1. Introduction
Cancer has plagued multi-cellular organisms since their conception. Recently, I wrote a paper about how many cancer patients may have one or more mutations that are ubiquitous throughout their tumor(s) [
1,
2,
3]. The rest may at least have a small set of subclonal mutations that together cover all sequenced regions of their tumors. These mutations could be targeted by an oncolytic vector with the broadest tropism possible that only replicates and becomes hyper-virulent after detecting said mutations. I called this strategy, “Oncolytic Vector Efficient Replication Contingent on Omnipresent Mutation Engagement” (OVERCOME).
To identify these mutations, multiregion, multi-sample sequencing should be employed for each patient. However, tumors in certain anatomical regions are not easy to biopsy - especially in a multiregional fashion. Such regions include the brain, the spinal cord, the liver, and the lungs.
Here, I explore another way of acquiring multiregion biopsies from tumors that are hard to reach via traditional means.
2. Mechanisms
2.1. Macrophage phagocytosis
Bioengineered macrophages could be used for this purpose. They could be loaded with magnetic nanoparticles (MNPs) and steered into the heart or lung tumors using an MRI machine [
4]. Perhaps the macrophages should be induced via small molecule to chemorepel [
5] each other once they have reached the target site or sites - in order to spread out more evenly throughout the tumor(s). A gene circuit possibly involving ARHGEF[
6] or Cdccould be employed as well - to allow for vigorous, random movement within the tumor in addition to their chemorepulsion from each other.
In either case, once there, they could be induced via small molecule to express a chimeric antigen receptor for phagocytosis (CAR-P) [
7]. Alternatively, they can be heated via an alternating magnetic field to induce gene expression of the CAR-P [
8]. The CAR-P would target a ubiquitously expressed cell surface protein [
9,
10]. Rapid, inducible depletion of SIRPα [
11] in the macrophages via TEVp-mediated degree exposure [
12], for example, might also be of use.
Realistically, multiple CAR-Ps should probably be induced via small molecule or heating in order to target a variety of ubiquitously expressed cell surface proteins and thereby help ensure phagocytosis of cancer cells throughout the tumor(s), as some cancer cells may have lost or suppressed the expression of one or more ubiquitous cell surface proteins.
Inhibiting maturation of and lysosomal fusion with the specific phagosome carrying the target cell could be achieved in a variety of ways [
13]. After giving the macrophages some time to collect a target - they would be drawn via magnetism or chemotaxis to an extraction point in the body. Perhaps they can be magnetically drawn to the peritoneal cavity [
14] and withdrawn via needle.
Importantly, it was shown that whole cancer cells can be engulfed via this CAR-P method, as opposed to trogocytosis. However, trogocytosis was still more common. Thus, more work may be necessary before this strategy can be employed.
Perhaps if necessary, the CAR-P macrophages employed in this setting could be programmed so that trogocytosis is not possible [
15]. They could also potentially be bioengineered so that frustrated phagocytosis [
16] both leads to the chemoattraction of other CAR-P macrophages and ensures homotypic fusion once in close proximity to create multinucleated giant cells that can engulf entire, large cancer cells [
17].
