, M.-S., Moon, P.-G., Baek, M.-C., Choi, I.-Y., Kim, W.-K., et al. (2010). α-Synuclein Activates Microglia by Inducing the Expressions of Matrix Metalloproteinases and the Subsequent Activation of Protease-Activated Receptor-1. The Journal of Immunology 185, 615–623. [CrossRef]
- Lehenkari, P. P., Kellinsalmi, M., Näpänkangas, J. P., Ylitalo, K. V., Mönkkönen, J., Rogers, M. J., et al. (2002). Further Insight into Mechanism of Action of Clodronate: Inhibition of Mitochondrial ADP/ATP Translocase by a Nonhydrolyzable, Adenine-Containing Metabolite. Mol Pharmacol 61, 1255–1262. [CrossRef]
- Lei, F., Cui, N., Zhou, C., Chodosh, J., Vavvas, D. G., and Paschalis, E. I. (2020). CSF1R inhibition by a small-molecule inhibitor is not microglia specific; affecting hematopoiesis and the function of macrophages. Proceedings of the National Academy of Sciences 117, 23336–23338. [CrossRef]
- Li, Q., Cheng, Z., Zhou, L., Darmanis, S., Neff, N. F., Okamoto, J., et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207-223.e10. [CrossRef]
- Liang, K. J., Lee, J. E., Wang, Y. D., Ma, W., Fontainhas, A. M., Fariss, R. N., et al. (2009). Regulation of Dynamic Behavior of Retinal Microglia by CX3CR1 Signaling. Investigative Ophthalmology & Visual Science 50, 4444–4451. [CrossRef]
- Lin, S.-S., Tang, Y., Illes, P., and Verkhratsky, A. (2020). The Safeguarding Microglia: Central Role for P2Y12 Receptors. Front Pharmacol 11, 627760. [CrossRef]
- Lindberg, R. L., De Groot, C. J., Montagne, L., Freitag, P., van der Valk, P., Kappos, L., et al. (2001). The expression profile of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in lesions and normal appearing white matter of multiple sclerosis. Brain 124, 1743–1753. [CrossRef]
- Littlewood, T. D., Hancock, D. C., Danielian, P. S., Parker, M. G., and Evan, G. I. (1995). A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23, 1686–1690. [CrossRef]
- Lituma, P. J., Woo, E., O’Hara, B. F., Castillo, P. E., Sibinga, N. E. S., and Nandi, S. (2021). Altered synaptic connectivity and brain function in mice lacking microglial adapter protein Iba1. Proc. Natl. Acad. Sci. U.S.A. 118, e2115539118. [CrossRef]
- Louveau, A., Nerrière-Daguin, V., Vanhove, B., Naveilhan, P., Neunlist, M., Nicot, A., et al. (2015). Targeting the CD80/CD86 costimulatory pathway with CTLA4-Ig directs microglia toward a repair phenotype and promotes axonal outgrowth: CTLA4-Ig Regulates Microglia and Axon Growth. Glia 63, 2298–2312. [CrossRef]
- Lowery, R. L., Mendes, M. S., Sanders, B. T., Murphy, A. J., Whitelaw, B. S., Lamantia, C. E., et al. (2021). Loss of P2Y12 Has Behavioral Effects in the Adult Mouse. International Journal of Molecular Sciences 22, 1868. [CrossRef]
- Maco, B., Holtmaat, A., Jorstad, A., Fua, P., and Knott, G. W. (2014). Correlative in vivo 2-photon imaging and focused ion beam scanning electron microscopy: 3D analysis of neuronal ultrastructure. Methods Cell Biol 124, 339–361. [CrossRef]
- Marino Lee, S., Hudobenko, J., McCullough, L. D., and Chauhan, A. (2021). Microglia depletion increase brain injury after acute ischemic stroke in aged mice. Exp Neurol 336, 113530. [CrossRef]
- Martin, E., El-Behi, M., Fontaine, B., and Delarasse, C. (2017). Analysis of Microglia and Monocyte-derived Macrophages from the Central Nervous System by Flow Cytometry. J Vis Exp, 55781. [CrossRef]
- Marzan, D. E., Brügger-Verdon, V., West, B. L., Liddelow, S., Samanta, J., and Salzer, J. L. (2021). Activated microglia drive demyelination via CSF1R signaling. Glia 69, 1583–1604. [CrossRef]
- Masuda, T., Amann, L., Sankowski, R., Staszewski, O., Lenz, M., D Errico, P., et al. (2020). Novel Hexb-based tools for studying microglia in the CNS. Nat Immunol 21, 802–815. [CrossRef]
- McKinsey, G. L., Lizama, C. O., Keown-Lang, A. E., Niu, A., Santander, N., Larpthaveesarp, A., et al. (2020). A new genetic strategy for targeting microglia in development and disease. Elife 9, e54590. [CrossRef]
- McQuade, A., Kang, Y. J., Hasselmann, J., Jairaman, A., Sotelo, A., Coburn, M., et al. (2020). Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer’s disease. Nat Commun 11, 5370. [CrossRef]
- Meighan, P. C., Meighan, S. E., Davis, C. J., Wright, J. W., and Harding, J. W. (2007). Effects of matrix metalloproteinase inhibition on short- and long-term plasticity of schaffer collateral/CA1 synapses. J Neurochem 102, 2085–2096. [CrossRef]
- Meighan, S. E., Meighan, P. C., Choudhury, P., Davis, C. J., Olson, M. L., Zornes, P. A., et al. (2006). Effects of extracellular matrix-degrading proteases matrix metalloproteinases 3 and 9 on spatial learning and synaptic plasticity. J Neurochem 96, 1227–1241. [CrossRef]
- Metzger, D., Clifford, J., Chiba, H., and Chambon, P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A 92, 6991–6995. [CrossRef]
- Milior, G., Lecours, C., Samson, L., Bisht, K., Poggini, S., Pagani, F., et al. (2016). Fractalkine receptor deficiency impairs microglial and neuronal responsiveness to chronic stress. Brain, Behavior, and Immunity 55, 114–125. [CrossRef]
