Submitted:
23 July 2023
Posted:
24 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
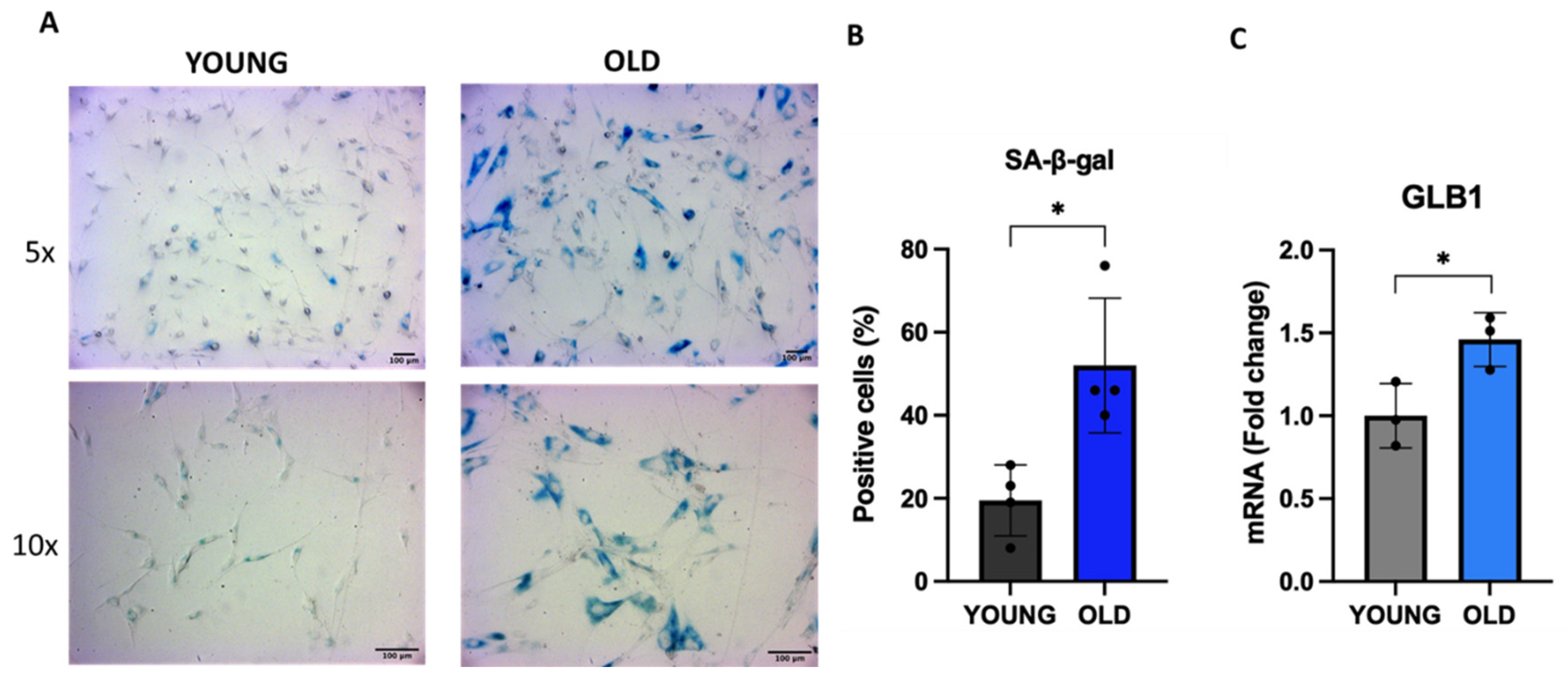