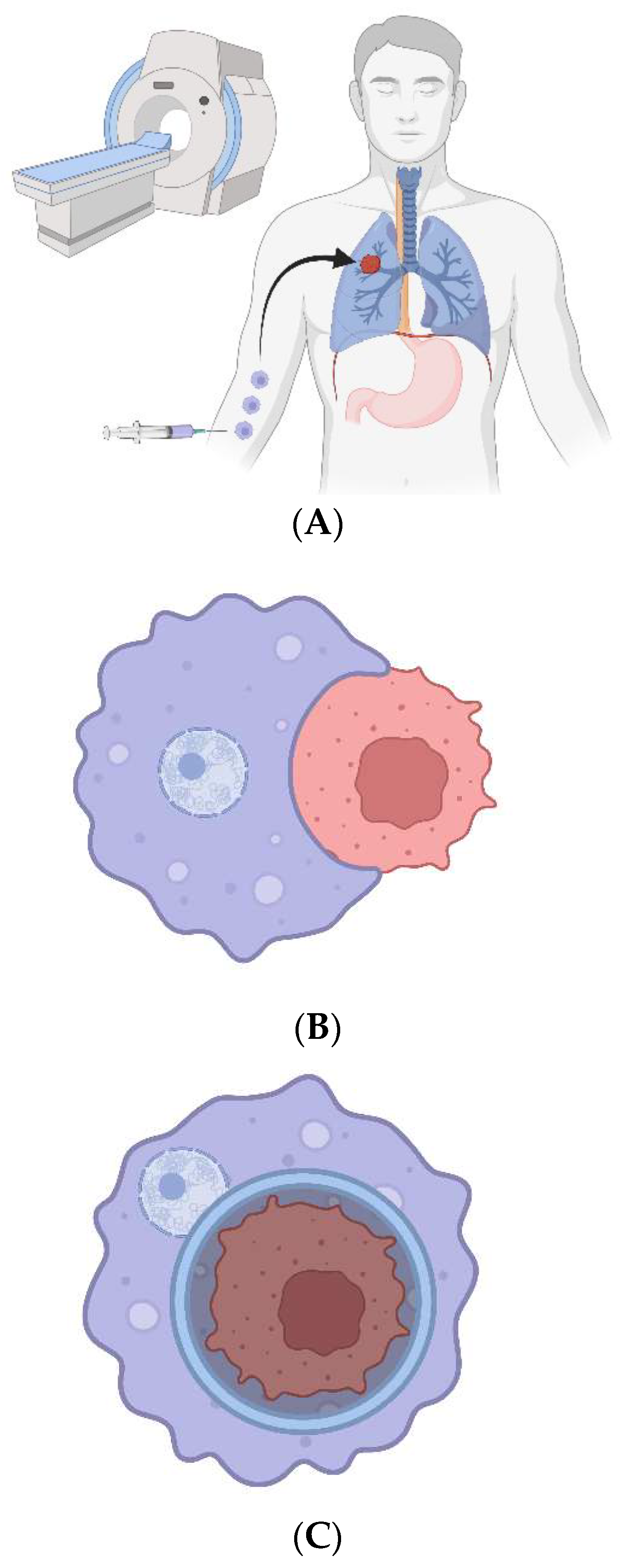
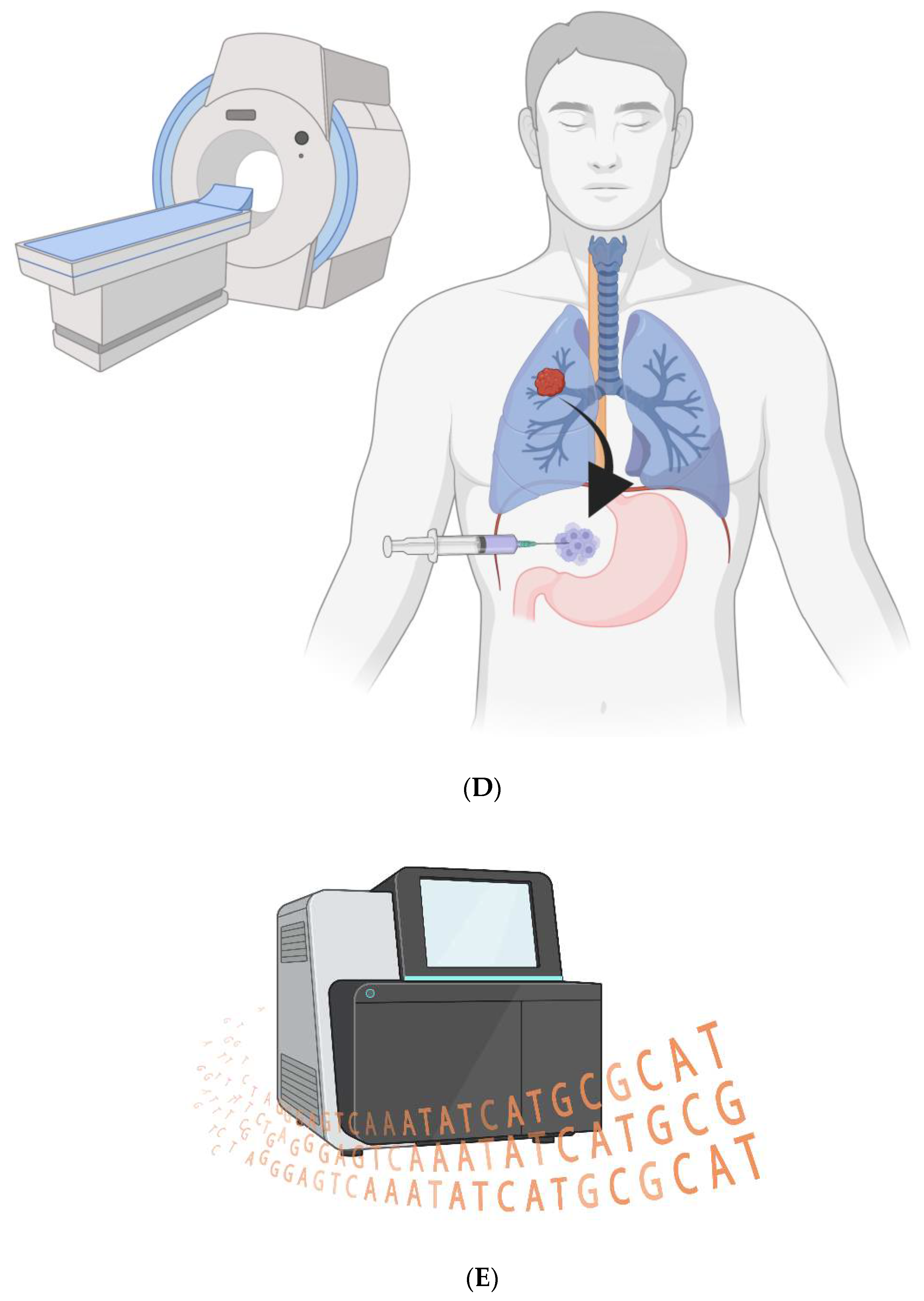
Figure 1.
CAR-Ps. A) The bioengineered macrophages are magnetically steered into the tumor. B) The chimeric antigen receptor is induced, and macrophages phagocytose cancer cells in a patient’s tumor. C) The cancer cell resides within a non-acidified phagosome that does not fuse with a lysosome. D) The macrophages are magnetically drawn to the peritoneal cavity; they are withdrawn via needle. E) The cancer cell genomes are sequenced to look for ubiquitous mutations.
Figure 1.
CAR-Ps. A) The bioengineered macrophages are magnetically steered into the tumor. B) The chimeric antigen receptor is induced, and macrophages phagocytose cancer cells in a patient’s tumor. C) The cancer cell resides within a non-acidified phagosome that does not fuse with a lysosome. D) The macrophages are magnetically drawn to the peritoneal cavity; they are withdrawn via needle. E) The cancer cell genomes are sequenced to look for ubiquitous mutations.
An alternative solution would be to install the T-cell lytic granule system in the CAR-P macrophages [
18,
19]. They would lack granzyme B, but be able to rapidly and directionally secrete perforin to lyse the target cell while sparing the nucleus [
20,
21]. Then, they could phagocytose debris until they bind the nuclear envelope - and selectively phagocytose that and inhibit the maturation of the phagosome and phagosome-lysosome fusion. (The nucleus would be a smaller target than the cell as a whole.)
2.2. Bacterial phagocytosis
Yet another strategy would be for carrier, Irf8(
-/-) [
22] macrophages to ferry non-replicating [
23,
24] facultative intracellular, phagocytic [
25], and potentially magnetosome [
26]-bearing bacteria to cancer cells. The bacteria would lyse the carrier macrophages once in the tumor region, invade tumor cells, and phagocytose their nuclei. Of course, the bacteria would also have to be programmed not to digest their cargo. Then, they would lyse the cancer cell - and could be directed chemotactically or magnetotactically to an extraction point.
Specifically, a species of phagocytic bacteria was recently identified, known as ’
Candidatus Uab amorphum’ [xxiii]. It is flat, spherical, or ovoid, and has a diameter of ~4-microns. Notably, only two genes are required for an extracellular bacterium to be able to invade target cells - an invasin gene and listeriolysin O [
27].
However, smaller bacteria (~1-microns in diameter) may have a higher invasion efficiency - and all currently known facultative intracellular bacteria that have evolved to invade and survive within human cells are of the smaller variety.