- Mitrasinovic, O. M., Grattan, A., Robinson, C. C., Lapustea, N. B., Poon, C., Ryan, H., et al. (2005). Microglia overexpressing the macrophage colony-stimulating factor receptor are neuroprotective in a microglial-hippocampal organotypic coculture system. J Neurosci 25, 4442–4451. [CrossRef]
- Montero, M., González, B., and Zimmer, J. (2009). Immunotoxic depletion of microglia in mouse hippocampal slice cultures enhances ischemia-like neurodegeneration. Brain Res 1291, 140–152. [CrossRef]
- Moreno, S. G. (2018). Depleting Macrophages In Vivo with Clodronate-Liposomes. Methods Mol Biol 1784, 259–262. [CrossRef]
- Mrdjen, D., Pavlovic, A., Hartmann, F. J., Schreiner, B., Utz, S. G., Leung, B. P., et al. (2018). High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48, 380-395.e6. [CrossRef]
- Nahirney, P. C., and Tremblay, M.-E. (2021). Brain Ultrastructure: Putting the Pieces Together. Front Cell Dev Biol 9, 629503. [CrossRef]
- Nguyen, P. T., Dorman, L. C., Pan, S., Vainchtein, I. D., Han, R. T., Nakao-Inoue, H., et al. (2020). Microglial Remodeling of the Extracellular Matrix Promotes Synapse Plasticity. Cell 182, 388-403.e15. [CrossRef]
- Nikodemova, M., and Watters, J. J. (2012). Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J Neuroinflammation 9, 147. [CrossRef]
- Nimmerjahn, A., Kirchhoff, F., and Helmchen, F. (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. [CrossRef]
- Nonnenmacher, M., and Weber, T. (2012). Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther 19, 649–658. [CrossRef]
- Nutma, E., Ceyzériat, K., Amor, S., Tsartsalis, S., Millet, P., Owen, D. R., et al. (2021a). Cellular sources of TSPO expression in healthy and diseased brain. Eur J Nucl Med Mol Imaging 49, 146–163. [CrossRef]
- Nutma, E., Gebro, E., Marzin, M. C., van der Valk, P., Matthews, P. M., Owen, D. R., et al. (2021b). Activated microglia do not increase 18 kDa translocator protein (TSPO) expression in the multiple sclerosis brain. Glia 69, 2447–2458. [CrossRef]
- Ohsawa, K., Imai, Y., Sasaki, Y., and Kohsaka, S. (2004). Microglia/macrophage-specific protein Iba1 binds to fimbrin and enhances its actin-bundling activity. J Neurochem 88, 844–856. [CrossRef]
- O’Koren, E. G., Mathew, R., and Saban, D. R. (2016). Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci Rep 6, 20636. [CrossRef]
- O’Koren, E. G., Yu, C., Klingeborn, M., Wong, A. Y. W., Prigge, C. L., Mathew, R., et al. (2019). Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity 50, 723-737.e7. [CrossRef]
- Olah, M., Menon, V., Habib, N., Taga, M. F., Ma, Y., Yung, C. J., et al. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat Commun 11, 6129. [CrossRef]
- Olmos-Alonso, A., Schetters, S. T. T., Sri, S., Askew, K., Mancuso, R., Vargas-Caballero, M., et al. (2016). Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain 139, 891–907. [CrossRef]
- Opperman, K. S., Vandyke, K., Clark, K. C., Coulter, E. A., Hewett, D. R., Mrozik, K. M., et al. (2019). Clodronate-Liposome Mediated Macrophage Depletion Abrogates Multiple Myeloma Tumor Establishment In Vivo. Neoplasia 21, 777–787. [CrossRef]
- Orban, P. C., Chui, D., and Marth, J. D. (1992). Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A 89, 6861–6865. [CrossRef]
- Pagani, F., Paolicelli, R. C., Murana, E., Cortese, B., Di Angelantonio, S., Zurolo, E., et al. (2015). Defective microglial development in the hippocampus of Cx3cr1 deficient mice. Front Cell Neurosci 9, 111. [CrossRef]
- Paolicelli, R. C., Sierra, A., Stevens, B., Tremblay, M.-E., Aguzzi, A., Ajami, B., et al. (2022). Microglia states and nomenclature: A field at its crossroads. Neuron 110, 3458–3483. [CrossRef]
- Parkhurst, C. N., Yang, G., Ninan, I., Savas, J. N., Yates, J. R., Lafaille, J. J., et al. (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609. [CrossRef]
- Patnaik, S., Rai, M., Jalali, S., Agarwal, K., Badakere, A., Puppala, L., et al. (2021). An interplay of microglia and matrix metalloproteinase MMP9 under hypoxic stress regulates the opticin expression in retina. Sci Rep 11, 7444. [CrossRef]
- Peng, J., Wang, K., Xiang, W., Li, Y., Hao, Y., and Guan, Y. (2019). Rosiglitazone polarizes microglia and protects against pilocarpine-induced status epilepticus. CNS Neurosci Ther 25, 1363–1372. [CrossRef]
- Penninger, J. M., Irie-Sasaki, J., Sasaki, T., and Oliveira-dos-Santos, A. J. (2001). CD45: new jobs for an old acquaintance. Nat Immunol 2, 389–396. [CrossRef]
- Perego, C., Fumagalli, S., and De Simoni, M.-G. (2011). Temporal pattern of expression and colocalization of microglia/macrophage phenotype markers following brain ischemic injury in mice. Journal of Neuroinflammation 8, 174. [CrossRef]
- Pons, V., Lévesque, P., Plante, M.-M., and Rivest, S. (2021). Conditional genetic deletion of CSF1 receptor in microglia ameliorates the physiopathology of Alzheimer’s disease. Alzheimers Res Ther 13, 8. [CrossRef]