3.1. Senescent VSMCs show increased SA-β-gal activity
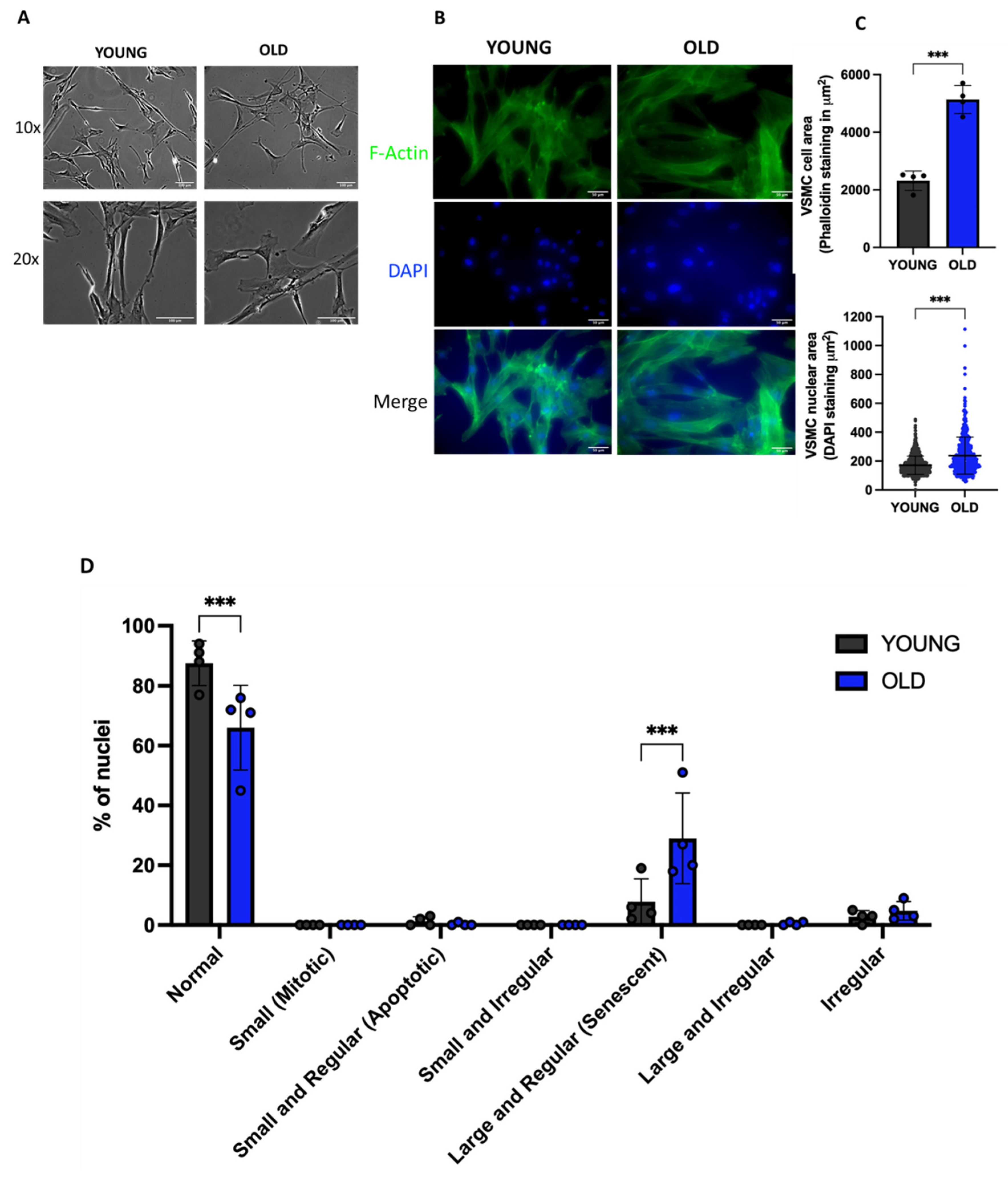
3.2. Senescent VSMCs display altered cell and nuclei morphology
