1. Introduction
The skin is the outermost layer of the human body. The largest organ in the human integument structure, it comprises multiple layers. It also involves immune cells and cells that generate melanin to keep the body healthy from the carcinogenic potential of ultraviolet radiation [
1]. Skin cancer arises when skin cells become disorganized and proliferate uncontrollably, potentially migrating to other body parts. Skin cancer is the most common type of cancer worldwide. Melanoma and non-melanoma pigmented lesions are the two main forms of skin cancer. Melanoma is associated with melanocytes, influencing the color of malignant cells [
2].
In 2022, the American Cancer Society predicts that there will be about 99,780 new melanomas diagnosed in the country [
3]. Melanoma is predicted to be fatal approximately 7,650 people (about 5,080 men and 2,570 women). Rates of melanoma have considerably increased in recent decades. On the other hand, it is more deadly since it has the ability to spread to many other areas of the body if it is not detected and treated promptly. Besides, it is associated with melanocytes, which cause malignant cells to change color, thus accounting for the bulk of pigmented skin cancer deaths [
4].
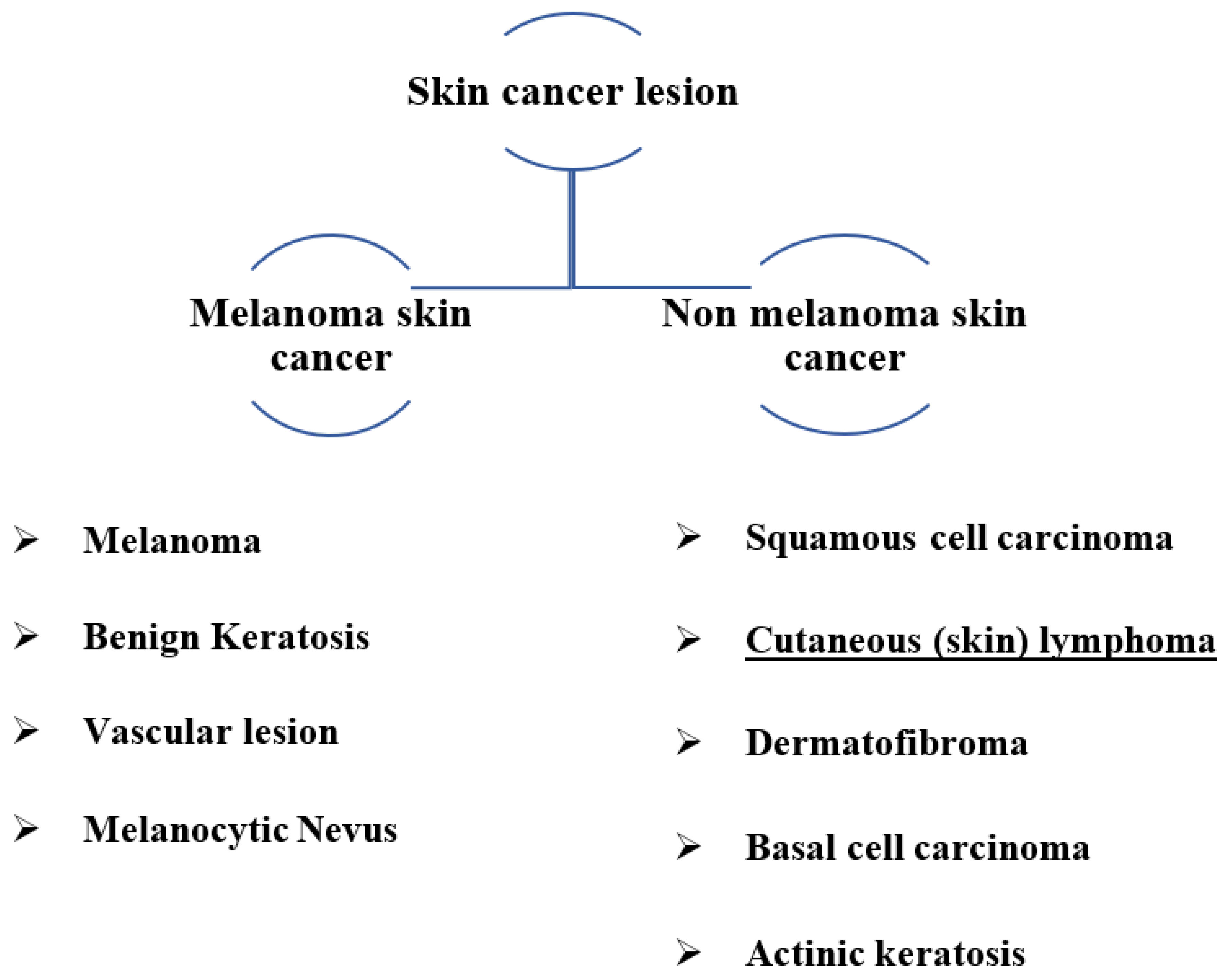
Figure 1 depicts various types of skin cancer lesions, including common types detected.
Due to the substantial similarities between the many forms of skin lesions, visual analysis is challenging, which can lead to incorrect choices [
5]. The ABCD (asymmetry, border irregularity, color variation, and diameter) examination is commonly used to diagnose malignant melanoma. Patients with melanoma who are detected early have a greater chance of survival [
6]. As digital computing capabilities progress, some researchers have developed computer-aided diagnosis (CAD) systems that integrate image processing, pattern recognition, and artificial neural networks to support physicians in diagnosing [
7].
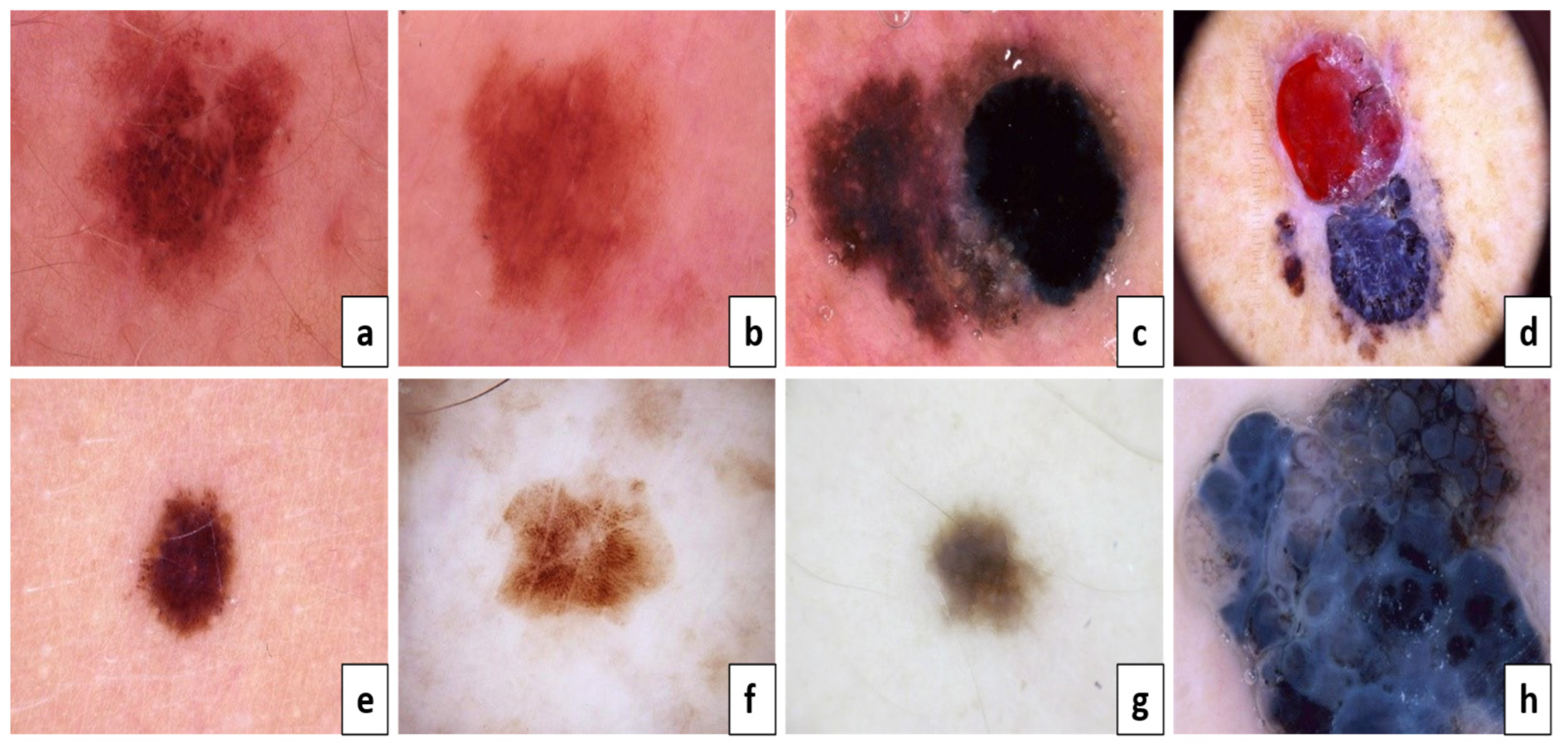
According to
Figure 2, (a, c, e, g) are considered benign, but (b, d, f, h) are melanoma skin cancer. Because of the result of the ABCD examination:
Asymmetry: (A) both sides match the other, and (B) one side does not match the other.
Border: (C) Regular edges, (D) Irregular or Blurred.
Color: (E) Consistent shades, (F) Different shades.
Diameter: (G) lesion is smaller than 6mm, (H) lesion is larger than 6mm.
The ABCD approach was utilized to distinguish malignant melanoma from benign lesions based on its ability to extract distinguishing morphological features. As a result, it is usually utilized in automated systems. This method's efficiency ranges from 85.0% to 91.0%. These low percentages inspire researchers to provide another approach, either by altering an existing method or developing a new approach to improve performance.
Deep learning (DL) techniques have to be the most effective, supervised, time and cost-efficient machine learning method. DL techniques have made substantial advances in automatically extracting characteristics across several deep layers, generating significant benefits [
8,
9]. DL has recently been successfully employed in visual tasks and object recognition by academics worldwide.
characteristics of lesions.