- Popova, G., Soliman, S. S., Kim, C. N., Keefe, M. G., Hennick, K. M., Jain, S., et al. (2021). Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell 28, 2153-2166.e6. [CrossRef]
- Prater, K. E., Aloi, M. S., Pathan, J. L., Winston, C. N., Chernoff, R. A., Davidson, S., et al. (2021). A Subpopulation of Microglia Generated in the Adult Mouse Brain Originates from Prominin-1-Expressing Progenitors. J Neurosci 41, 7942–7953. [CrossRef]
- Quan, Y., Möller, T., and Weinstein, J. R. (2009). Regulation of Fcgamma receptors and immunoglobulin G-mediated phagocytosis in mouse microglia. Neurosci Lett 464, 29–33. [CrossRef]
- Rangaraju, S., Raza, S. A., Li, N. X., Betarbet, R., Dammer, E. B., Duong, D., et al. (2018). Differential Phagocytic Properties of CD45low Microglia and CD45high Brain Mononuclear Phagocytes—Activation and Age-Related Effects. Front Immunol 9, 405. [CrossRef]
- Ransohoff, R. M., and El Khoury, J. (2015). Microglia in Health and Disease. Cold Spring Harb Perspect Biol 8, a020560. [CrossRef]
- Reifschneider, A., Robinson, S., van Lengerich, B., Gnörich, J., Logan, T., Heindl, S., et al. (2022). Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J 41, e109108. [CrossRef]
- Rice, R. A., Pham, J., Lee, R. J., Najafi, A. R., West, B. L., and Green, K. N. (2017). Microglial repopulation resolves inflammation and promotes brain recovery after injury. Glia 65, 931–944. [CrossRef]
- Rojo, R., Raper, A., Ozdemir, D. D., Lefevre, L., Grabert, K., Wollscheid-Lengeling, E., et al. (2019). Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nat Commun 10, 3215. [CrossRef]
- Rooijen, N. V., and Sanders, A. (1994). Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. Journal of Immunological Methods 174, 83–93. [CrossRef]
- Rosell, A., Ortega-Aznar, A., Alvarez-Sabín, J., Fernández-Cadenas, I., Ribó, M., Molina, C. A., et al. (2006). Increased Brain Expression of Matrix Metalloproteinase-9 After Ischemic and Hemorrhagic Human Stroke. Stroke 37, 1399–1406. [CrossRef]
- Rosito, M., Sanchini, C., Gosti, G., Moreno, M., De Panfilis, S., Giubettini, M., et al. (2023). Microglia reactivity entails microtubule remodeling from acentrosomal to centrosomal arrays. Cell Reports 42, 112104. [CrossRef]
- Ruan, C., Sun, L., Kroshilina, A., Beckers, L., De Jager, P., Bradshaw, E. M., et al. (2020). A novel Tmem119-tdTomato reporter mouse model for studying microglia in the central nervous system. Brain Behav Immun 83, 180–191. [CrossRef]
- Rubino, S. J., Mayo, L., Wimmer, I., Siedler, V., Brunner, F., Hametner, S., et al. (2018). Acute microglia ablation induces neurodegeneration in the somatosensory system. Nat Commun 9, 4578. [CrossRef]
- Rustenhoven, J., Park, T. I.-H., Schweder, P., Scotter, J., Correia, J., Smith, A. M., et al. (2016). Isolation of highly enriched primary human microglia for functional studies. Sci Rep 6, 19371. [CrossRef]
- Salman, H., Shuai, X., Nguyen-Lefebvre, A. T., Giri, B., Ren, M., Rauchman, M., et al. (2018). SALL1 expression in acute myeloid leukemia. Oncotarget 9, 7442–7452. [CrossRef]
- Sanchez Mejia, R. O., Ona, V. O., Li, M., and Friedlander, R. M. (2001). Minocycline reduces traumatic brain injury-mediated caspase-1 activation, tissue damage, and neurological dysfunction. Neurosurgery 48, 1393–1399; discussion 1399-1401. [CrossRef]
- Sasaki, Y., Ohsawa, K., Kanazawa, H., Kohsaka, S., and Imai, Y. (2001). Iba1 is an actin-cross-linking protein in macrophages/microglia. Biochem Biophys Res Commun 286, 292–297. [CrossRef]
- Satoh, J., Kino, Y., Asahina, N., Takitani, M., Miyoshi, J., Ishida, T., et al. (2016a). TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49. [CrossRef]
- Satoh, J., Kino, Y., Asahina, N., Takitani, M., Miyoshi, J., Ishida, T., et al. (2016b). TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49. [CrossRef]
- Savage, J. C., St-Pierre, M.-K., Carrier, M., El Hajj, H., Novak, S. W., Sanchez, M. G., et al. (2020a). Microglial physiological properties and interactions with synapses are altered at presymptomatic stages in a mouse model of Huntington’s disease pathology. J Neuroinflammation 17, 98. [CrossRef]
- Savage, J. C., St-Pierre, M.-K., Carrier, M., El Hajj, H., Novak, S. W., Sanchez, M. G., et al. (2020b). Microglial physiological properties and interactions with synapses are altered at presymptomatic stages in a mouse model of Huntington’s disease pathology. J Neuroinflammation 17, 98. [CrossRef]
- Savchenko, V. L., Nikonenko, I. R., Skibo, G. G., and McKanna, J. A. (1997). Distribution of microglia and astrocytes in different regions of the normal adult rat brain. Neurophysiology 29, 343–351. [CrossRef]
- Saxena, S., Kruys, V., Vamecq, J., and Maze, M. (2021). The Role of Microglia in Perioperative Neuroinflammation and Neurocognitive Disorders. Frontiers in Aging Neuroscience 13. Available at:.