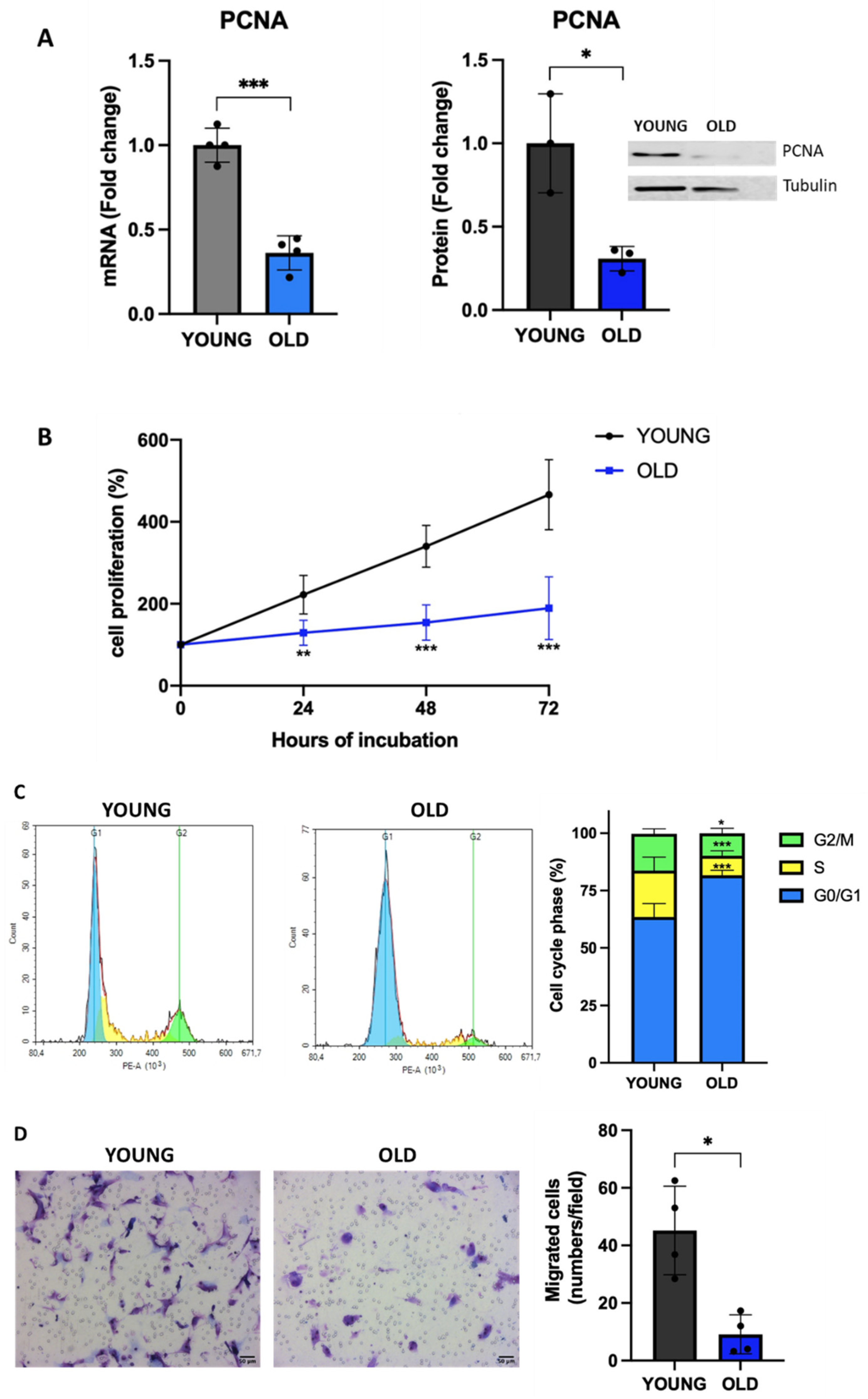
3.3. Senescent VSMCs display reduced proliferation and cell cycle arrest
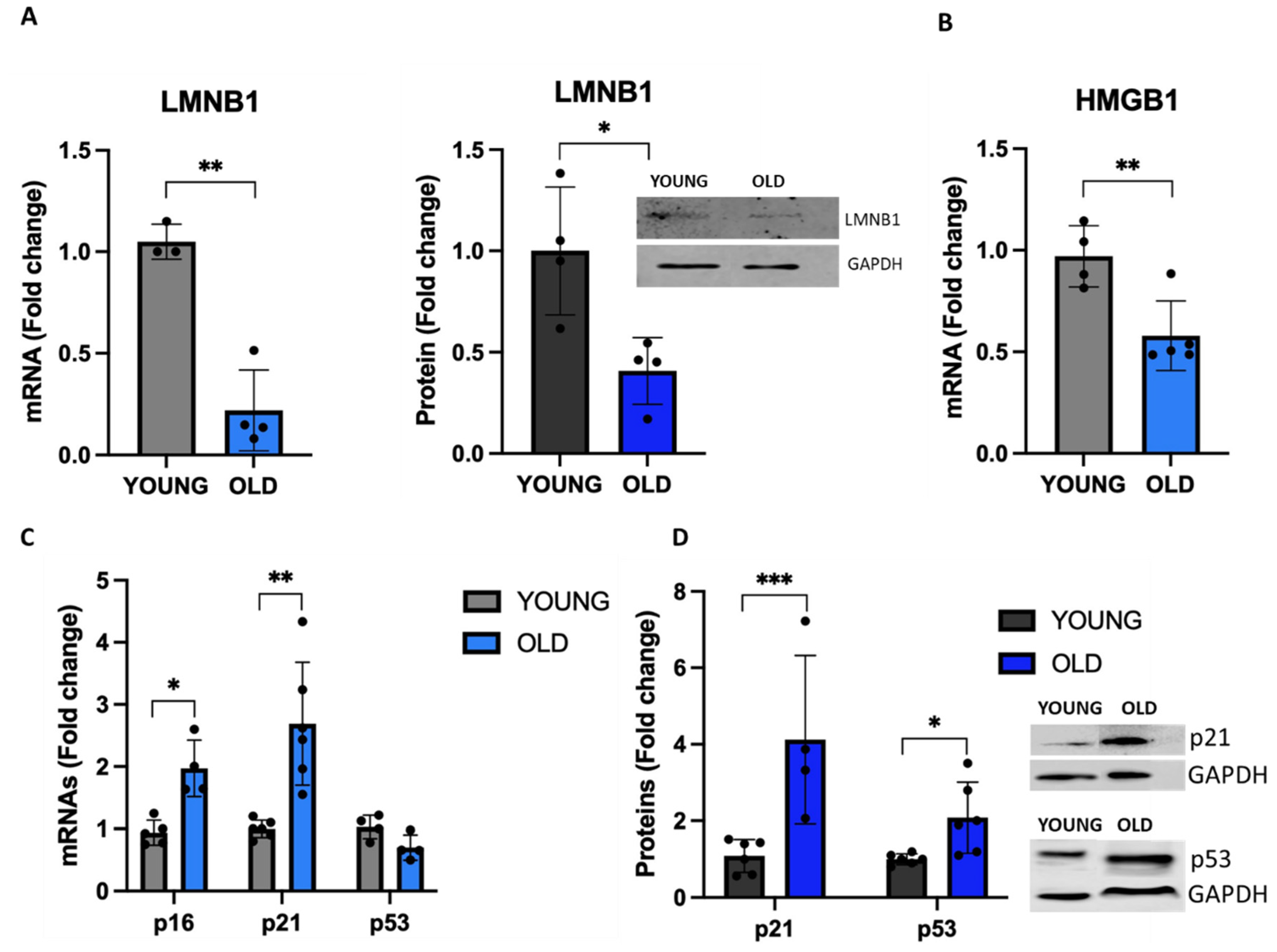