The characteristics of melanoma have been used to construct certain machine learning approaches to identify the disease. CAD models involve effective algorithms to categorize and forecast melanoma. Algorithms like the adaptive histogram equalization approach, contrast stretching, and a median filter are used to improve the pictures. Following that, there are a variety of segmentation algorithms, including normalized Otsu's segmentation (NOS), which separates the damaged skin lesion from the normal skin and solves the issue of fluctuating illumination [
6]. The segmented images are used to construct and extract features, which are then given to the various classifiers, including hybrid Adaboost, support vector machine (SVM), and DL neural networks [10-14]. Various architectures such as ResNet, Dense Net, and Senet were used. Various methods are applied to deal with each class's unequal quantity of images, such as balanced batch sampling and loss weighting.
In order to get an improved consistent skin lesions classification technique, the proposed system is pre-trained more than once with various hyperparameter settings. The suggested architecture uses a single stage to combine detection and classification instead of more conventional approaches. The proposed system has three significant benefits over previous computer-assisted skin cancer screening approaches:
The proposed method applies to any image (dermoscopy or photographic) of pigmented skin lesions using you only look once (YOLOv5) and ResNet50.
The suggested system classifies samples and determines each class with probability.
It interacted directly with the skin-color images that were obtained with different sizes.
This study developed, implemented, and effectively assessed a novel DL-based skin lesion classification algorithm against a publicly available dermoscopy dataset based on utilizing seven categories of skin lesions (the HAM10000 dataset) [
15,
16].
The arrangement of the paper is outlined below. The second section offers a review of the related studies.
Section 3 goes into detail about the suggested method and the datasets utilized.
Section 4 outlines the experimental setup and performance assessment of the suggested technique, and
Section 5, in the end, discusses the conclusions.
2. Related Work
Early detection and treatment of melanoma frequently result in a cure. It becomes more dangerous and difficult to treat if it penetrates deeper into the skin or other body parts. Most melanoma classification algorithms currently in use contain custom-made characteristics such as measurements of lesion shape, distribution, and color, as well as measurements of texture and border irregularity [
17]. After feature extraction, machine learning techniques such as artificial neural networks (ANNs), K-nearest neighbors (KNN) classification, SoftMax classification, SVM, and logistic regression can be used to successfully solve the classification problem [
18]. Examples of relevant DL research include the following, which are summarized in
Table 1.
Using a contrast-constrained adaptive histogram equalization strategy, Premaladha et al. [
6] improved the melanoma classification system. The images were enhanced before segmenting the filtered grayscale image using the Otsu normalized method. DL achieved 92.8% classification accuracy. A deep convolutional neural network (DCNN) divided color images of skin cancer into three groups: atypical nevus, melanoma, and typical nevus from Med node and PH2 datasets. This proposed system needs to add more classes for more accuracy.
Codella et al. [
10] established a hybrid technique for melanoma categorization. A help vector was used in this method. A support vector machine (SVM), deep learning, and sparse coding were all used. 2624 clinical cases from the International Skin Imaging Collaboration were used as a dataset. When all the results were added together, the categorization efficiency was 93.1 percent. There is a need to deepen feature extraction and add more cases for the diagnosis of melanoma.
Gessert et al. [
11] used a huge ensemble of state-of-the-art convolutional neural network (CNN) models to classify skin lesions. Various architectures such as ResNet, Dense net, and Senet were used. To deal with the unequal quantity of images for each class, they applied various methods, such as balanced batch sampling and loss weighting. Finally, the ensemble of multiple convolutional neural network architectures was fine-tuned utilizing their dataset. This task was correctly classified 85.1% of the time.
Waheed et al. [
12] constructed a model using machine learning for diagnosing melanoma depending on dermoscopy images from HAM10000 dataset.. It was based on distinguishing attributes such as the appearance and texture of many skin lesions. SVM was used in their research to distinguish melanoma images from all other classes. Their model correctly classified objects 96.0% of the time. There is a need for more attributes of skin lesions classification.
In order to categorize skin lesion images into five diagnostic groups, Hekler et al. [
19] used CNN. They applied their method to 300 test images from the HAM10000 dataset and discovered that it was 82.9 percent accurate (60 percent for each of the five illness classifications). An invasive technique is demonstrated with a small number of low-resolution pictures. Their method's binary distinction between melanoma and nevi is another drawback.
DCNN evaluation for melanoma categorization was given by Pham et al. [
20]. Additionally, they helped with data improvement. Using CNN level layers at different levels, each feature was retrieved. Additionally, the dataset was probably altered in some way. The analysis was tested using the ISBI dataset, which had an area under the curve (AUC) of 89.2%. Because skin lesion images are identical, it was necessary to reuse network weights to increase sensitivity.
Yu et al. [
21] published a two-stage melanoma detection approach. They used a deep residual network (DRN) for classifying and a DCNN network with more than 50 layers for segmentation. Segmentation was performed using a fully convolutional residual network (FCRN), while classification was performed using a DRN. The ISBI dataset was used to evaluate the findings, and the AUC was 80.4%. It is vital to explore techniques to include Bayesian learning, particularly probabilistic graphical models, into networks in order to further increase the discrimination capability of the very deep CNNs and address the issue of insufficient training data.
Li and Shen [
22] proposed an automated melanoma detection method based on two deep learning techniques. They employed two FCRN simultaneously for a more thorough classification. The lesion feature network was used to extract dermoscopy features. The model used the International Skin Imaging Collaboration (ISIC 2017) dataset to test the performance. It produced 2357 photos of both malignant and harmless oncological illnesses for this collection (ISIC). All pictures were sorted based on the categories were identified using ISIC, and the same number of photos were used to divide each subgroup. Model segmentation and classification results were 75.3% and 91.2%, respectively. There is overfitting in AUC, and the results of segmentation are low.
Seeja and Suresh [
23] presented a DCNN for precise skin lesion segmentation using the U-net technique. To obtain their findings, they combined CNN and FCNN. The color, texture, and shape attributes were selected from the segmented images of ISBI 2016 dataset. The method used for texture analysis was local binary pattern (LBP). Form features were extracted using the edge histogram, Gabor, and histogram of oriented gradients (HOG) approaches. For classification, SVM, random forest (RF), K-nearest neighbor (KNN), and naïve Bayes (NB) classifiers were selected. The test results showed that the Dice co-efficiency value for image segmentation was 77.5%. The classification accuracy of the SVM classifier was 85.1%, 82.2 percent for RF, 79.2 percent for KNN, and 65.9 percent for NB. The proposed system needs to improve the result for classification.
For the purpose of melanoma early detection, Nasiri et al. [
24] created a method of case-based reasoning. DL algorithms were used to categorize skin lesions in their strategy. This investigation is a follow-up to their case-based learning assistant system study that looked at how to detect and predict melanoma from ISIC dataset. A 19-layer model of CNN, a deep learning method, was used in this work to categorize skin lesions. Three fully connected layers, three max-pooling layers, and eleven convolutional layers form the model. The proposed approach has a 75.0% success rate in the ISIC Melanoma Project, where it was tested.
S. Inthiyaz
et al. [
25] recently created an approach based on the combination of CNN and Softmax. Skin photographs were first filtered to remove unwanted noise from the image before being processed to improve the overall quality of the image. The presented work was based on extracting features from skin images, which were then classified using the Softmax classifier. It was accurate to 87.0%. This model should be improved for greater accuracy by including more skin cancer classes and employing DL methods.
M. Mohammed
et al. [
26] suggested model investigated the various forms of cancer. By examining, categorizing, and manipulating the multi-omics dataset in a fog cloud network. The study created hybrid cancer detection systems based on
state action reward state action (SARSA) on-policy and multi-omics workload learning, which were made achievable by reinforcement learning. It has several layers, including the collection of clinical data through laboratories. To improve performance, this model needs many widely dispersed clinics for cancer categorization and prediction.
In light of the importance of early melanoma identification, the visible similarity among melanoma and non-melanoma tumors, the absence of contrast between lesions and skin, and other considerations. Therefore, accurate automatic diagnosis of skin tumors is crucial to improving the precision and effectiveness of pathologists.
According to previous studies, there are limitations in melanoma detection due to the small number of classes used and the need to determine the degree of classes. There is a need to deepen feature extraction and add more cases for the diagnosis of melanoma. The similarities between classes can't be determined. Recently, work has been based on two stages to determine melanoma (segmentation and classification). But there is overfitting in AUC, and the segmentation results are low. A system that relies on DL must be created in to obtain reliable melanoma classification. YOLOv5 relies on a single step for identifying and classifying skin lesions to determine the type of melanoma, in contrast to earlier DL studies for skin lesion classification that concentrated on employing specific layers. The core concept behind the YOLO technique is to employ an end-to-end convolutional neural network to predict the target's class and position. It used bounding boxes for detection and probabilities to determine an object's probability percent. The design is composed of an input layer, a convolutional layer, a layer for pooling, a layer for fully connecting, and a layer for the output. In the primary step, YOLO splits the input image into S × S grids. Each grid is diagnosed to check if it has any class of skin lesions. It then classifies each object and gives it its probability. Then, Resnet network is used to prevent gradient explosion issues. Because we have seven classes with multiple scales, it is employed as the image classification network.
3. Proposed Melanoma Detection Technique
3.1. Preprocessing
The preprocessing step of melanoma detection aims to provide a suitable source for the model's appropriateness in actual time. Data augmentation is provided to increase the sample size for those imbalanced classes and prevent this imbalanced dataset from producing a biased or skewed prediction. Since an image could contain noise, preprocessing is necessary for detection algorithms [
27,
28]. Pictures of skin lesions often have uneven lighting, skin surface light reflection, and hair. These kinds of noises need to be reduced because they can impair segmentation performance.