Of course, on the other hand, human cell nuclei can be ~5-microns in diameter. Thus, while the bacteria should remain as small as possible for invasion, growth should be induced upon entering a target cancer cell’s cytoplasm. If ‘Ca. Uab amorphum’ cannot readily be engineered to be smaller and grow upon cytoplasmic entry, perhaps the genes required for said bacterium to perform phagocytosis can be transferred to a comparatively small bacterial species.
For example, a probiotic
E. Coli strain could be genetically engineered for a spherical shape, which may be a prequisite for phagocytosis [
28].
It could be given the genes required for phagocytosis, as well as multiple invasins against various ubiquitous cell surface proteins and listeriolysin O.
Growth and division could initially be suppressed via truncated RelA overexpression [
29,
30] and FtsZ inhibition [xxiv]. FtsZ-deficient bacteria are more susceptible to minor stressors than wild-type bacteria, but an inactive form of the protein can prevent this [xxiv].
Growth upon cytoplasmic entry could possibly be achieved by combining the ActA promoter [
31] with induced overexpression of a “useless” protein [
32] and inhibition of the truncated RelA protein. Some amount of RelA may be required for cellular survival, so inhibiting the non-truncated, endogenous gene copy might not be the best idea [
33].
An automated timer mechanism via a temporal promoter cascade involving the expression of early, intermediate, then late genes could be implemented for growth in the cytoplasm of target cancer cells. At the end of this time, the useless protein would be inhibited permanently and truncated RelA would be permanently re-expressed - via recombinase-mediated gene inversion.
Proteins could also potentially be secreted by the bacteria to decrease the size of the host cell nucleus or nuclei [
34].
Motility [
35] should likely be enabled during this throughout this process and after to help evade autophagy and at the end of the growth period for the bacteria to be able to search for its nuclear target. (Phagocytosis receptors should be expressed at the end of the growth period that recognize multiple ubiquitously expressed nuclear outer membrane proteins with domains on present on the cytosolic leaflet of said membrane.)
2.3. Remaining strategies
While it is true that traditional needle biopsies may increase the risk of causing metastasis, the following two strategies may involve an unacceptably high risk of causing metastasis.
The macrophages perhaps could simply attach to the cancer cells and magnetotactically “tow” them to an extraction point. Matrix metalloprotease secretion by the macrophages could help to detach cancer cells from the extracellular matrix beforehand. It would be very important that they do not drop their cargo along the way.
However, the cancer cell could theoretically replicate while being towed. To prevent this, the T-cell lytic granule system mentioned before could be employed to deliver recombinases that excise genes required for division prior to towing.
Finally, cell-cell fusion [
36] could potentially be locally induced in the tumor, provided the macrophage were equipped with many chromosomally-encoded, small molecule-controlled kill switches - and could inhibit the nuclear activity of the cancer cell after fusion. Perhaps this could be achieved by rapidly surrounding the cancer cell’s nucleus or nuclei with autophagosomes that are prohibited from becoming acidic or otherwise maturing.
2.4. Brain and spinal cord tumors:
With regard to the brain and spinal cord, perhaps magnetism is all that’s required to cross the blood-brain and blood-spinal cord barrier - but perhaps not. If not, the easiest solution would probably be an intrathecal injection. If the tumor or tumors are in the brain, the MNP-loaded macrophages could be magnetically dragged up the spinal cord into the brain. This would be considered invasive, of course, but certainly not as much as drilling a burr hole in the patient’s skull.
Alternatively, magnetic resonance-guided motorized transcranial ultrasound [
37] could be used to open the blood-brain barrier at the tumor locale(s). The same technology is applicable to the spinal cord as well; MRI-guided ultrasound has already been tested in that context [
38].
Instead of magnetism, perhaps a prodrug version of deschloroclozapine [
39] can be employed that is blood-brain and blood-spinal cord barrier—permeable. It would be activated by an extracellular enzyme that is abundant in the brain and/or spinal cord [
40] and serve as a chemoattractant for the macrophages [
41]. This could be superior to magnetism because it might induce more crawling along the barrier endothelial cells until an infiltration point can be found.
The question of egress from the central nervous system is also an issue. Chemotaxis and/or magnetotaxis may again be sufficient. Or, they could also simply be directed to a site in the spinal cord for lumbar puncture-based withdrawal - again, a relatively invasive procedure. Maybe in the future, the macrophages or bacteria can somehow be directed to the lymphatic system in the brain and spinal cord and drain into a more easily accessible lymph node [
42] for needle-based extraction.
3. Conclusion
This magnetism-based approach is also suitable for the macrophage-mediated delivery of oncolytic vectors to tumors that would be difficult to reach via needle, of course. This has already been accomplished in mice [iv]. With regard to OVERCOME, vector replication in the carrier macrophages would be induced via small molecule, heat, or even magneto-mechanical actuation [
43] once they have reached the tumor(s). Excess iron oxide nanoparticle deposition in the body from carrier macrophage lysis may not be ideal, but they should be degraded eventually [
44].
Acknowledgments
The figure in this piece was created with BioRender.com.
References
- Renteln, M. Conditional replication of oncolytic viruses based on detection of oncogenic mRNA. Gene Ther. 2018, 25, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Renteln, M. Correction: Conditional replication of oncolytic viruses based on detection of oncogenic mRNA. Gene Ther. 2021, 28, 469–469. [Google Scholar] [CrossRef] [PubMed]
- Renteln, M.A. Promoting oncolytic vector replication with switches that detect ubiquitous mutations. Curr. Cancer Ther. Rev. 2023. [Google Scholar] [CrossRef]
- Muthana, M.; Kennerley, A.J.; Hughes, R.; Fagnano, E.; Richardson, J.; Paul, M.; Murdoch, C.; Wright, F.; Payne, C.; Lythgoe, M.F.; et al. Directing cell therapy to anatomic target sites in vivo with magnetic resonance targeting. Nat. Commun. 2015, 6, 8009. [Google Scholar] [CrossRef]
- Dowdell, A.; Paschke, P.I.; Thomason, P.A.; Tweedy, L.; Insall, R.H. Competition between chemoattractants causes unexpected complexity and can explain negative chemotaxis. Curr. Biol. 2023, 33, 1704–1715. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, L.; Wu, J.; Feng, R.; Chen, Y.; Li, R.; Wu, M.; Zheng, M.; Wu, X.G.; Luo, W.; et al. ARHGEF37 overexpression promotes extravasation and metastasis of hepatocellular carcinoma via directly activating Cdc42. J. Exp. Clin. Cancer Res. 2022, 41, 1–16. [Google Scholar] [CrossRef]
- Morrissey, M.A.; Williamson, A.P.; Steinbach, A.M.; Roberts, E.W.; Kern, N.; Headley, M.B.; Vale, R.D. Chimeric antigen receptors that trigger phagocytosis. eLife 2018, 7, e36688. [Google Scholar] [CrossRef]
- Ito, A.; Teranishi, R.; Kamei, K.; Yamaguchi, M.; Ono, A.; Masumoto, S.; Sonoda, Y.; Horie, M.; Kawabe, Y.; Kamihira, M. Magnetically triggered transgene expression in mammalian cells by localized cellular heating of magnetic nanoparticles. J. Biosci. Bioeng. 2019, 128, 355–364. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Hofmann, A.; Bock, T.; Frei, A.P.; Cerciello, F.; Jacobs, A.; Moest, H.; Omasits, U.; Gundry, R.L.; Yoon, C.; et al. A Mass Spectrometric-Derived Cell Surface Protein Atlas. PLOS ONE 2015, 10, e0121314–e0121314. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Goldmann, U.; Müller, S.; van Oostrum, M.; Müller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. 2018, 115, E10988–E10997. [Google Scholar] [CrossRef]
- Yang, H.; Shao, R.; Huang, H.; Wang, X.; Rong, Z.; Lin, Y. Engineering macrophages to phagocytose cancer cells by blocking the CD47/SIRPɑ axis. Cancer Med. 2019, 8, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Jungbluth, M.; Renicke, C.; Taxis, C. Targeted protein depletion in Saccharomyces cerevisiae by activation of a bidirectional degron. BMC Syst. Biol. 2010, 4, 176–176. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, J.M.; Ravichandran, K.S. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 2008, 9, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Maxwell, K.G.; Wang, K.; Bowers, D.T.; Flanders, J.A.; Liu, W.; Wang, L.-H.; Liu, Q.; Liu, C.; Naji, A.; et al. A nanofibrous encapsulation device for safe delivery of insulin-producing cells to treat type 1 diabetes. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- A Gilmartin, A.; Ralston, K.S.; A Petri, W. Inhibition of Amebic Cysteine Proteases Blocks Amebic Trogocytosis but Not Phagocytosis. J. Infect. Dis. 2020, 221, 1734–1739. [Google Scholar] [CrossRef]
- Underhill, D.M.; Goodridge, H.S. Information processing during phagocytosis. Nat. Rev. Immunol. 2012, 12, 492–502. [Google Scholar] [CrossRef]
- Milde, R.; Ritter, J.; Tennent, G.A.; Loesch, A.; Martinez, F.O.; Gordon, S.; Pepys, M.B.; Verschoor, A.; Helming, L. Multinucleated Giant Cells Are Specialized for Complement-Mediated Phagocytosis and Large Target Destruction. Cell Rep. 2015, 13, 1937–1948. [Google Scholar] [CrossRef]
- Li, H.; Pohler, U.; Strehlow, I.; Hertig, S.; Baccarini, M.; Emmendörffer, A.; Tschopp, J.; Lohmann-Matthes, M.-L. Macrophage precursor cells produce perforin and perform Yac-1 lytic activity in response to stimulation with interleukin-2. J. Leukoc. Biol. 1994, 56, 117–123. [Google Scholar] [CrossRef]
- Tamang, D.L.; Alves, B.N.; Elliott, V.; Redelman, D.; Wadhwa, R.; Fraser, S.A.; Hudig, D. Regulation of perforin lysis: Implications for protein disulfide isomerase proteins. Cell. Immunol. 2009, 255, 82–92. [Google Scholar] [CrossRef]
- Heusel, J.W.; Wesselschmidt, R.L.; Shresta, S.; Russell, J.H.; Ley, T.J. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell 1994, 76, 977–987. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Smyth, M.J.; Trapani, J.A. Perforin-mediated target-cell death and immune homeostasis. Nat. Rev. Immunol. 2006, 6, 940–952. [Google Scholar] [CrossRef]
- Gupta, M.; Shin, D.-M.; Ramakrishna, L.; Goussetis, D.J.; Platanias, L.C.; Xiong, H.; Iii, H.C.M.; Ozato, K. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Yoon, Y.G.; Koob, M.D. Nonreplicating Intracellular Bacterial Vector for Conjugative DNA Transfer into Mitochondria. Pharm. Res. 2012, 29, 1040–1045. [Google Scholar] [CrossRef]
- Sánchez-Gorostiaga, A.; Palacios, P.; Martínez-Arteaga, R.; Sánchez, M.; Casanova, M.; Vicente, M. Life without Division: Physiology of Escherichia coli FtsZ-Deprived Filaments. mBio 2016, 7. [Google Scholar] [CrossRef]
- Shiratori, T.; Suzuki, S.; Kakizawa, Y.; Ishida, K.-I. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Kolinko, I.; Lohße, A.; Borg, S.; Raschdorf, O.; Jogler, C.; Tu, Q.; Pósfai, M.; Tompa, É.; Plitzko, J.M.; Brachmann, A.; et al. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat. Nanotechnol. 2014, 9, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Grillot-Courvalin, C.; Goussard, S.; Huetz, F.; Ojcius, D.M.; Courvalin, P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 1998, 16, 862–866. [Google Scholar] [CrossRef]
- Shiomi, D.; Sakai, M.; Niki, H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008, 27, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Metzger, S.; Aizenman, E.; Roza, S.; Cashel, M.; Glaser, G. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 1991, 266, 3760–3767. [Google Scholar] [CrossRef] [PubMed]
- Büke, F.; Grilli, J.; Lagomarsino, M.C.; Bokinsky, G.; Tans, S.J. ppGpp is a bacterial cell size regulator. Curr. Biol. 2022, 32, 870–877. [Google Scholar] [CrossRef]
- Reniere, M.L.; Whiteley, A.T.; Hamilton, K.L.; John, S.M.; Lauer, P.; Brennan, R.G.; Portnoy, D.A. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 2015, 517, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Basan, M.; Zhu, M.; Dai, X.; Warren, M.; Sévin, D.; Wang, Y.; Hwa, T. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 2015, 11, 836. [Google Scholar] [CrossRef]
- Taylor, C.M.; Beresford, M.; Epton, H.A.S.; Sigee, D.C.; Shama, G.; Andrew, P.W.; Roberts, I.S. Listeria monocytogenes relA and hpt Mutants Are Impaired in Surface-Attached Growth and Virulence. J. Bacteriol. 2002, 184, 621–628. [Google Scholar] [CrossRef]
- Kume, K.; Cantwell, H.; Burrell, A.; Nurse, P. Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.B.; A Theriot, J. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. . 1995, 92, 6572–6576. [Google Scholar] [CrossRef]
- Xu, C.; Ren, X.-H.; Han, D.; Peng, Y.; Lei, J.-J.; Yu, L.-X.; Liu, L.-J.; Xu, W.-C.; Cheng, S.-X. Precise Detection on Cell–Cell Fusion by a Facile Molecular Beacon-Based Method. Anal. Chem. 2022, 94, 17334–17340. [Google Scholar] [CrossRef] [PubMed]
- Magnin, R.; Rabusseau, F.; Salabartan, F.; Mériaux, S.; Aubry, J.-F.; Le Bihan, D.; Dumont, E.; Larrat, B. Magnetic resonance-guided motorized transcranial ultrasound system for blood-brain barrier permeabilization along arbitrary trajectories in rodents. J. Ther. Ultrasound 2015, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Weber-Adrian, D.; Thévenot, E.; O'Reilly, M.A.; Oakden, W.; Akens, M.K.; Ellens, N.; Markham-Coultes, K.; Burgess, A.; Finkelstein, J.; Yee, A.J.; et al. Gene delivery to the spinal cord using MRI-guided focused ultrasound. Gene Ther. 2015, 22, 568–577. [Google Scholar] [CrossRef]
- Nagai, Y.; Miyakawa, N.; Takuwa, H.; Hori, Y.; Oyama, K.; Ji, B.; Takahashi, M.; Huang, X.-P.; Slocum, S.T.; DiBerto, J.F.; et al. Deschloroclozapine, a potent and selective chemogenetic actuator enables rapid neuronal and behavioral modulations in mice and monkeys. Nat. Neurosci. 2020, 23, 1157–1167. [Google Scholar] [CrossRef]
- Yevtodiyenko, A.; Bazhin, A.; Khodakivskyi, P.; Godinat, A.; Budin, G.; Maric, T.; Pietramaggiori, G.; Scherer, S.S.; Kunchulia, M.; Eppeldauer, G.; et al. Portable bioluminescent platform for in vivo monitoring of biological processes in non-transgenic animals. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Park, J.S.; Rhau, B.; Hermann, A.; McNally, K.A.; Zhou, C.; Gong, D.; Weiner, O.D.; Conklin, B.R.; Onuffer, J.; Lim, W.A. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc Natl Acad Sci USA 2014, 11, 5896–5901. [Google Scholar] [CrossRef]
- Lu, H.-B.; Cao, Y.; Hu, J.-Z.; Xu, J.-Q.; Liu, Q.-Q.; Huang, S.-Y.; Duan, C.-Y. The lymphatic system: a therapeutic target for central nervous system disorders. Neural Regen. Res. 2023, 18, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Beltran-Huarac, J.; Yamaleyeva, D.N.; Dotti, G.; Hingtgen, S.; Sokolsky-Papkov, M.; Kabanov, A.V. Magnetic Control of Protein Expression via Magneto-mechanical Actuation of ND-PEGylated Iron Oxide Nanocubes for Cell Therapy. ACS Appl. Mater. Interfaces 2023, 15, 19877–19891. [Google Scholar] [CrossRef] [PubMed]
- Mazuel, F.; Espinosa, A.; Luciani, N.; Reffay, M.; Le Borgne, R.; Motte, L.; Desboeufs, K.; Michel, A.; Pellegrino, T.; Lalatonne, Y.; et al. Massive Intracellular Biodegradation of Iron Oxide Nanoparticles Evidenced Magnetically at Single-Endosome and Tissue Levels. ACS Nano 2016, 10, 7627–7638. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).