3.4. Senescent VSMCs express well-known senescence biomarkers
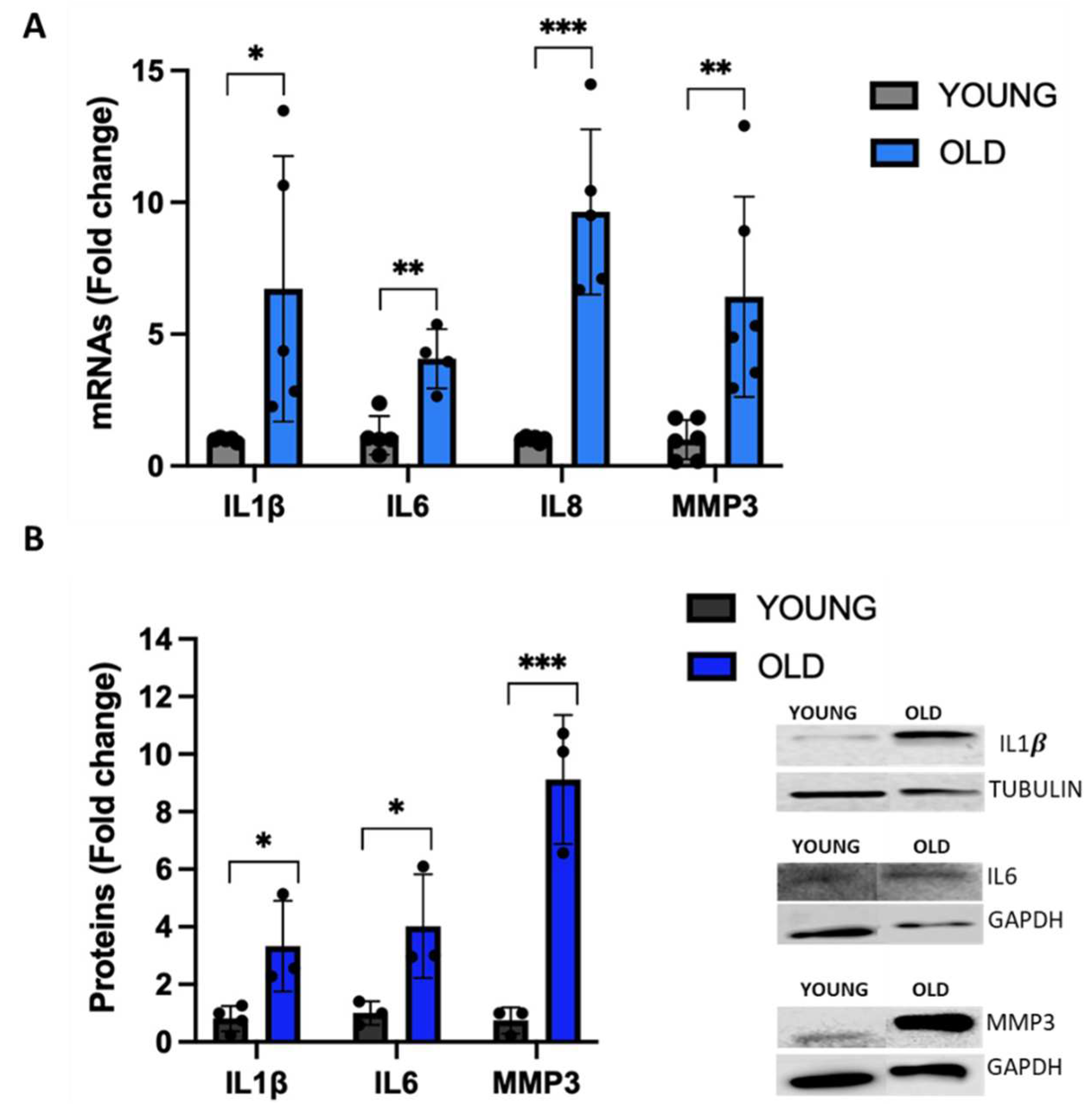
3.5. Senescent VSMCs exhibit a change in inflammatory biomarkers
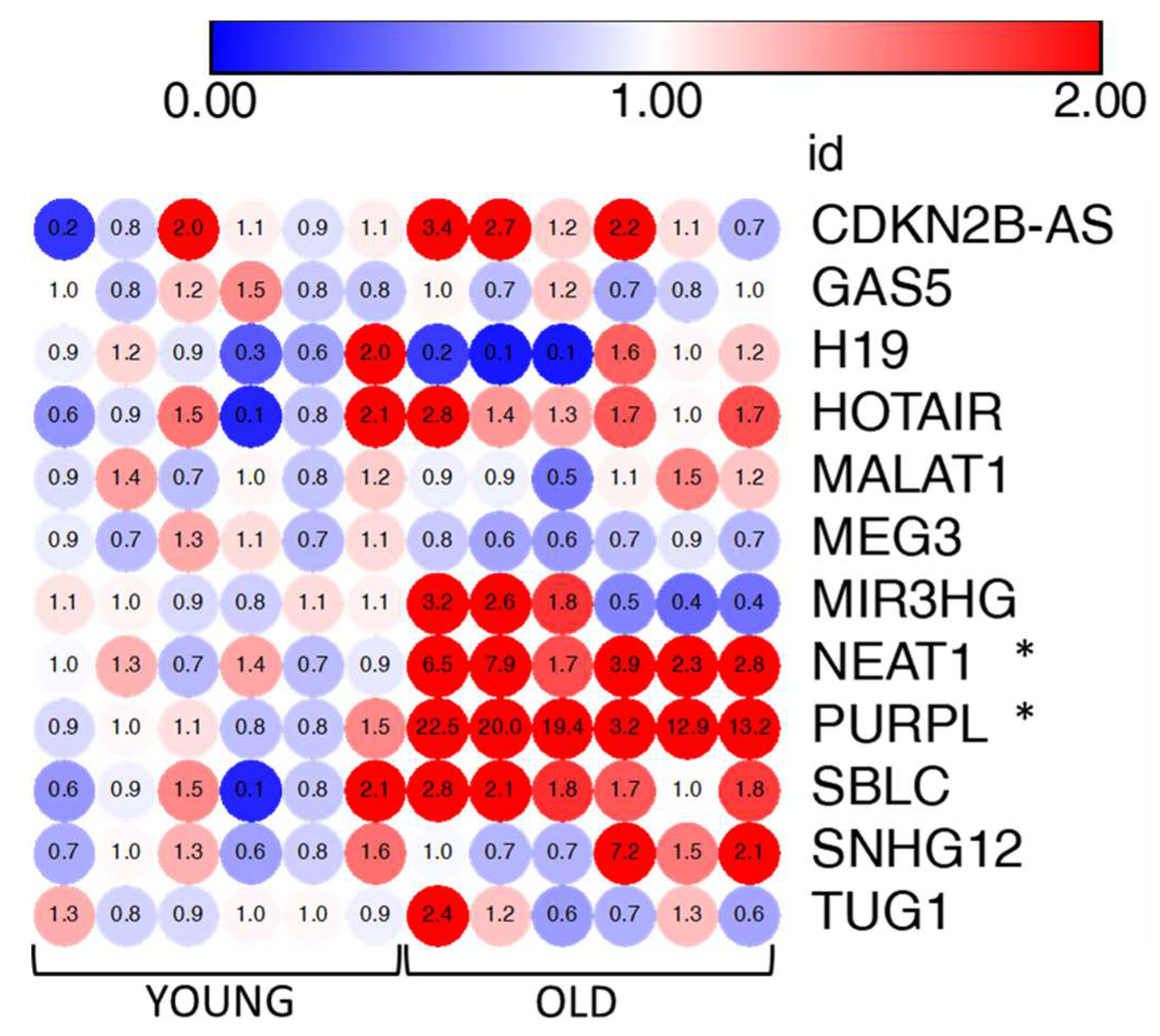
3.6. LncRNAs PURPL and NEAT1 are markers of senescent VMSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Chaib, S.; Tchkonia, T.; Kirkland, J.L. Cellular Senescence and Senolytics: The Path to the Clinic. