Submitted:
07 August 2023
Posted:
09 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
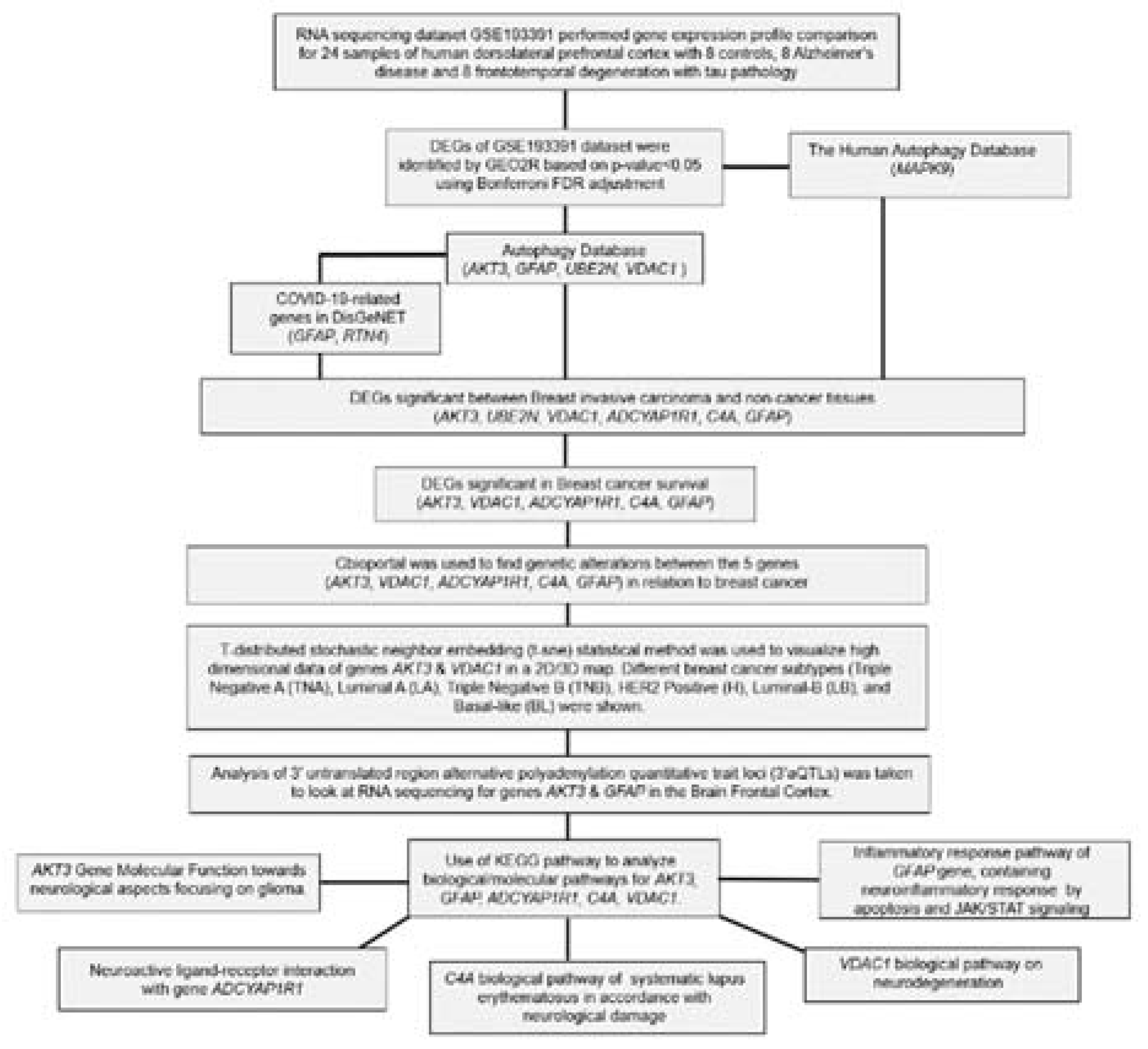
2.1. Differential expression analysis of FTD genes
2.2. Analysis of DEGs involved in autophagy
2.3. Analysis of the genes involved in Covid-19
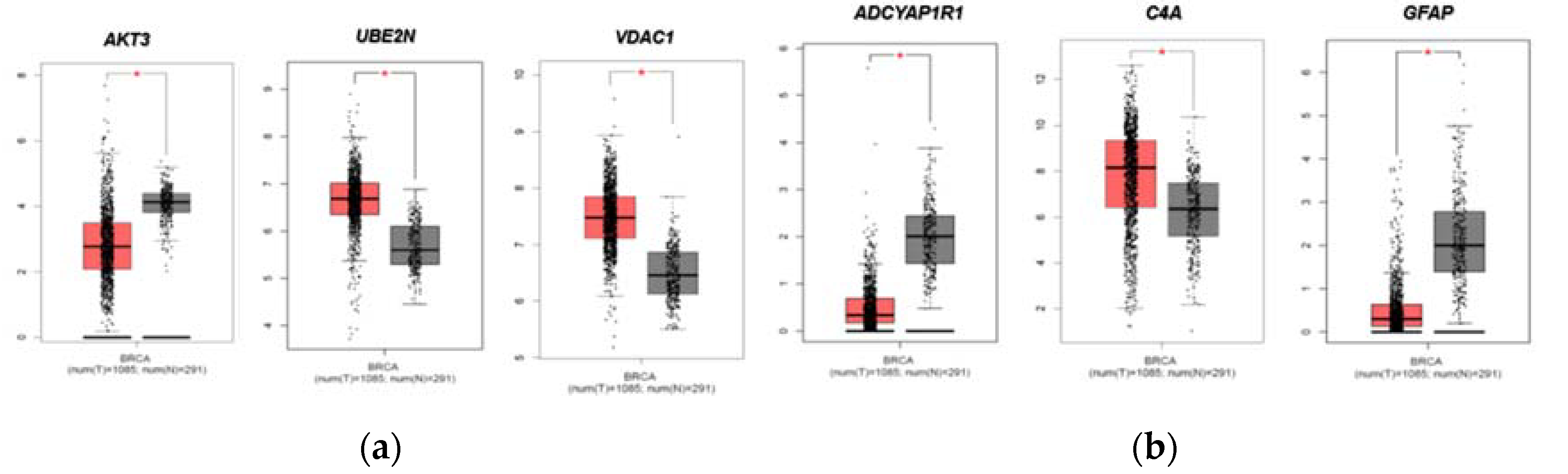
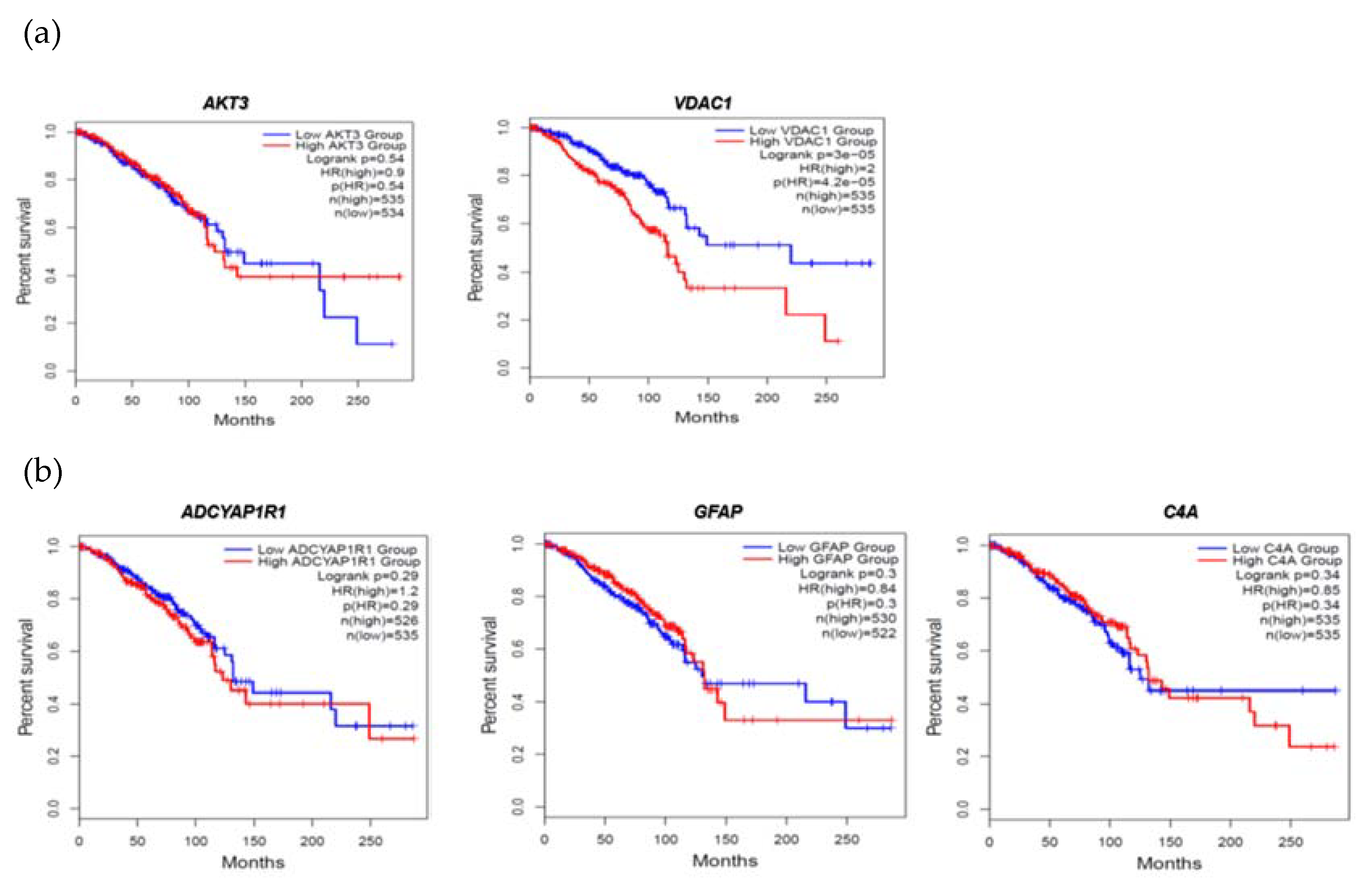
2.4. Evaluation of FTD DEGs expression in Breast Invasive Carcinoma and survival analysis
2.5. Pathogenicity analysis of FTD-Breast cancer comorbid genes
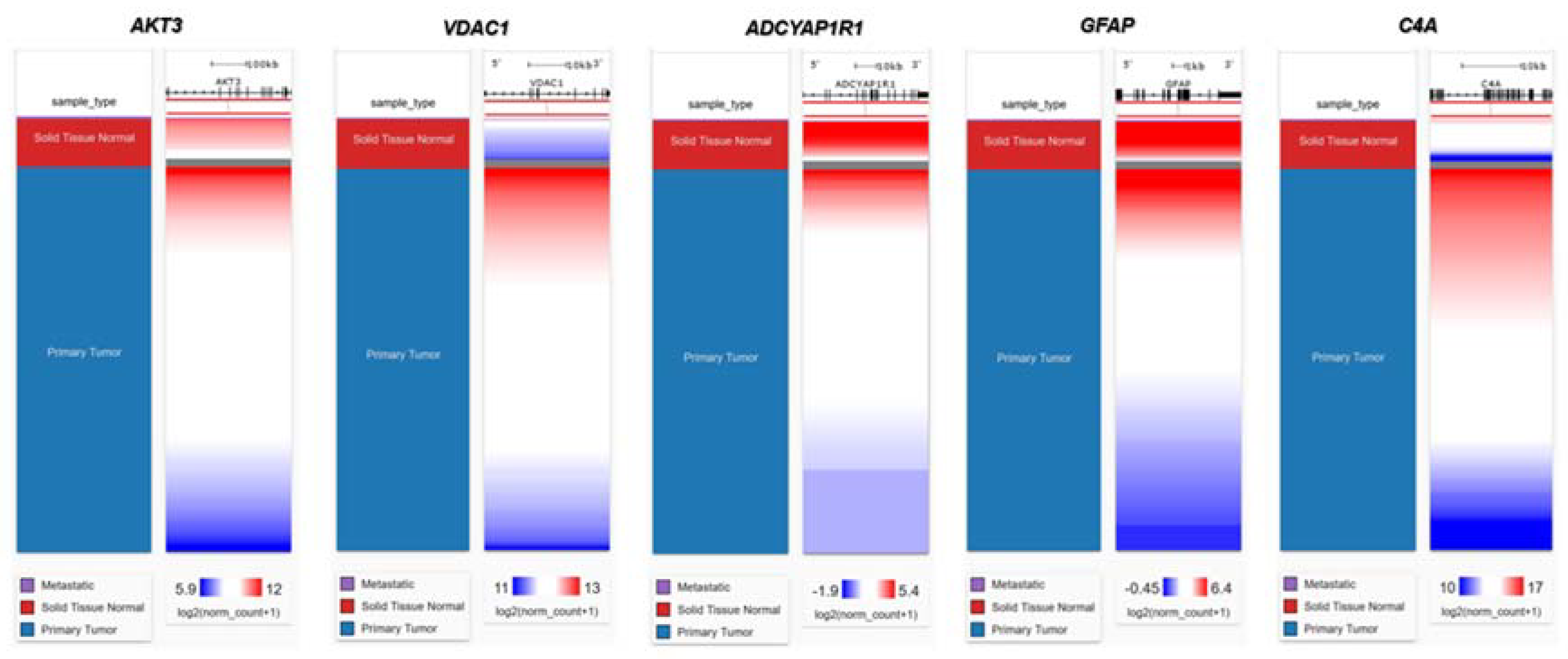
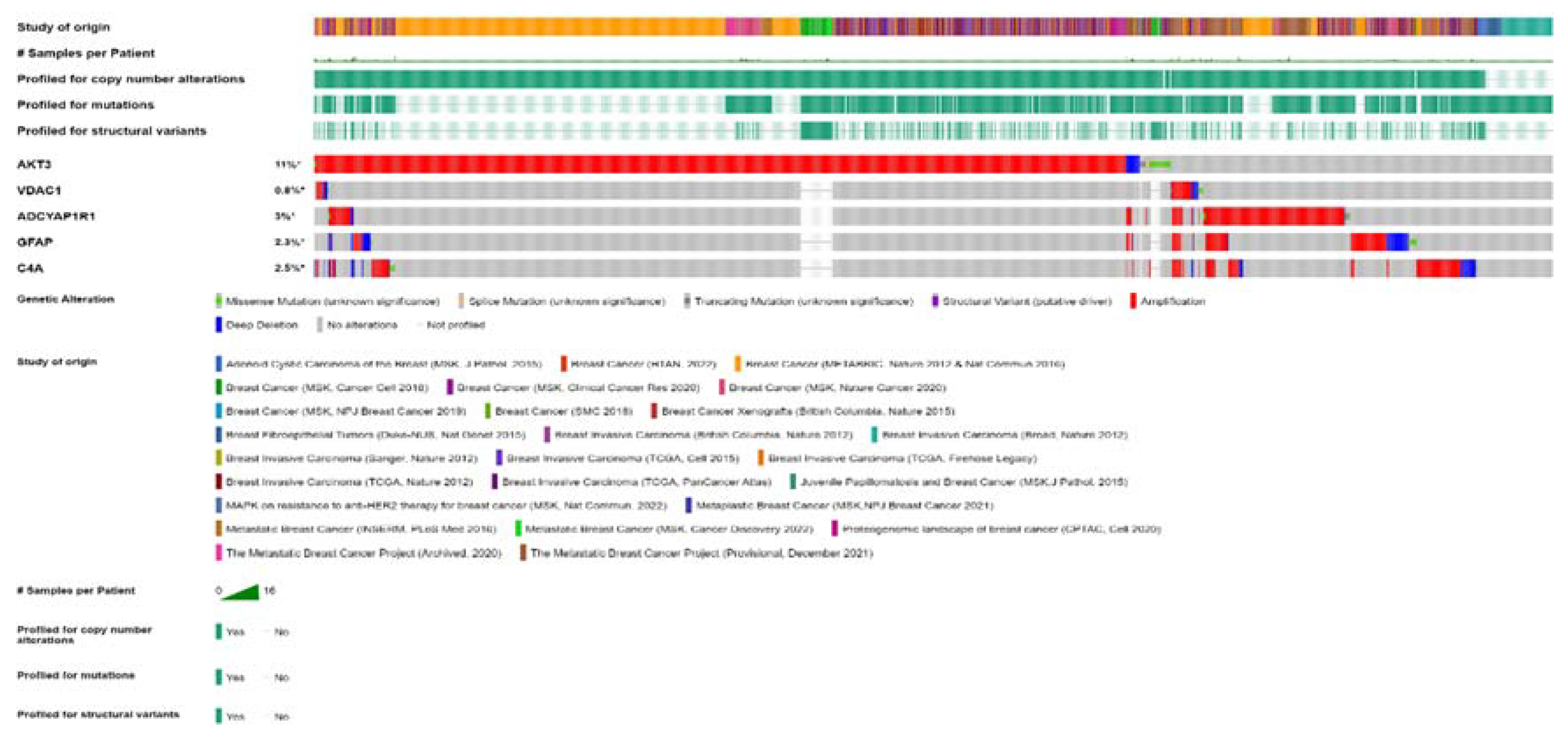
2.6. Genetic Alteration analysis in FTD-Breast cancer comorbid genes
2.7. Clinical association of FTD-Breast comorbid genes
2.8. Pathway enrichment analysis
3. Discussion
4. Materials and Methods
4.1. Data collection and identification of differentially expressed genes in FTD
4.2. Identification of Autophagy genes
4.3. Identification of COVID-19-associated genes
4.4. Verification of FTD DEGs in breast cancer
4.5. Identification of the alteration in the DEGs
4.6. Expression analysis of FTD-breast cancer comorbid genes
4.7. Identification of RNA interactions for the FTD-breast cancer comorbid genes
4.8. Single-cell transcriptome profiling of FTD-breast cancer comorbid genes
4.9. Prediction of potential therapeutic drugs
4.10. Pathogenicity Gene Score Analysis
4.11. Interpretation of genetic variants
4.12. Enrichment of functional pathways
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Khan I, De Jesus O: Frontotemporal Lobe Dementia. In: StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC.; 2023.
- Onyike, C.U.; Diehl-Schmid, J. The epidemiology of frontotemporal dementia. Int Rev Psychiatry 2013, 25, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Magrath Guimet N, Zapata-Restrepo LM, Miller BL: Advances in Treatment of Frontotemporal Dementia. J Neuropsychiatry Clin Neurosci 2022, 34, 316–327. [CrossRef] [PubMed]
- Zanella I, Zacchi E, Piva S, Filosto M, Beligni G, Alaverdian D, Amitrano S, Fava F, Baldassarri M, Frullanti E et al.: C9orf72 Intermediate Repeats Confer Genetic Risk for Severe COVID-19 Pneumonia Independently of Age. Int J Mol Sci 2021, 22.
- Smeyers J, Banchi EG, Latouche M: C9ORF72: What It Is, What It Does, and Why It Matters. Front Cell Neurosci 2021, 15, 661447. [CrossRef]
- Rohrer JD, Warren JD, Fox NC, Rossor MN: Presymptomatic studies in genetic frontotemporal dementia. Rev Neurol (Paris) 2013, 169, 820–824. [CrossRef]
- Katisko K, Haapasalo A, Koivisto A, Krüger J, Hartikainen P, Korhonen V, Helisalmi S, Herukka SK, Remes AM, Solje E: Low Prevalence of Cancer in Patients with Frontotemporal Lobar Degeneration. J Alzheimers Dis 2018, 62, 789–794.
- Ng KP, Chiew HJ, Hameed S, Ting SKS, Ng A, Soo SA, Wong BYX, Lim L, Yong ACW, Mok VCT et al. : Frontotemporal dementia and COVID-19: Hypothesis generation and roadmap for future research. Alzheimers Dement (N Y) 2020, 6, e12085. [Google Scholar] [CrossRef]
- Bottero V, Alrafati F, Santiago JA, Potashkin JA: Transcriptomic and Network Meta-Analysis of Frontotemporal Dementias. Front Mol Neurosci 2021, 14, 747798. [CrossRef]
- Edgar R, Domrachev M, Lash AE: Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002, 30, 207–210. [CrossRef]
- Homma K, Suzuki K, Sugawara H: The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res 2011, 39, D986–D990. [CrossRef]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI: DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017, 45, D833–d839. [CrossRef] [PubMed]
- Tang Z, Li C, Kang B, Gao G, Zhang Z: GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017, 45, W98–w102. [CrossRef] [PubMed]
- Lánczky A, Győrffy B: Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res 2021, 23, e27633. [CrossRef] [PubMed]
- Gambardella G, Viscido G, Tumaini B, Isacchi A, Bosotti R, di Bernardo D: A single-cell analysis of breast cancer cell lines to study tumour heterogeneity and drug response. Nat Commun 2022, 13, 1714. [CrossRef]
- Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN et al. : Visualizing and interpreting cancer genomics data via the Xena platform. In: Nat Biotechnol. 2020, 38, 675–678. [Google Scholar]
- Kr Z: An encyclopedia of RNA interactomes in ENCORI. In. Edited by Huang JH LS, Li B, Liu SR, Zheng WJ, cai L, et al.
- Freshour SL, Kiwala S, Cotto KC, Coffman AC, McMichael JF, Song JJ, Griffith M, Griffith OL, Wagner AH: Integration of the Drug-Gene Interaction Database (DGIdb 4. 0) with open crowdsource efforts. Nucleic Acids Res 2021, 49, D1144–d1151. [Google Scholar] [CrossRef]
- Chen Z, Zhong Z, Zhang W, Su G, Yang P: Integrated Analysis of Key Pathways and Drug Targets Associated With Vogt-Koyanagi-Harada Disease. Front Immunol 2020, 11, 587443. [CrossRef]
- Cui Y, Peng F, Wang D, Li Y, Li JS, Li L, Li W: 3'aQTL-atlas: an atlas of 3'UTR alternative polyadenylation quantitative trait loci across human normal tissues. Nucleic Acids Res 2022, 50, D39–d45. [CrossRef]
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W: DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022, 50, W216–w221. [CrossRef]
- Kanehisa M, Goto S: KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28, 27–30. [CrossRef]
- Gojobori T, Ikeo K, Katayama Y, Kawabata T, Kinjo AR, Kinoshita K, Kwon Y, Migita O, Mizutani H, Muraoka M et al. : VaProS: a database-integration approach for protein/genome information retrieval. J Struct Funct Genomics 2016, 17, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Wolfsberg TG, Primakoff P, Myles DG, White JM: ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol 1995, 131, 275–278. [CrossRef] [PubMed]
- Finger EC: Frontotemporal Dementias. Continuum (Minneap Minn) 2016, 22, 464–489.
- Devenney EM, Ahmed RM, Hodges JR: Frontotemporal dementia. Handb Clin Neurol 2019, 167, 279–299.
- DeSantis C, Siegel R, Bandi P, Jemal A: Breast cancer statistics, 2011. CA Cancer J Clin 2011, 61, 409–418.
- Palluzzi F, Ferrari R, Graziano F, Novelli V, Rossi G, Galimberti D, Rainero I, Benussi L, Nacmias B, Bruni AC et al. : A novel network analysis approach reveals DNA damage, oxidative stress and calcium/cAMP homeostasis-associated biomarkers in frontotemporal dementia. PLoS One 2017, 12, e0185797. [Google Scholar]
- Martín-Guerrero SM, Markovinovic A, Mórotz GM, Salam S, Noble W, Miller CCJ: Targeting ER-Mitochondria Signaling as a Therapeutic Target for Frontotemporal Dementia and Related Amyotrophic Lateral Sclerosis. Front Cell Dev Biol 2022, 10, 915931.
- Heller C, Foiani MS, Moore K, Convery R, Bocchetta M, Neason M, Cash DM, Thomas D, Greaves CV, Woollacott IO et al. : Plasma glial fibrillary acidic protein is raised in progranulin-associated frontotemporal dementia. J Neurol Neurosurg Psychiatry 2020, 91, 263–270. [Google Scholar] [CrossRef]
- Reus LM, Pasaniuc B, Posthuma D, Boltz T, Pijnenburg YAL, Ophoff RA: Gene Expression Imputation Across Multiple Tissue Types Provides Insight Into the Genetic Architecture of Frontotemporal Dementia and Its Clinical Subtypes. Biol Psychiatry 2021, 89, 825–835. [CrossRef]
- Nagatsu T: The catecholamine system in health and disease -Relation to tyrosine 3-monooxygenase and other catecholamine-synthesizing enzymes. Proc Jpn Acad Ser B Phys Biol Sci 2007, 82, 388–415.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