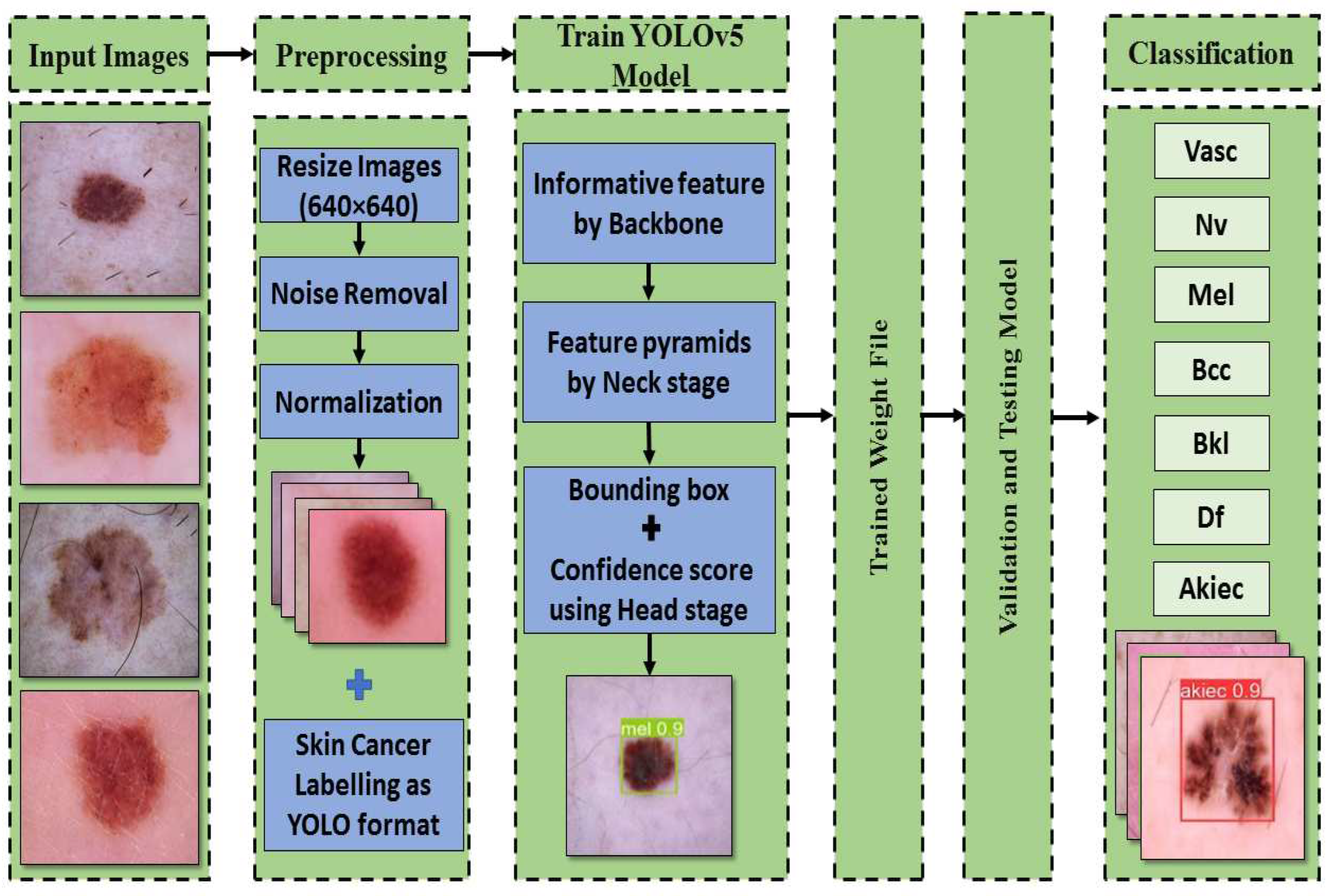
Additionally, each of these photographs has been carefully scaled and smoothed. Normalizing the original image speeds up detection without sacrificing any data. This is an essential technique to guarantee that every image is annotated and to increase performance accuracy. For the necessary computation parameters, as well as for later implementation and detection, it is done. The DL architecture scales image pixels before the training process. In experiments, photos are scaled to (224, 224, 3) using the ImageDataGenerator class and scaling methods. The image pixels are normalized to standardize the image samples. The [0, 255] range of possible values is replaced with the [0, 1] range. If images are not scaled, they will receive a lot of votes to update weights due to their wide pixel range. The YOLO model provides the output from the processed stage [
29,
30]. After combining the detection model's anchor box size for classification, the preparation methods would be completed according to
Figure 3. There are many models of YOLOv5.
The proposed framework utilized a small model with only 7MB of memory. Here is a brief explanation of all the versions of YOLOv5, which is used for model configuration. First, YOLOv5n (the nano edition) is the smallest in the series, intended for Internet-of-Things data, and it also supports OpenCV Dl. In INT8 format, it weighs less than 2.5 MB, and in FP32 format, it is about 4 MB. It's perfect for software devices. Second, YOLOv5s (the small edition) is the family's smallest model, with about 7.2 million parameters, making it perfect for inference to run on the CPU. Third, unlike YOLOv5m (the medium edition), which has 21.2 million parameters, this medium-sized version is of interest. Given that it offers a pretty good balance between speed and efficiency, it may be the model that is most suitable for many datasets and training. Fourth, YOLOv5l (the large edition) contains 46.5 million components; it is the biggest version in the YOLOv5 group. For datasets that necessitate us finding tiny things, it works perfectly. Finally, YOLOv5x (the extra-large edition) is the largest of the five and also has the greatest MAP, although it has 86.7 million parameters and therefore is slower than the others.
3.2. The Structure of the YOLOv5-S Model
The YOLO model is a target detection method that uses regression. A regression model is created from the target detection problem. When photos of skin cancer are entered into a DNN by YOLO, the technique predicts the classification and localization information of the various skin lesion classes based on the computation of the loss function [
31]. YOLOv5 is based on the YOLO detection architecture. It employs top-notch algorithm optimization techniques developed in recent years in convolutional neural networks, including auto mosaic data augmentation, learning bounding box anchors, Leaky Rectified Linear Unit (Leaky Relu) activation function, and others. They are in charge of various tasks in various parts of the YOLOv5 architecture.
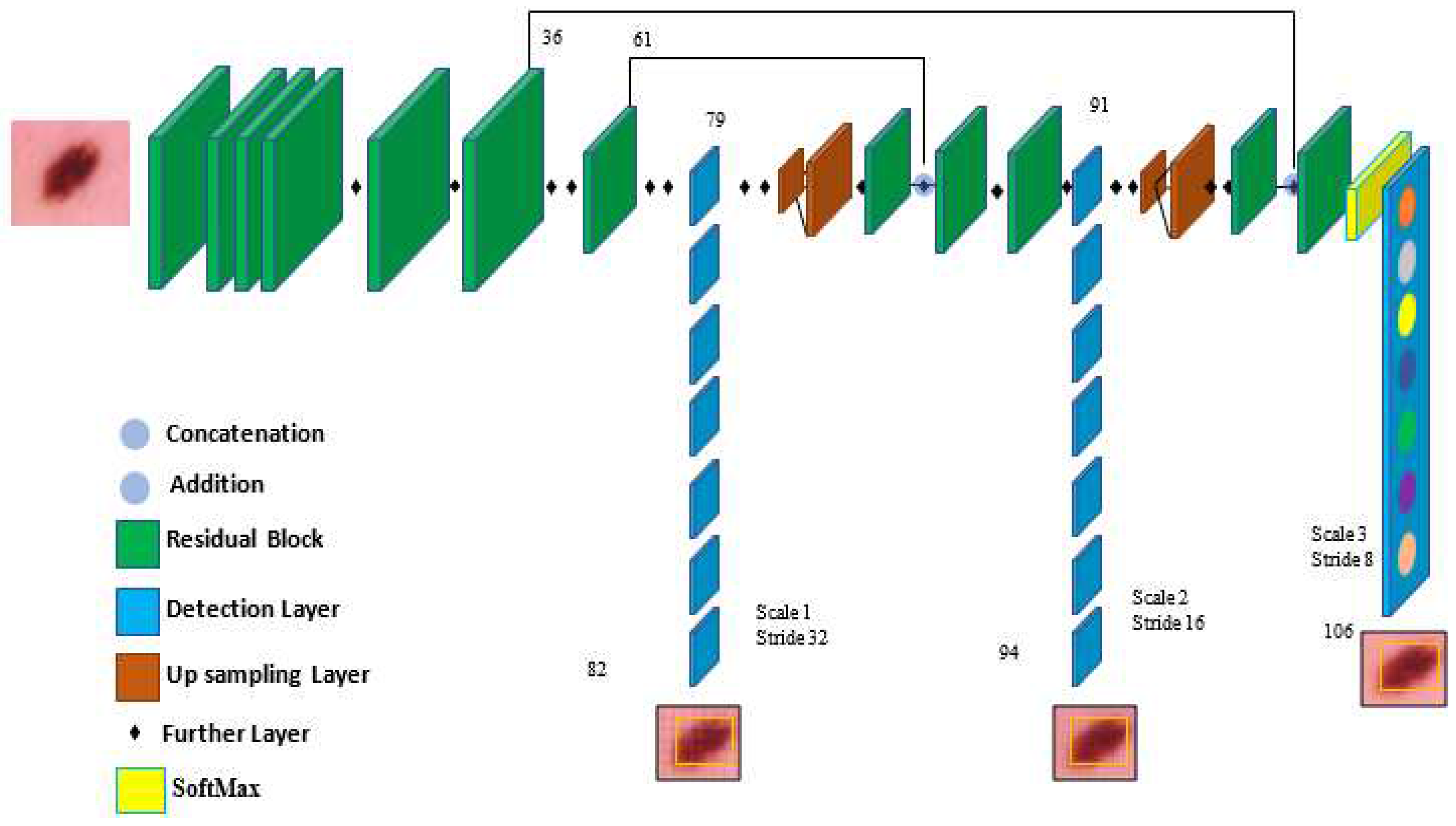
In the architecture, YOLOv5 comprises the dataset images, backbone, neck, and detection output components, as shown in
Figure 4. The input is the preprocessing output according to the previous subsection. The second component is the backbone. This backbone eliminates the redundant gradient information present in large backbones. Gradient change is incorporated into feature maps, which speeds up inference, improves accuracy, and shrinks the model's size by reducing the number of hyperparameters. It is a CNN that uses the Cross Stage Partial network (CSP) and focus interlaced sampling splicing structure as its core to produce significant features from provided pictures. The problem of recurring gradient information in large-scale networks is addressed by CSPNET [
32]. Lowering model hyperparameters and FLOPS (floating-point operations per second) decreases calculations while increasing the speed and precision of inference and shrinking the model's size.