- Schmid, M. C., Khan, S. Q., Kaneda, M. M., Pathria, P., Shepard, R., Louis, T. L., et al. (2018). Integrin CD11b activation drives anti-tumor innate immunity. Nat Commun 9, 5379. [CrossRef]
- Scholz, R., Sobotka, M., Caramoy, A., Stempfl, T., Moehle, C., and Langmann, T. (2015). Minocycline counter-regulates pro-inflammatory microglia responses in the retina and protects from degeneration. J Neuroinflammation 12, 209. [CrossRef]
- Schulz, C., Gomez Perdiguero, E., Chorro, L., Szabo-Rogers, H., Cagnard, N., Kierdorf, K., et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. [CrossRef]
- Scott, E. P., Breyak, E., Nishinakamura, R., and Nakagawa, Y. (2022). The zinc finger transcription factor Sall1 is required for the early developmental transition of microglia in mouse embryos. Glia 70, 1720–1733. [CrossRef]
- Scott-Hewitt, N., Perrucci, F., Morini, R., Erreni, M., Mahoney, M., Witkowska, A., et al. (2020). Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia. The EMBO Journal 39, e105380. [CrossRef]
- Seelaar, H., and Van Swieten, J. C. (2021). In vivo PET imaging of neuroinflammation in familial frontotemporal dementia. J Neurol Neurosurg Psychiatry 92, 231. [CrossRef]
- Segal, B. M., and Giger, R. J. (2016). Stable biomarker for plastic microglia. Proc Natl Acad Sci U S A 113, 3130–3132. [CrossRef]
- Serganova, I., and Blasberg, R. G. (2019). Molecular Imaging with Reporter Genes: Has Its Promise Been Delivered? J Nucl Med 60, 1665–1681. [CrossRef]
- Shah, S., Wong, L. M., Ellis, K., Bodnar, B., Saribas, S., Ting, J., et al. (2022). Microglia-Specific Promoter Activities of HEXB Gene. Front Cell Neurosci 16, 808598. [CrossRef]
- Shah, V. B., Huang, Y., Keshwara, R., Ozment-Skelton, T., Williams, D. L., and Keshvara, L. (2008). β-Glucan Activates Microglia without Inducing Cytokine Production in Dectin-1-Dependent Manner. The Journal of Immunology 180, 2777–2785. [CrossRef]
- Shahidehpour, R. K., Higdon, R. E., Crawford, N. G., Neltner, J. H., Ighodaro, E. T., Patel, E., et al. (2021). Dystrophic microglia are associated with neurodegenerative disease and not healthy aging in the human brain. Neurobiol Aging 99, 19–27. [CrossRef]
- Shapiro, L. A., Perez, Z. D., Foresti, M. L., Arisi, G. M., and Ribak, C. E. (2009). Morphological and ultrastructural features of Iba1-immunolabeled microglial cells in the hippocampal dentate gyrus. Brain Res 1266, 29–36. [CrossRef]
- Sharma, K., Bisht, K., and Eyo, U. B. (2021). A Comparative Biology of Microglia Across Species. Front Cell Dev Biol 9, 652748. [CrossRef]
- Sheng, J., Ruedl, C., and Karjalainen, K. (2015). Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity 43, 382–393. [CrossRef]
- Shi, F.-J., Xie, H., Zhang, C.-Y., Qin, H.-F., Zeng, X.-W., Lou, H., et al. (2021). Is Iba-1 protein expression a sensitive marker for microglia activation in experimental diabetic retinopathy? Int J Ophthalmol 14, 200–208. [CrossRef]
- Shi, L., Rocha, M., Zhang, W., Jiang, M., Li, S., Ye, Q., et al. (2020). Genome-wide transcriptomic analysis of microglia reveals impaired responses in aged mice after cerebral ischemia. J Cereb Blood Flow Metab 40, S49–S66. [CrossRef]
- Sierksma, A., Lu, A., Mancuso, R., Fattorelli, N., Thrupp, N., Salta, E., et al. (2020). Novel Alzheimer risk genes determine the microglia response to amyloid-β but not to TAU pathology. EMBO Mol Med 12, e10606. [CrossRef]
- Sierra, A., Paolicelli, R. C., and Kettenmann, H. (2019). Cien Años de Microglía: Milestones in a Century of Microglial Research. Trends Neurosci 42, 778–792. [CrossRef]
- Simpson, D., Gharehgazlou, A., Da Silva, T., Labrie-Cleary, C., Wilson, A. A., Meyer, J. H., et al. (2022). In vivo imaging translocator protein (TSPO) in autism spectrum disorder. Neuropsychopharmacology 47, 1421–1427. [CrossRef]
- Sims, R., van der Lee, S. J., Naj, A. C., Bellenguez, C., Badarinarayan, N., Jakobsdottir, J., et al. (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet 49, 1373–1384. [CrossRef]
- Sipe, G. O., Lowery, Tremblay, M.-È., Kelly, E. A., Lamantia, C. E., and Majewska, A. K. (2016). Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun 7, 10905. [CrossRef]
- Šišková, Z., and Tremblay, M.-È. (2013). Microglia and synapse: interactions in health and neurodegeneration. Neural Plast 2013, 425845. [CrossRef]
- Smith, J. A., Das, A., Ray, S. K., and Banik, N. L. (2012). Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res Bull 87, 10–20. [CrossRef]
- Sobue, A., Komine, O., Hara, Y., Endo, F., Mizoguchi, H., Watanabe, S., et al. (2021). Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol Commun 9, 1. [CrossRef]
- Song, L., Lee, C., and Schindler, C. (2011). Deletion of the murine scavenger receptor CD68[S]. Journal of Lipid Research 52, 1542–1550. [CrossRef]
- Song, W. M., and Colonna, M. (2018). The identity and function of microglia in neurodegeneration. Nat Immunol 19, 1048–1058. [CrossRef]
- Sousa, C., Golebiewska, A., Poovathingal, S. K., Kaoma, T., Pires-Afonso, Y., Martina, S., et al. (2018). Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep 19, e46171. [CrossRef]
- Spangenberg, E., Severson, P. L., Hohsfield, L. A., Crapser, J., Zhang, J., Burton, E. A., et al. (2019). Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat Commun 10, 3758. [CrossRef]
- Speicher, A. M., Wiendl, H., Meuth, S. G., and Pawlowski, M. (2019). Generating microglia from human pluripotent stem cells: novel in vitro models for the study of neurodegeneration. Molecular Neurodegeneration 14, 46. [CrossRef]
- Stojiljkovic, M. R., Schmeer, C., and Witte, O. W. (2022). Pharmacological Depletion of Microglia Leads to a Dose-Dependent Reduction in Inflammation and Senescence in the Aged Murine Brain. Neuroscience 488, 1–9. [CrossRef]
- St-Pierre, M.-K., Carrier, M., González Ibáñez, F., Šimončičová, E., Wallman, M.-J., Vallières, L., et al. (2022a). Ultrastructural characterization of dark microglia during aging in a mouse model of Alzheimer’s disease pathology and in human post-mortem brain samples. Journal of Neuroinflammation 19, 235. [CrossRef]
- St-Pierre, M.-K., Carrier, M., Lau, V., and Tremblay, M.-È. (2022b). Investigating Microglial Ultrastructural Alterations and Intimate Relationships with Neuronal Stress, Dystrophy, and Degeneration in Mouse Models of Alzheimer’s Disease. Methods Mol Biol 2515, 29–58. [CrossRef]
- St-Pierre, M.-K., Šimončičová, E., Bögi, E., and Tremblay, M.-È. (2020). Shedding Light on the Dark Side of the Microglia. ASN Neuro 12, 1759091420925335. [CrossRef]
- Strahan, J. A., Walker, W. H., Montgomery, T. R., and Forger, N. G. (2017). Minocycline Causes Widespread Cell Death and Increases Microglial Labeling in the Neonatal Mouse Brain. Dev Neurobiol 77, 753–766. [CrossRef]
- Stratoulias, V., Venero, J. L., Tremblay, M.-È., and Joseph, B. (2019). Microglial subtypes: diversity within the microglial community. EMBO J 38, e101997. [CrossRef]
- Streit, W. J., Braak, H., Xue, Q.-S., and Bechmann, I. (2009). Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer’s disease. Acta Neuropathol 118, 475–485. [CrossRef]
- Streit, W. J., Khoshbouei, H., and Bechmann, I. (2020). Dystrophic microglia in late-onset Alzheimer’s disease. Glia 68, 845–854. [CrossRef]
- Streit, W. J., Sammons, N. W., Kuhns, A. J., and Sparks, D. L. (2004). Dystrophic microglia in the aging human brain. Glia 45, 208–212. [CrossRef]
- Suzuki, Y., Shirai, M., Asada, K., Yasui, H., Karayama, M., Hozumi, H., et al. (2018). Macrophage mannose receptor, CD206, predict prognosis in patients with pulmonary tuberculosis. Sci Rep 8, 13129. [CrossRef]
- Swanson, M. E. V., Mrkela, M., Murray, H. C., Cao, M. C., Turner, C., Curtis, M. A., et al. (2023). Microglial CD68 and L-ferritin upregulation in response to phosphorylated-TDP-43 pathology in the amyotrophic lateral sclerosis brain. Acta Neuropathologica Communications 11, 69. [CrossRef]
- Swanson, M. E. V., Scotter, E. L., Smyth, L. C. D., Murray, H. C., Ryan, B., Turner, C., et al. (2020). Identification of a dysfunctional microglial population in human Alzheimer’s disease cortex using novel single-cell histology image analysis. Acta Neuropathologica Communications 8, 170. [CrossRef]
- Szepesi, Z., Hosy, E., Ruszczycki, B., Bijata, M., Pyskaty, M., Bikbaev, A., et al. (2014). Synaptically released matrix metalloproteinase activity in control of structural plasticity and the cell surface distribution of GluA1-AMPA receptors. PLoS One 9, e98274. [CrossRef]
- Szepesi, Z., Manouchehrian, O., Bachiller, S., and Deierborg, T. (2018a). Bidirectional Microglia–Neuron Communication in Health and Disease. Frontiers in Cellular Neuroscience 12. Available at:.
- Szepesi, Z., Manouchehrian, O., Bachiller, S., and Deierborg, T. (2018b). Bidirectional Microglia-Neuron Communication in Health and Disease. Front Cell Neurosci 12, 323. [CrossRef]
- Takagi, S., Murayama, S., Torii, K., Takemura-Morita, S., Kurganov, E., Nagaoka, S., et al. (2020). Depletion of microglia and macrophages with clodronate liposomes attenuates zymosan-induced Fos expression and hypothermia in the adult mouse. Journal of Neuroimmunology 344, 577244. [CrossRef]
- In Tan, S; discoverer of phagocytosis: Y., and Dee, M. K. (2009). Elie Metchnikoff (1845-1916); Tan, S. Y., and Dee, M. K. (2009). Elie Metchnikoff (1845-1916): discoverer of phagocytosis. Singapore Med J 50, 456–457.
- Tanaka, S., Ohgidani, M., Hata, N., Inamine, S., Sagata, N., Shirouzu, N., et al. (2021). CD206 Expression in Induced Microglia-Like Cells From Peripheral Blood as a Surrogate Biomarker for the Specific Immune Microenvironment of Neurosurgical Diseases Including Glioma. Front Immunol 12, 670131. [CrossRef]
- Tang, P. W., Chua, P. S., Chong, S. K., Mohamad, M. S., Choon, Y. W., Deris, S., et al. (2015). A Review of Gene Knockout Strategies for Microbial Cells. Recent Pat Biotechnol 9, 176–197. [CrossRef]
- Tang, Z., Gan, Y., Liu, Q., Yin, J.-X., Liu, Q., Shi, J., et al. (2014). CX3CR1 deficiency suppresses activation and neurotoxicity of microglia/macrophage in experimental ischemic stroke. Journal of Neuroinflammation 11, 26. [CrossRef]
- Tay, T. L., Mai, D., Dautzenberg, J., Fernández-Klett, F., Lin, G., Sagar, et al. (2017). A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci 20, 793–803. [CrossRef]
- Theocharis, A. D., Skandalis, S. S., Gialeli, C., and Karamanos, N. K. (2016). Extracellular matrix structure. Adv Drug Deliv Rev 97, 4–27. [CrossRef]
- Thomas, M., and Le, W. D. (2004). Minocycline: neuroprotective mechanisms in Parkinson’s disease. Curr Pharm Des 10, 679–686. [CrossRef]
- Tremblay, M.-È., Lowery, R. L., and Majewska, A. K. (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. [CrossRef]
- Tremblay, M.-È., and Majewska, A. K. (2011). A role for microglia in synaptic plasticity? Commun Integr Biol 4, 220–222. [CrossRef]
- Tremblay, M.-È., and Majewska, A. K. (2019). Ultrastructural Analyses of Microglial Interactions with Synapses. Methods Mol Biol 2034, 83–95. [CrossRef]
- Unger, M. S., Schernthaner, P., Marschallinger, J., Mrowetz, H., and Aigner, L. (2018). Microglia prevent peripheral immune cell invasion and promote an anti-inflammatory environment in the brain of APP-PS1 transgenic mice. J Neuroinflammation 15, 274. [CrossRef]
- Vainio, S. K., Dickens, A. M., Matilainen, M., López-Picón, F. R., Aarnio, R., Eskola, O., et al. (2022). Dimethyl fumarate decreases short-term but not long-term inflammation in a focal EAE model of neuroinflammation. EJNMMI Res 12, 6. [CrossRef]
- Van Hove, H., Martens, L., Scheyltjens, I., De Vlaminck, K., Pombo Antunes, A. R., De Prijck, S., et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 22, 1021–1035. [CrossRef]
- van Wageningen, T. A., Vlaar, E., Kooij, G., Jongenelen, C. A. M., Geurts, J. J. G., and van Dam, A.-M. (2019). Regulation of microglial TMEM119 and P2RY12 immunoreactivity in multiple sclerosis white and grey matter lesions is dependent on their inflammatory environment. Acta Neuropathol Commun 7, 206. [CrossRef]
- VanderZwaag, J., Halvorson, T., Dolhan, K., Šimončičová, E., Ben-Azu, B., and Tremblay, M.-È. (2023). The Missing Piece? A Case for Microglia’s Prominent Role in the Therapeutic Action of Anesthetics, Ketamine, and Psychedelics. Neurochem Res 48, 1129–1166. [CrossRef]
- von Kügelgen, I. (2019). Pharmacology of P2Y receptors. Brain Res Bull 151, 12–24. [CrossRef]
- Waller, R., Baxter, L., Fillingham, D. J., Coelho, S., Pozo, J. M., Mozumder, M., et al. (2019). Iba-1-/CD68+ microglia are a prominent feature of age-associated deep subcortical white matter lesions. PLoS One 14, e0210888. [CrossRef]
- Wang, C., and Sun, H. (2019). [Progress in gene knockout mice]. Sheng Wu Gong Cheng Xue Bao 35, 784–794. [CrossRef]
- Wang, Q., Davis, P. B., Qi, X., Chen, S. G., Gurney, M. E., Perry, G., et al. (2021). Gut–microbiota–microglia–brain interactions in Alzheimer’s disease: knowledge-based, multi-dimensional characterization. Alzheimer’s Research & Therapy 13, 177. [CrossRef]
- Wang, S., Sudan, R., Peng, V., Zhou, Y., Du, S., Yuede, C. M., et al. (2022). TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell 185, 4153-4169.e19. [CrossRef]
- Wang, Y., Cella, M., Mallinson, K., Ulrich, J. D., Young, K. L., Robinette, M. L., et al. (2015). TREM2 Lipid Sensing Sustains the Microglial Response in an Alzheimer’s Disease Model. Cell 160, 1061–1071. [CrossRef]
- Wang, Y., Szretter, K. J., Vermi, W., Gilfillan, S., Rossini, C., Cella, M., et al. (2012). IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 13, 753–760. [CrossRef]
- Weisser, S. B., van Rooijen, N., and Sly, L. M. (2012). Depletion and reconstitution of macrophages in mice. J Vis Exp, 4105. [CrossRef]
- Wieghofer, P., Hagemeyer, N., Sankowski, R., Schlecht, A., Staszewski, O., Amann, L., et al. (2021). Mapping the origin and fate of myeloid cells in distinct compartments of the eye by single-cell profiling. EMBO J 40, e105123. [CrossRef]
- Wieghofer, P., Knobeloch, K.-P., and Prinz, M. (2015). Genetic targeting of microglia. Glia 63, 1–22. [CrossRef]
- Wolf, Y., Yona, S., Kim, K.-W., and Jung, S. (2013). Microglia, seen from the CX3CR1 angle. Front Cell Neurosci 7, 26. [CrossRef]
- Woo, M.-S., Park, J.-S., Choi, I.-Y., Kim, W.-K., and Kim, H.-S. (2008). Inhibition of MMP-3 or -9 suppresses lipopolysaccharide-induced expression of proinflammatory cytokines and iNOS in microglia. J Neurochem 106, 770–780. [CrossRef]
- Wu, W., Li, Y., Wei, Y., Bosco, D. B., Xie, M., Zhao, M.-G., et al. (2020). Microglial depletion aggravates the severity of acute and chronic seizures in mice. Brain, Behavior, and Immunity 89, 245–255. [CrossRef]
- Wu, X., Saito, T., Saido, T. C., Barron, A. M., and Ruedl, C. (2021). Microglia and CD206+ border-associated mouse macrophages maintain their embryonic origin during Alzheimer’s disease. Elife 10, e71879. [CrossRef]
- Xia, Y. 2290; Xia, Y., Vetvicka, V., Yan, J., Hanikýrová, M., Mayadas, T., and Ross, G. D. (1999). The beta-glucan-binding lectin site of mouse CR3 (CD11b/CD18) and its function in generating a primed state of the receptor that mediates cytotoxic activation in response to iC3b-opsonized target cells. J Immunol 162, 2281–2290. [Google Scholar]
- Xu, Z.-J., Gu, Y., Wang, C.-Z., Jin, Y., Wen, X.-M., Ma, J.-C., et al. (2020). The M2 macrophage marker CD206: a novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 9, 1683347. [CrossRef]
- Yaghmoor, F., Noorsaeed, A., Alsaggaf, S., Aljohani, W., Scholtzova, H., Boutajangout, A., et al. (2014). The Role of TREM2 in Alzheimer’s Disease and Other Neurological Disorders. J Alzheimers Dis Parkinsonism 4, 160. [CrossRef]
- Yang, B., Wang, F., and Zheng, G. (2021). Transmembrane protein TMEM119 facilitates the stemness of breast cancer cells by activating Wnt/β-catenin pathway. Bioengineered 12, 4856–4867. [CrossRef]
- Yankam Njiwa, J., Costes, N., Bouillot, C., Bouvard, S., Fieux, S., Becker, G., et al. (2017). Quantitative longitudinal imaging of activated microglia as a marker of inflammation in the pilocarpine rat model of epilepsy using [11C]-( R)-PK11195 PET and MRI. J Cereb Blood Flow Metab 37, 1251–1263. [CrossRef]
- Yao, Y., Echeverry, S., Shi, X. Q., Yang, M., Yang, Q. Z., Wang, G. Y. F., et al. (2016). Dynamics of spinal microglia repopulation following an acute depletion. Sci Rep 6, 22839. [CrossRef]
- Yeo, H.-G., Hong, J. J., Lee, Y., Yi, K. S., Jeon, C.-Y., Park, J., et al. (2019). Increased CD68/TGFβ Co-expressing Microglia/ Macrophages after Transient Middle Cerebral Artery Occlusion in Rhesus Monkeys. Exp Neurobiol 28, 458–473. [CrossRef]
- Yi, S. Y., Barnett, B. R., Torres-Velázquez, M., Zhang, Y., Hurley, S. A., Rowley, P. A., et al. (2019). Detecting Microglial Density With Quantitative Multi-Compartment Diffusion MRI. Frontiers in Neuroscience 13. Available at:.
- Yona, S., Kim, K.-W., Wolf, Y., Mildner, A., Varol, D., Breker, M., et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [CrossRef]
- Yrjänheikki, J. 1577; Yrjänheikki, J., Keinänen, R., Pellikka, M., Hökfelt, T., and Koistinaho, J. (1998). Tetracyclines inhibit microglial activation and are neuroprotective in global brain ischemia. Proc Natl Acad Sci U S A 95, 15769–15774. [Google Scholar]
- Yusuying, S., Yusuyin, S., and Cheng, X. (2022). Translocator Protein Regulate Polarization Phenotype Transformation of Microglia after Cerebral Ischemia–reperfusion Injury. Neuroscience 480, 203–216. [CrossRef]
- Zhan, L., Fan, L., Kodama, L., Sohn, P. D., Wong, M. Y., Mousa, G. A., et al. (2020). A MAC2-positive progenitor-like microglial population is resistant to CSF1R inhibition in adult mouse brain. Elife 9, e51796. [CrossRef]
- Zhang, Y., Zhao, L., Wang, X., Ma, W., Lazere, A., Qian, H.-H., et al. (2018). Repopulating retinal microglia restore endogenous organization and function under CX3CL1-CX3CR1 regulation. Sci Adv 4, eaap8492. [CrossRef]
- Zhao, J.-F., Ren, T., Li, X.-Y., Guo, T.-L., Liu, C.-H., and Wang, X. (2022). Research Progress on the Role of Microglia Membrane Proteins or Receptors in Neuroinflammation and Degeneration. Front Cell Neurosci 16, 831977. [CrossRef]
- Zhong, L., Chen, X.-F., Wang, T., Wang, Z., Liao, C., Wang, Z., et al. (2017). Soluble TREM2 induces inflammatory responses and enhances microglial survival. J Exp Med 214, 597–607. [CrossRef]
- Zhou, X., Ji, B., Seki, C., Nagai, Y., Minamimoto, T., Fujinaga, M., et al. (2021). PET imaging of colony-stimulating factor 1 receptor: A head-to-head comparison of a novel radioligand, 11C-GW2580, and 11C-CPPC, in mouse models of acute and chronic neuroinflammation and a rhesus monkey. J Cereb Blood Flow Metab 41, 2410–2422. [CrossRef]
- Zuccaro, M. V., Xu, J., Mitchell, C., Marin, D., Zimmerman, R., Rana, B., et al. (2020). Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell 183, 1650-1664.e15. [CrossRef]
| Protein | Location | Main function | Other cell types expressing the protein | References |
|---|---|---|---|---|
| CD11b | Membrane protein | Adhesion and inflammatory processes of the complement system | Monocytes, neutrophils, NK cells, granulocytes, macrophages, DCs | (Fischer and Reichmann, 2001; Greter et al., 2015; Martin et al., 2017; Agalave et al., 2020) |
| CD86 | Membrane protein | T cell activation | DCs, Langerhans cells, macrophages, and B cells | (Hellström Erkenstam et al., 2016; Bai et al., 2017; Peng et al., 2019; Yusuying et al., 2022) |
| CD45 | Membrane protein | Protein tyrosine phosphatase, T-cell activation | Leukocyte common antigen | (Martin et al., 2017; Sousa et al., 2018) |
| CD68 | Membrane protein | Innate inflammatory response, possible role in phagocytosis; regulation of antigen processing | Monocytic phagocytes, osteoclasts, Kupffer cells | (Rice et al., 2017; Yeo et al., 2019; Swanson et al., 2023) |
| CD206 | Membrane protein | Endocytosis and phagocytosis | Astrocytes, macrophages, DCs, and endothelial cells | (Hellström Erkenstam et al., 2016; Wu et al., 2021, 206; Yusuying et al., 2022) |
| TMEM119 | Membrane protein | Proliferation, migration and genetic stability | DCs, fibroblasts, peritubular cells | (Bennett et al., 2016; Satoh et al., 2016a; Ibanez et al., 2019; Kaiser and Feng, 2019) |
| P2RY12 | Membrane protein | Detects ATP-derived particles; motility | Vascular smooth muscle cells, brown adipocytes, cholangiocyte primary cilia, osteoblasts, osteoclasts, DCs, lymphocytes. | (Avignone et al., 2008; van Wageningen et al., 2019; Bisht et al., 2021) |
| CX3CR1 | Membrane protein | Microglia adhesion and migration; neural communication | Monocytes, macrophages, T helper cells, CD8+ effector/memory T cells, NK cells, γδ T cells, DCs | (Liang et al., 2009; Tang et al., 2014; Pagani et al., 2015; González-Prieto et al., 2021) |
| CLEC7a | Membrane protein | Glucan receptor; immune response via reactive oxygen species | Monocytes, macrophages, DCs, neutrophils, B cells | (Bisht et al., 2016; Shi et al., 2020; Wang et al., 2022) |
| TREM2 | Membrane protein | Mediates transcription factors; synaptic pruning | Macrophages, DCs | (Bisht et al., 2016; Krasemann et al., 2017; Filipello et al., 2018; Reifschneider et al., 2022; Gao et al., 2023) |
| IBA1 | Intracellular protein | Microglial cytoskeleton reorganization | Macrophages, monocytes, Hofbauer cells, Kupffer cells, Langerhans cells | (Bisht et al., 2016; Ibanez et al., 2019; Shi et al., 2021) |
| SALL1 | Intracellular protein | Transcriptional regulator in homeostasis | Stem cells, oligodendrocytes, hepatocytes, astrocytes | (Buttgereit et al., 2016; Salman et al., 2018; Scott et al., 2022) |
| HEXB | Intracellular protein | Lysosomal processes, ganglioside degradation | Adipose progenitor cells, fibroblasts, thyroid glandular cells | (Masuda et al., 2020; Sierksma et al., 2020; Jia et al., 2021) |
| MMP-9 and MMP-3 | Extracellular proteins | Cytokine activation in inflammatory processes | Neutrophils, macrophages, and fibroblasts | (Woo et al., 2008; Lee et al., 2014; Kim et al., 2021a) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