Second, feature pyramids are created using the Neck model. Models can achieve good object scaling generalization with the help of feature pyramids. It helps with object identification when it appears in different scales and sizes. The neck model of YOLOv5 is based on spatial pyramid pooling (SPP) and path aggregation network (PANET) [
33,
34]. It increases the utilization of precise location signals at lower layers and information flow, increasing the accuracy of object location. The spatial scale of the convolved information is decreased in this version due to the employment of a subsampling layer. By lowering dimensionality, the amount of computing required to process the data should be minimized.
The YOLO layer completes the last detection step, which is the head of YOLOv5. The method can simulate small, medium-sized, and large objects according to the generation of a multi-scale prediction for the anchor boxes. To optimize the overlap between the ground truth and the anticipated bounding box of the detected class, generalized intersection over union (GIOU-loss) is utilized [
35]. Stochastic gradient descent (SGD) is used by default in the original version of YOLOv5 [
36]. It is a straightforward yet highly effective method for fitting linear classifiers with convex loss functions. SGD produced divergence at the specified learning rate of 0.0001 pretty rapidly. After training started, loss parameters grew significantly and eventually reached infinity after around ten batches. So, Adam is used as an optimizer in the training step since it consistently converges. With ADAM serving as the adaptive learning rate, it begins with an initial learning rate.
In order to explain how a certain class is used, the network's final layer employs SoftMax. The training stage receives the detection stage's results after being altered. These findings include the bounding box coordinates
and
is the class of the detected class and represented in vector as following [
as well as the probability of each class
for each detection. The intersection over union (IOU) represents the accuracy of the target skin cancer class anticipated and the actual skin cancer class. When there is no object in the target image, the probability of detecting an object is 0. When there is a complete object, the probability is equal to 1, and the IOU is calculated using the predicted target (p) and the real target (t) as Equation. (1).
The real target boundary is box t, whereas the expected target is box p. As observed, the IOU is the ratio of the intersection of the predicted and real target frames to their union. Then each image's annotations are recorded in YOLO format in txt files, with each line containing a skin lesion bounding box description. The training stage is then conducted using YOLOv5. Once the Yolov5 original model has been applied, a particular additional layer is applied to increase efficiency. A new scale includes convolutional layers with activation functions to enhance object detection.
Target classification uses Resnet as a tool. The issue of the network advancing in a deeper manner without gradient explosion is resolved by the formation of the ResNet network. As is well known, DCNN excels at extracting low-, medium-, and high-level characteristics from images. Accuracy is improved by stacking more layers. The residual module, which is made up of two dense layers and a skip connection, is the main point of ResNet. Each of the two dense layers has a different RELU function that activates it.
4. Experimental Results
4.1. Dataset
If the dataset is limited and the data does not contain various images, i.e., photos of different classes, training neural networks on them is extremely difficult. If the dataset is heavily skewed, it will not meet the goal and may give us an incorrect impression of accomplishment. Fortunately, the HAM10000 dataset is utilized. The HAM10000 dataset has been made available to the general public to aid dermatoscopic image recognition research. 10015 dermatoscopic images from the ISIC collection make up the HAM dataset. Multiple procedures are used to obtain HAM dermatoscopic images from varied populations. This dataset can be used to identify benign keratosis lesions (BKL), melanoma (MEL), vascular lesions (VASC), basal cell carcinoma (BCC), actinic keratosis (AKIEC), dermatofibroma (DF), and melanocytic nevi (NV). These classes are added to the data configuration file, and the number of classes is changed to seven. Most of these lesions are confirmed by histopathology. The dataset is split into two sets, as listed in
Table 2.
4.3. Performance Metrics
The evaluation outcomes of the trained model are computed using several performance indicators. Precision, often referred to as positive predictive value, is defined in Equation. (2). It is a metric used in this study to show how well the model detects skin lesions. The recall rate in Equation. (3) is also called sensitivity. It is determined by dividing the number of skin lesions genuinely detected by the sum of skin lesions that are both indeed discovered and missed in each image. The harmonic mean of recall and precision is used to determine the dice similarity coefficient (DSC) using Equation. (4). Specificity is also calculated, which is the true negative rate using Equation. (5) and accuracy by using Equation. (6). As shown below, false negatives (FN), false positives (FP), true negatives (TN) and true positives (TP) are all used to calculate them.
By averaging the average precision (AP) of each class, the mean average precision (MAP) is obtained using Equation. (7). It is frequently used to assess how well object detection algorithm’s function. The MAP formula comprises various submatrices, including the confusion matrix, GIOU, recall, and precision. The detection model's ground truth bounding boxes overlap the anticipated and actual objects, and GIOU measures this overlap. Each IOU threshold value results in a unique MAP. Therefore, this value needs to be provided. An IOU is compared to a defined threshold, and either a correct or incorrect detection is generated. The performance of the trained algorithm is assessed using GIOU criteria of 0.5 and 0.5:0.95 to examine the efficiency of the skin lesions model in a set of experiments.
4.4. Results
Images are given learnable weights and biases in this stage of preprocessing. The YOLOv5 algorithm is then utilized with the initial configuration as presented in
Table 4. Because of its quick execution, it is possible to use the YOLO-trained model in real-time with a prediction in a split second. In the first run, the 12519 dermoscopy photos from seven different types of skin cancer in the HAM10000 dataset were split into training and testing sets, with training sets accounting for 80% of the dataset's total data and testing sets for 20%. The HAM dataset is tested over seven classes (BKL, AKIEC, VASC, BCC, DF, NV, and MEL) using 9514 dermoscopy images from the original training run and 3005 from the testing run. The network is trained for a total of 300 epochs. Six performance measures are generated separately for each class to assess the proposed method's performance. As a result, the average of these values is computed. The performance metrics average is 98.1%, 97.5%, 97.7%, 98.9%, 97.5%, 97.1%, and 96.3% for precision, recall, DSC, specificity, accuracy, MAP from 0.0 to 0.5, and MAP from 0.5 to 0.95, respectively. With the help of the settings in
Table 4 for the first experiment run,
Table 5 displays the metrics findings for the HAM10000 dataset.
The same splitting ratio of 80% training and 20% testing is used for the second run, which uses new parameters. The new parameters are 100, 32, and 0.0001 for epochs, batch size, and learning rate, respectively. The greatest results are achieved with a batch size of 32. So, the performance metrics average is 99.0%, 98.6%, 98.8%, 99.5%, 99.8%, 98.3%, and 98.7% for precision, recall, DSC, accuracy, specificity, MAP from 0.0 to 0.5, and MAP from 0.5 to 0.95, respectively as in
Table 6.
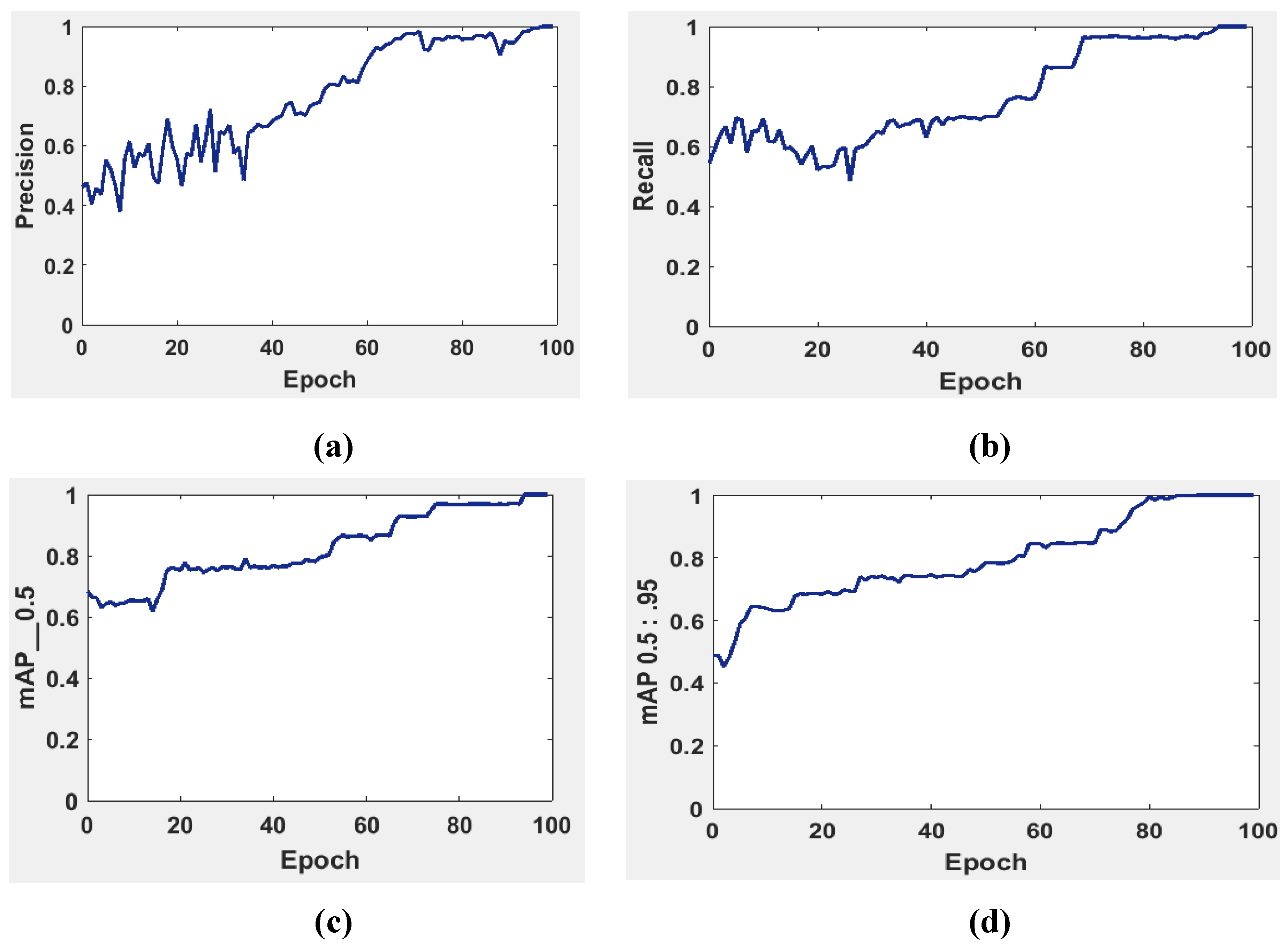
It is possible to determine the sensitivity of the neural network in
Figure 5 using the MAP to summarize the study's findings.
The precision and recall curves in
Figure 5a and
Figure 5b are implemented at network size and evaluated at the GIOU threshold range from 0.5 to 0.95. The model worked well throughout, with the greatest MAP from 0.5 to 0.95 value of 98.7% occurring for network size 224 with a threshold value equal to 0.5. Additionally, the weights produced by YOLOv5 (S-Model) require 14 MB.
The MAP determines the area under the precision-recall curve, making it a useful tool for comparing various models regardless of confidence level.
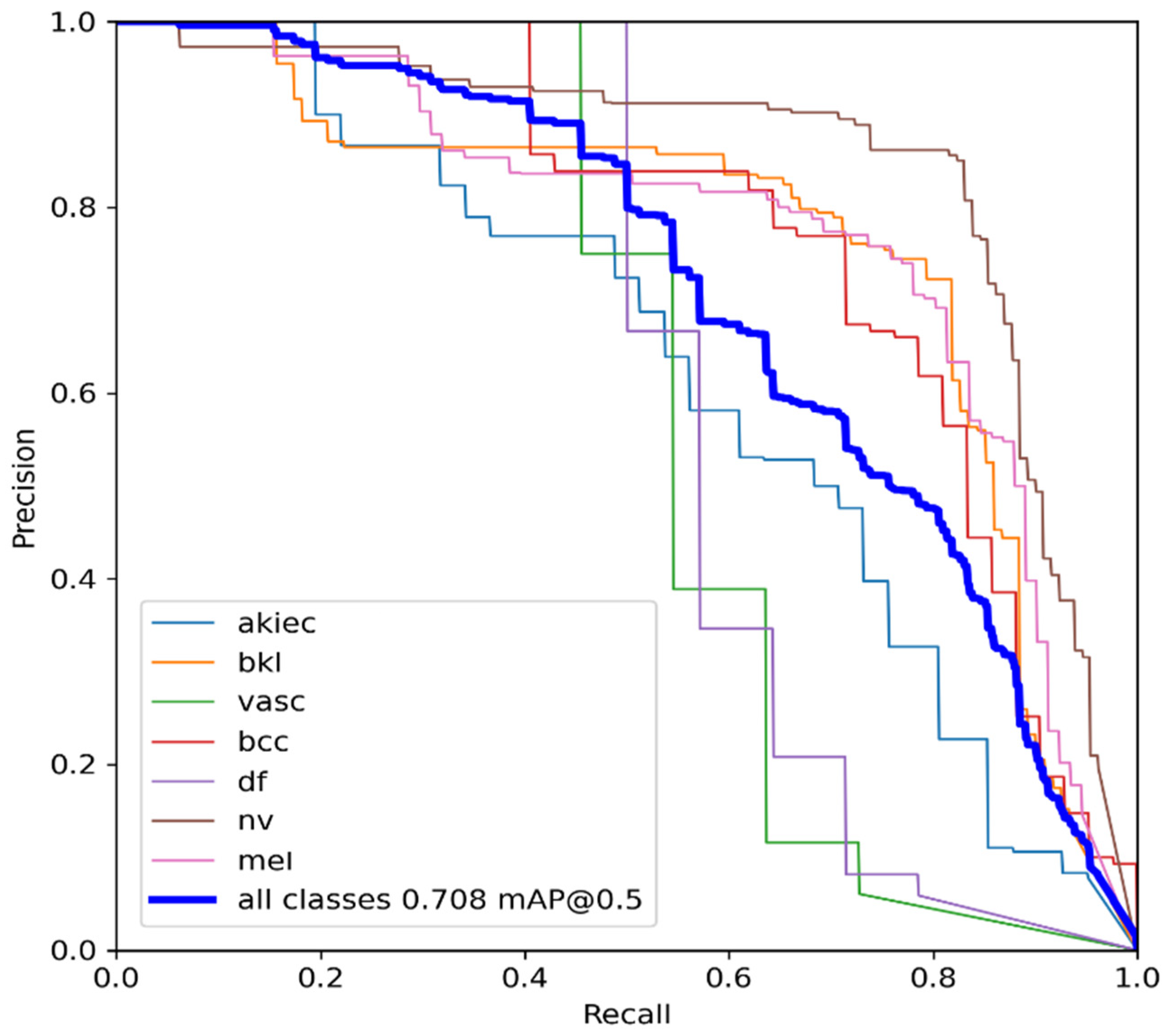
Figure 5a and
Figure 5b illustrate how recall and precision grow with increasing epochs. Additionally, when the confidence score for each class differs, the effectiveness of the melanoma detection performed by the YOLOv5 models is evaluated by looking at the precision-recall curve. When precision retains a considerable contribution to growth in the recall, it is easier to evaluate the capacity to predict melanoma. The goal is to find the confidence level that maximizes F1 across all classes. In this case, the results are shown in
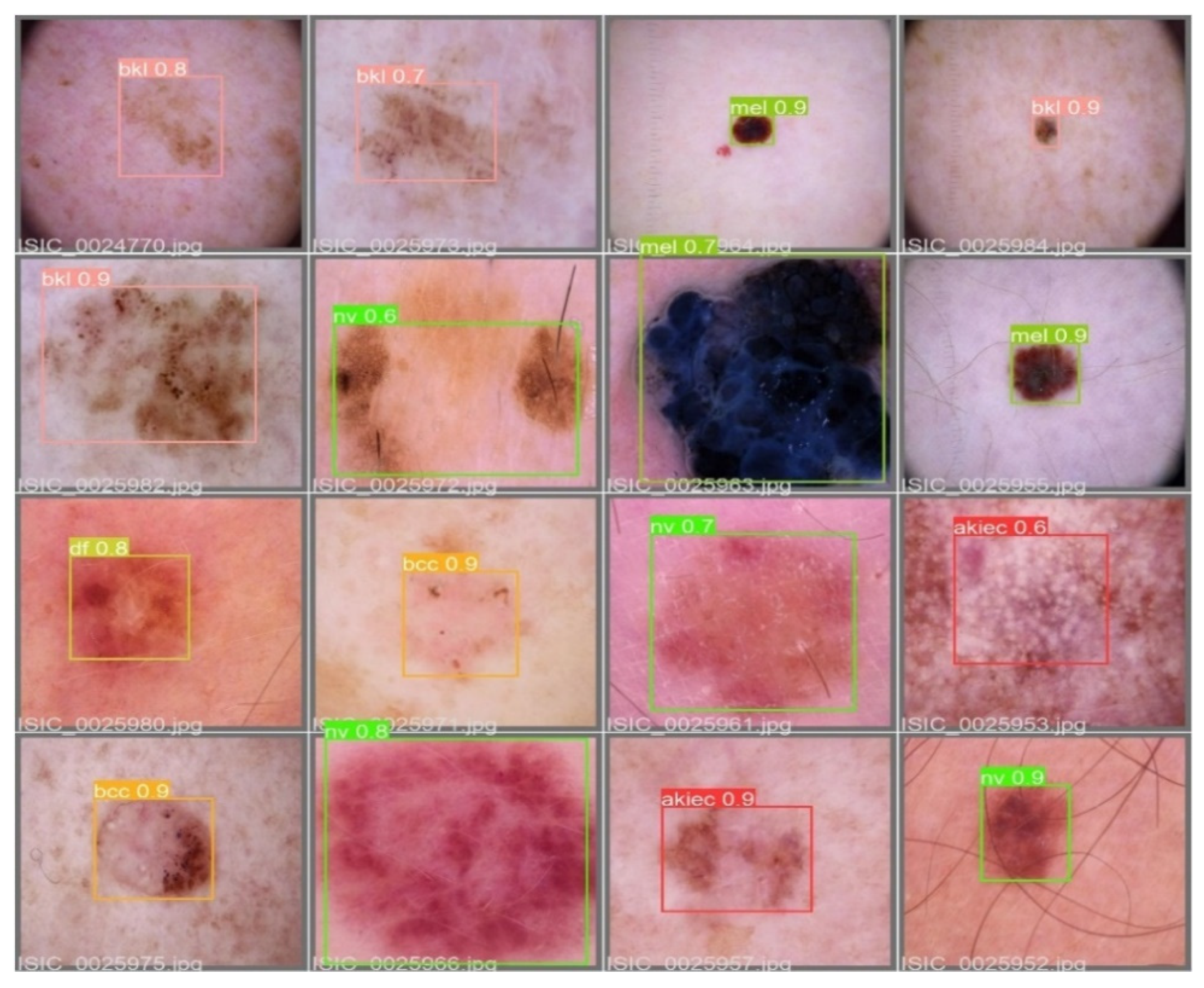
Figure 6 with a confidence of 70.8%, a precision of 96.0%, and a recall of 91.0%. In the illustration in
Figure 7, batch selections are made from the testing set to display the bounding box for each class with each probability.
5. Discussion
In order to assess the performance of the suggested model as a melanoma detection approach utilizing modified YOLOv5 and ResNet techniques, a comparison analysis is presented in this section. In
Table 7, our model is compared with other models. Despite the similarities between classes, the suggested YOLOv5 has detected melanoma using adequate coordinates, including the bounding box. The comparison results allow us to conclude that the suggested YOLOv5 model is reliable for melanoma detection in real-time photos that have been gathered.
The results show that our model achieves more accuracy, better performance, and a more accurate network. In the comparison, studies used different methods with different and the same dataset. For the HAM1000 dataset, Ali
et al. [
38] achieved 91.9% using CNN, and Khaledyan
et al. [
39] achieved 83.6% using Ensemble Bayesian Networks for the precision measure. In addition to these references, Alsaade
et al.[
37] produced a model using CNN based on the PH2 dataset, which contains 40 melanomas, 80 normal nevi, and 80 abnormal nevi as in
Table 8. The model achieves 97.5% for accuracy. Chang
et al. [
40] accomplished 94.1% using the XBG classifier using 10-fold cross validation. Despite this, our model achieves its best performance using two-fold cross-validation. Kawahara
et al. [
41] used 1700 photos from the ISIC-ISBI 2017 skin analysis challenge, which were used to train our network, and 300 images were utilized to assess the network's efficiency using various hyperparameters. It exhibited a fully convolutional neural network that could extract clinical dermoscopic features from photos of dermoscopy skin lesions. It redefined the segmentation process for categorizing clinical dermoscopic characteristics within super pixels. This model achieved 98.0% for accuracy.
A mask RCNN-based model was proposed by Khan et al. [
42]. The decorrelation formulation algorithm was used to perform the initial preprocessing of the dermoscopy images. Following that, it forwarded the obtained pictures to the MASK-RCNN for lesion segmentation. In this step, the segmented RGB pictures are produced from the ground truth images of the ISIC datasets, the MASK RCNN model was trained. The DenseNet deep model was given the segmented images as a response to extract features from [
43]. It used a Mobile Net model that was transfer-learned and fine-tuned on 10,015 dermoscopy pictures from the HAM10000 dataset after being pre-trained on roughly 1,280,000 images from the 2014 ImageNet Challenge. In
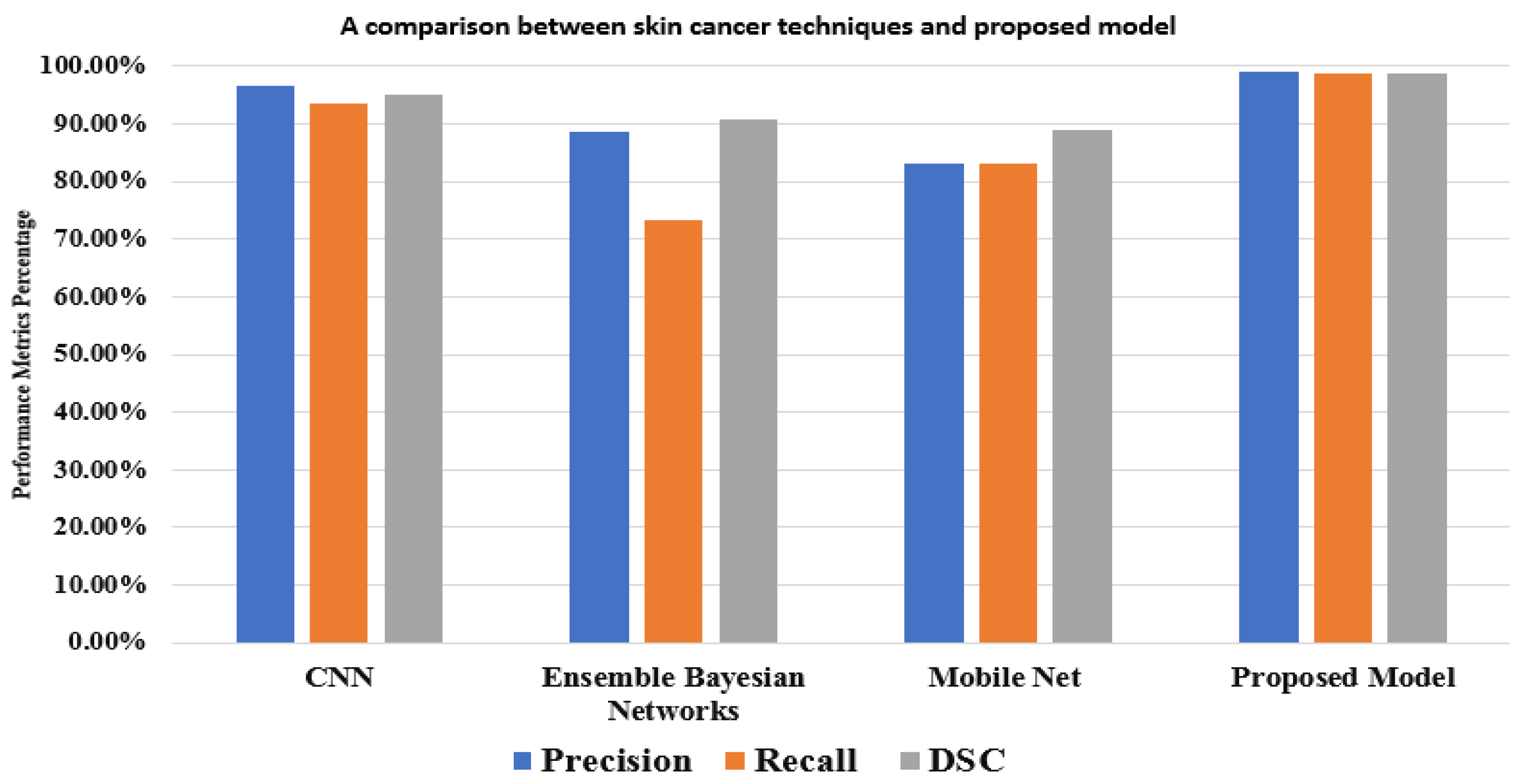
Figure 8, the outcomes of the proposed model and various models based on the same dataset (HAM10000) are displayed. These comparisons demonstrate that, in performance matrices, the proposed model outperforms CNN, Ensemble Bayesian, and Mobile Net approaches.
6. Conclusion
Early detection and drastic treatment of melanoma are challenging for professionals, and sometimes, even when presented with identical dermoscopy photos, different experts may reach different results. As a result, the study of skin cancer classification significantly impacts skin cancer secondary diagnosis. In order to analyze the skin lesion image data, this paper primarily examined the categorization of skin lesion images using the HAM10000 database. It contains many challenges, such as the similarity between classes of skin lesions, low contrast, and hair, which appears in some images. The proposed model is based on a small model from the YOLOv5 and Resnet networks. To classify seven skin lesions and detect melanoma using a bounding box provided with probability. The model consists of three stages to get the best categorization accuracy possible: preprocessing, hyperparameters, additional layers, previewing, and annotating images. The third stage assigns labels with probability classes to each image for diagnosis. Finally, the average performance metrics are 99.0%, 98.6%, 98.8%, 98.3%, and 98.7% for precision, recall, DSC, MAP from 0.0 to 0.5, and MAP from 0.5 to 0.95, respectively. Along with the recent studies for skin cancer diagnosis, the researchers hope to increase their success in future work by enhancing the model with patients' individualized data like genes and color. Additional melanoma types and bigger datasets are desperately needed. Additionally, generalizable outcomes are required to test the model against a broader range of skin conditions and make the application practical in most health organizations. It helps doctors, especially undertrained doctors, with guidelines to determine which classes are found and each probability to determine the degree of disease. It also saves time compared with traditional methods. It assists patients in making self-examinations for guidance and follows their status and treatment.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Figure S1: title; Table S1: title; Video S1: title.
Author Contributions
Conceptualization, M.E., A.Eln., and M.M.E.; methodology, M.E., A.Eln, and M.M.E.; software, M.E. and M.S.; validation, A.Elg., A.G.S. and M.O.; formal analysis, M.E., M.O. and M.M.E.; investigation, A.Elg., A.G.S. and M.O.; resources, M.M.E., M.S. and A.Elg.; data curation, M.E. and A.Eln.; writing—original draft preparation, M.E., A.Eln., and M.M.E.; writing—review and editing, A.G.S., M.O., M.M.E., and A.Elg.; visualization, M.M.E; supervision, M.M.E. and M.S.; project administration, M.M.E. and M.S.; funding acquisition, A.G.S. and M.O. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Park, S. Biochemical, Structural and Physical Changes in Aging Human Skin, and Their Relationship. 2022,
23, 275-288,. [CrossRef]
- Liu, L.; Tsui, Y.Y.; Mandal, M. Skin Lesion Segmentation Using Deep Learning with Auxiliary Task. 2021,
7. [CrossRef]
- Islami, F.; Guerra, C.E.; Minihan, A.; Yabroff, K.R.; Fedewa, S.A.; Sloan, K.; Wiedt, T.L.; Thomson, B.; Siegel, R.L.; Nargis, N. American Cancer Society's Report on the Status of Cancer Disparities in the United States, 2021. 2022, 72, 112–143.
- Saleem, S.M.; Abdullah, A.; Ameen, S.Y.; Sadeeq, M.A.M.; Zeebaree, S.R.M. Multimodal Emotion
Recognition Using Deep Learning. 2021, 2. [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Skin Cancer Classification Using Deep Learning and Transfer Learning.; IEEE: Cairo, Egypt, July 3, 2018. [Google Scholar] [CrossRef]
- Premaladha, J.; Ravichandran, K. Novel Approaches for Diagnosing Melanoma Skin Lesions Through Supervised and Deep Learning Algorithms. 2016, 40, 1–12.
- Lee, H.; Chen, Y.-P.P. Image Based Computer Aided Diagnosis System for Cancer Detection. 2015, 42. [CrossRef]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. 2019, 8, 4171–4181.
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A Survey of Deep Learning and Its Applications: A New Paradigm to Machine Learning. 2020; 27. [Google Scholar] [CrossRef]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep Learning, Sparse Coding, and
SVM for Melanoma Recognition in Dermoscopy Images. In Proceedings of the International workshop on
machine learning in medical imaging; Springer: Munich, Germany, July 3, 2015. [CrossRef]
- Gessert, N.; Sentker, T.; Madesta, F. ; Schmitz,. digger.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin Lesion Diagnosis Using Ensembles, Unscaled Multi-crop Evaluation and Loss Weighting.. 2018.
- Waheed, Z.; Waheed, A.; Zafar, M.; Riaz, F. An Efficient Machine Learning Approach for the Detection of Melanoma Using Dermoscopic Images.; IEEE: Islamabad, Pakistan, July 3 2017. [Google Scholar]
- Roy, S.; Meena, T.; Lim, S.-J. Demystifying Supervised Learning in Healthcare 4. 0: A New Reality of Transforming Diagnostic Medicine.. 2022, 12, 2549. [Google Scholar]
- Srivastava, V.; Kumar, D.; Roy, S. A Median Based Quadrilateral Local Quantized Ternary Pattern Technique for the Classification of Dermatoscopic Images of Skin Cancer. 2022, 102, 108259.
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 Dataset, a Large Collection of Multi-source
Dermatoscopic Images of Common Pigmented Skin Lesions. 2018, 5, 1–9. [CrossRef]
- The HAM10000 Dataset, a Large Collection of Multi-sources Dermatoscopic Images of Common Pigmented Skin Lesions Available online: https://github.com/ptschandl/HAM10000_dataset.
- Romero Lopez, A.; Giro Nieto, X.; Burdick, J.; Marques, O. Skin Lesion Classification from Dermoscopic Images Using Deep Learning Techniques.; ACTA Press: Innsbruck, Austria, January 1 2017. [Google Scholar]
- Dreiseitl, S.; Ohno-Machado, L.; Kittler, H.; Vinterbo, S.A.; Billhardt, H.; Binder, M. A Comparison of
Machine Learning Methods for the Diagnosis of Pigmented Skin Lesions. 2001, 34, 28–36. [CrossRef]
- Hekler, A.; Utikal, J.; Utikal, J.; Enk, A.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C. Deep Learning Outperformed 11 Pathologists in the Classification of Histopathological
Melanoma Images. 2019, 118. [CrossRef]
- Pham, T.-C.; Luong, C.-M.; Visani, M.; Hoang, V.-D. Deep CNN and Data Augmentation for Skin Lesion Classification. In Proceedings of the Intelligent Information and Database Systems; Springer: Dong Hoi City, Vietnam, July 3 2018. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.-A. Automated Melanoma Recognition in Dermoscopy Images
via Very Deep Residual Networks. 2017, 36. [CrossRef]
- Li, Y.; Shen, L. Skin Lesion Analysis Towards Melanoma Detection Using Deep Learning Network. 2018,
18. [CrossRef]
- Seeja, R.D.; Suresh, A. Deep Learning Based Skin Lesion Segmentation and Classification of Melanoma
Using Support Vector Machine (SVM). 2019, 20. [CrossRef]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. Depict Melanoma Deep-class: A Deep Convolutional Neural
Networks Approach to Classify Skin Lesion Images. 2020, 21. [CrossRef]
- Inthiyaz, S.; Altahan, B.R.; Ahammad, S.H.; Rajesh, V.; Kalangi, R.R.; Smirani, L.K.; Hossain, M.A.; Rashed, A.N.Z. Skin Disease Detection Using Deep Learning. In Proceedings of the Advances in Engineering Software; Elsevier, July 3 2023; Vol. 175. [Google Scholar]
- Mohammed, M.A.; Lakhan, A.; Abdulkareem, K.H. ; Bego, ; Garcia-Zapirain,. A Hybrid Cancer Prediction Based on Multi-omics Data and Reinforcement Learning State Action Reward State Action (SARSA). 2023, 154, 106617. [Google Scholar]
- Wu, X.; Sahoo, D.; Hoi, S.C.H.; Hoi, S.C.H. Recent Advances in Deep Learning for Object Detection. 2020,
396. [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. Efficientdet: Scalable and Efficient Object Detection. In Proceedings of the
Computer Vision and Pattern Recognition; IEEE: Seattle, Washington, USA, July 3, 2020.
- Thuan, D. Evolution of Yolo Algorithm and Yolov5: The State-of-the-art Object Detection Algorithm, 2021.
- Jung, H.-K.; Choi, G.-S. Improved Yolov5: Efficient Object Detection Using Drone Images Under Various Conditions. 2022, 12, 7255.
- Liu, K.; Tang, H.; He, S.; Yu, Q.; Xiong, Y.; Wang, N. Performance Validation of YOLO Variants for Object Detection; Proceedings of the 2021 International Conference on bioinformatics and intelligent computing: Harbin, China, 2021. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Liao, H.-Y.M.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. Cspnet: A New Backbone That Can Enhance Learning Capability of CNN.; IEEE Computer Society: Seattle, WA, USA, June 14 2020. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual
Recognition. 2015, 37. [CrossRef]
- Hafiz, A.M.; Bhat, G.M. A Survey on Instance Segmentation: State of the Art. 2020, 9. [CrossRef]
- Redmon, J.; Farhadi, A. Yolov3: An Incremental Improvement. 2018.
- Gao, H.; Huang, H. Stochastic Second-order Method for Large-scale Nonconvex Sparse Learning Models.; International Joint Conferences on Artificial Intelligence Organization: Cincinnati, Ohio, USA, July 1, 2021. [Google Scholar]
- Alsaade, F.W.; Aldhyani, T.H.; Al-Adhaileh, M.H. Developing a Recognition System for Diagnosing Melanoma Skin Lesions Using Artificial Intelligence Algorithms. 2021, 1–20.
- Ali, S.; Miah, S.; Miah, S.; Haque, J.; Rahman, M.; Islam, K. An Enhanced Technique of Skin Cancer
Classification Using Deep Convolutional Neural Network with Transfer Learning Models. 2021, 5. [CrossRef]
- Khaledyan, D.; Tajally, A.; Sarkhosh, A.; Shamsi, A.; Asgharnezhad, H.; Khosravi, A.; Nahavandi, S. Confidence Aware Neural Networks for Skin Cancer Detection. 2021. [Google Scholar] [CrossRef]
- Chang, C.-C.; Li, Y.-Z.; Wu, H.-C.; Tseng, M.-H. Melanoma Detection Using XGB Classifier Combined with Feature Extraction and K-means SMOTE Techniques. 2022, 12, 1747.
- Kawahara, J.; Hamarneh, G. Fully Convolutional Neural Networks to Detect Clinical Dermoscopic
Features. 2019, 23. [CrossRef]
- Khan, M.A.; Akram, T.; Zhang, Y.; Sharif, M. Attributes Based Skin Lesion Detection and Recognition: A
Mask RCNN and Transfer Learning-based Deep Learning Framework. 2021, 143. [CrossRef]
- Chaturvedi, S.S.; Gupta, K.; Prasad, P.S. Skin Lesion Analyzer: An Efficient Seven-way Multi-class Skin Cancer Classification Using Mobilenet; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
Figure 1.
Skin lesions classification with common types.
Figure 1.
Skin lesions classification with common types.
Figure 2.
The ABCD lesion diagnosis criteria focus on identifying specific.
Figure 2.
The ABCD lesion diagnosis criteria focus on identifying specific.
Figure 3.
The proposed framework for categorizing seven skin lesions.
Figure 3.
The proposed framework for categorizing seven skin lesions.
Figure 4.
The components of the YOLOv5 model used for melanoma classification.
Figure 4.
The components of the YOLOv5 model used for melanoma classification.
Figure 5.
The evaluation of precision curve, recall curve, and mean average precision at two thresholds for the second run using 100 epochs,.
Figure 5.
The evaluation of precision curve, recall curve, and mean average precision at two thresholds for the second run using 100 epochs,.
Figure 6.
YOLOv5 precision-recall curves for each class. The average precision for each class is the area under each curve for the HAM10000 dataset.
Figure 6.
YOLOv5 precision-recall curves for each class. The average precision for each class is the area under each curve for the HAM10000 dataset.
Figure 7.
An example batch of detection results on some test images using YOLOv5.
Figure 7.
An example batch of detection results on some test images using YOLOv5.
Figure 8.
A comparison between the proposed model and other techniques based on the HAM10000 dataset.
Figure 8.
A comparison between the proposed model and other techniques based on the HAM10000 dataset.
Table 1.
Some examples of recent studies on related topics.
Table 1.
Some examples of recent studies on related topics.
| Reference |
Proposed Technique |
Accuracy |
Limitation |
| Premaladha et al. [6] |
Segmentation using Otsu's normalized algorithm and then classification |
SVM (90.44), DCNN (92.89), and Hybrid AdaBoost (91.73) |
Uses only three classes of skin cancer lesions |
Codella et al.
[10] |
Melanoma recognition using DL, sparse coding, and SVM |
93.1% |
Need to deepen features and add more cases of melanoma. |
Waheed et al.
[12] |
Diagnosing melanoma using the color and texture of different types of lesions |
SVM (96.0%) |
Need more attributes of skin lesions |
Hekler et al.
[19] |
Classifying histopathologic melanoma using DCNN |
68.0% |
Uses low resolution and can't differentiate between melanoma and nevi classes |
| Pham et al. [20] |
Classification using DCNN |
AUC (89.2%) |
Less sensitivity |
| Yu et al. [21] |
Segmentation and classification using DCNN and FCRN |
AUC (80.4 %) |
Insufficient training data |
| Li and Shen [22] |
Two FCRN for melanoma segmentation and classification |
AUC (91.2%) |
Overfitting in AUC and low segmentation |
| Seeja and Suresh [23] |
Segmenting data using form, color, and texture variables, then classification using SVM, RF, KNN, and NB |
SVM (85.1%), RF (82.2%), KNN(79.2%) and NB (65.9%) |
Low classification accuracy |
Nasiri et al.
[24] |
Using the 19-layer model of CNN for melanoma classification |
75.0% |
Need to enhance accuracy |
Table 2.
The distribution of the HAM10000 dataset for training and testing sets.
Table 2.
The distribution of the HAM10000 dataset for training and testing sets.
| |
Vasc |
Nv |
Mel |
Df |
Bkl |
Bcc |
Akiec |
| All images |
142 |
6705 |
1113 |
115 |
1099 |
514 |
327 |
| Train |
115 |
5360 |
891 |
92 |
879 |
300 |
262 |
| Test |
27 |
1345 |
222 |
23 |
220 |
214 |
65 |
Table 3.
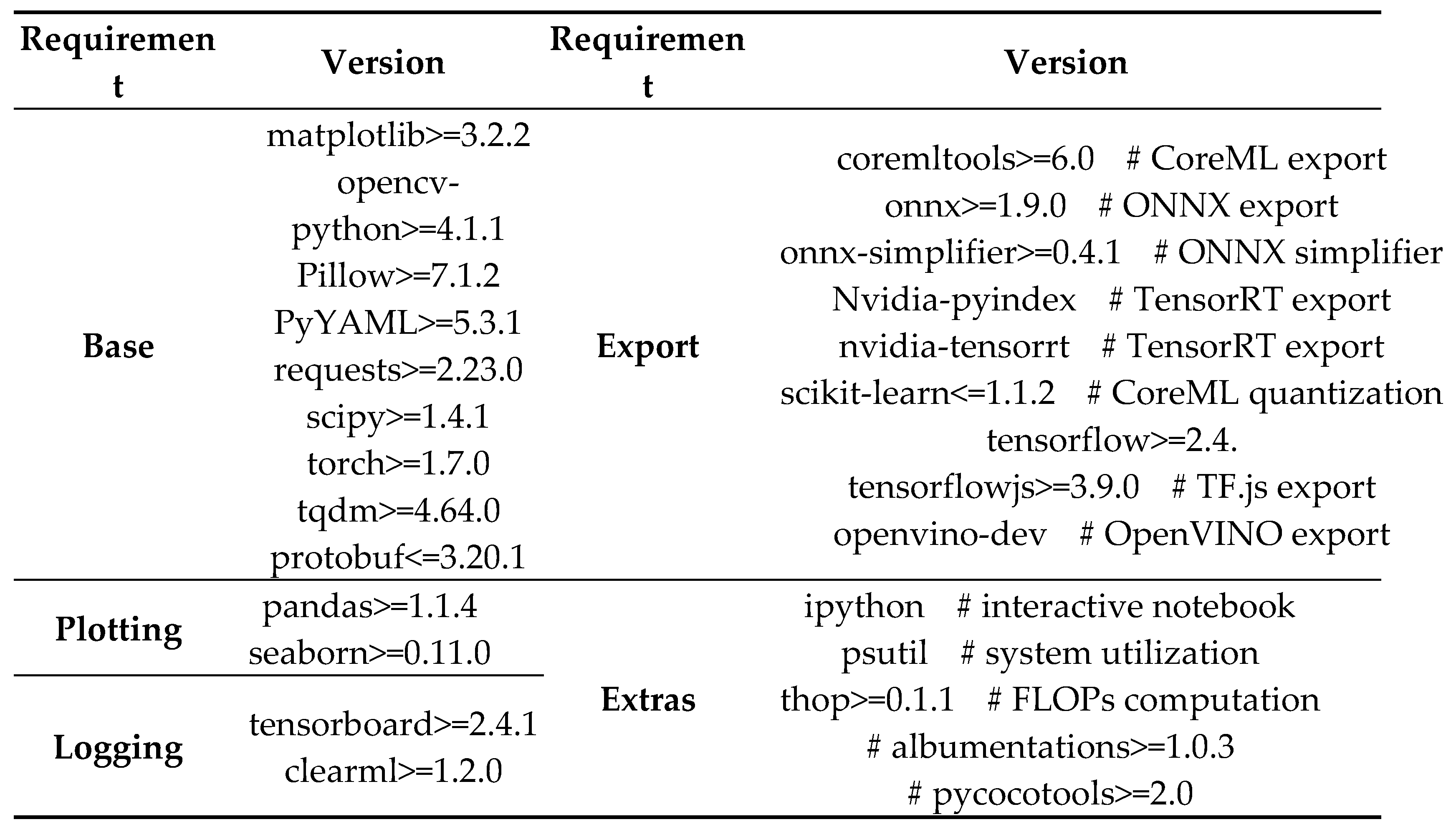
Google Colab software requirements and its versions.
Table 3.
Google Colab software requirements and its versions.
Table 4.
Training guidelines for the first and second experiments with 640 image size.
Table 4.
Training guidelines for the first and second experiments with 640 image size.
| Parameter |
First run |
Second run |
Definition |
| Epoch |
300 |
100 |
The frequency with which the learning algorithm |
| Batch_size |
16 |
32 |
how many training instances are used in a single iteration |
| lr0 |
0.001 |
0.001 |
Initial learning rate (SGD=1E-2, Adam=1E-3) |
| Lrf |
0.2 |
0.2 |
Final OneCycleLR learning rate (lr0 * lrf) |
| Momentum |
0.937 |
0.937 |
SGD momentum/Adam beta1 |
| warmup_epochs |
3.0 |
3.0 |
Warmup epochs (fractions ok) |
| weight_decay |
0.0005 |
0.0005 |
Optimizer weight decay 5e-4 |
| warmup_momentum |
0.8 |
0.8 |
Warmup initial momentum |
| warmup_bias_lr |
0.1 |
0.1 |
Warmup initial bias learning rate |
| Box |
0.05 |
0.05 |
Box loss gain |
| Cls |
0.5 |
0.5 |
Class loss gain |
| cls_pw |
1.0 |
1.0 |
Cls BCELoss positive_weight |
| Obj |
1.0 |
1.0 |
Obj loss gain (scale with pixels) |
| obj_pw |
1.0 |
1.0 |
Obj BCELoss positive_weight |
| anchor_t |
4.0 |
4.0 |
Anchor-multiple threshold |
| iou_t |
0.20 |
0.20 |
IOU training threshold |
| Scale |
0.5 |
0.5 |
Image scale (+/- gain) |
| Shear |
0.0 |
0.0 |
Image shear (+/- deg) |
| Perspective |
0.0 |
0.0 |
Image perspective (+/- fraction), range 0-0.001 |
Table 5.
Utilizing 300 epochs, the model YOLOv5s' results on the HAM10000 dataset.
Table 5.
Utilizing 300 epochs, the model YOLOv5s' results on the HAM10000 dataset.
| |
Precision (%) |
Recall (%) |
DSC (%) |
MAP 0.0:0.5 (%) |
MAP 0.5:0.95 (%) |
Accuracy (%) |
| AKIEC |
99.1 |
94.9 |
96.9 |
99.7 |
95.2 |
95.2 |
| BKL |
95.3 |
96.8 |
96.0 |
95.3 |
94.5 |
96.1 |
| VASC |
97.0 |
95.6 |
96.2 |
98.7 |
95.5 |
97.2 |
| BCC |
97.1 |
97.6 |
97.3 |
97.5 |
96.4 |
97.3 |
| DF |
98.7 |
99.5 |
99.0 |
94.3 |
94.8 |
98.8 |
| NV |
100.0 |
98.6 |
99.2 |
96.4 |
99.5 |
98.1 |
| MEL |
98.8 |
100.0 |
99.3 |
98.2 |
98.6 |
100.0 |
| Average |
98.1 |
97.5 |
97.7 |
97.1 |
96.3 |
97.5 |
Table 6.
Results of the model YOLOv5s for HAM10000 dataset utilizing 100 epochs.
Table 6.
Results of the model YOLOv5s for HAM10000 dataset utilizing 100 epochs.
| |
Precision (%) |
Recall (%) |
DSC (%) |
MAP 0.0:0.5 (%) |
MAP 0.5:0.95 (%) |
Accuracy(%) |
| AKIEC |
100.0 |
96.7 |
98.3 |
98.9 |
99.7 |
98.8 |
| BKL |
98.2 |
98.2 |
98.2 |
97.6 |
94.9 |
98.9 |
| VASC |
98.8 |
99.6 |
99.1 |
97.9 |
97.9 |
99.4 |
| BCC |
97.1 |
96.9 |
96.9 |
99.5 |
99.1 |
99.7 |
| DF |
99.6 |
98.9 |
99.2 |
98.6 |
96.2 |
100.0 |
| MV |
100.0 |
100.0 |
100.0 |
96.2 |
100.0 |
99.8 |
| MEL |
99.9 |
100.0 |
99.9 |
99.8 |
98.9 |
100.0 |
| Average |
99.0 |
98.6 |
98.8 |
98.3 |
98.7 |
99.5 |
Table 7.
The comparison of precision, recall, and DSC over some existing models.
Table 7.
The comparison of precision, recall, and DSC over some existing models.
| Reference |
Year |
Method |
Precision (%) |
Recall (%) |
DSC (%) |
Accuracy(%) |
Dataset |
|
Nasiri et al. [24] |
2020 |
KNN |
73.0 |
55.0 |
79.0 |
67.0 |
ISIC dataset |
| SVM |
58.0 |
47.0 |
66.0 |
62.0 |
| CNN |
77.0 |
73.0 |
78.0 |
75.0 |
|
Alsaade et al. [37] |
2021 |
CNN |
81.2 |
92.9 |
87.5 |
97.5 |
PH2 |
|
Ali et al. [38] |
2021 |
CNN |
96.5 |
93.6 |
95.0 |
91.9 |
HAM10000 |
|
Khaledyan et al. [39] |
2021 |
Ensemble Bayesian Networks |
88.6 |
73.4 |
90.7 |
83.6 |
HAM10000 |
|
Chang et al. [40] |
2022 |
XGB classifier |
97.4 |
87.8 |
90.5 |
94.1 |
ISIC |
|
Kawahara et al. [41] |
2019 |
FCNN |
97.6 |
81.3 |
93.0 |
98.0 |
ISIC |
|
Khan et al. [42] |
2021 |
Mask RCNN |
88.5% |
88.5% |
88.6% |
93.6 |
ISIC |
|
Chaturvedi et al. [43] |
2020 |
Mobile Net |
83.0% |
83.0% |
89.0% |
83.1 |
HAM10000 |
| Proposed model |
2022 |
YOLOv5+ResNet |
99.0 |
98.6 |
98.8 |
99.5 |
HAM10000 |
Table 8.
The details of datasets that are used for performance evaluation.
Table 8.
The details of datasets that are used for performance evaluation.
| Database |
Description |
| PH2 |
It contains the medical assessment, manual segmentation, and identification of a number of dermoscopic lesions. In a collection of 200 dermoscopic photos, it was completed by dermatologists with expertise. The photos have an 8-bit RGB color depth and a 768x560 pixel resolution. It has 40 melanomas, 80 atypical nevi, and 80 normal nevi. |
| ISIC |
2,000 dermoscopic pictures of cancers. It contains malignant and harmless oncological illnesses. 1,372 benign nevi, 254 seborrheic keratoses, and 374 melanomas. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).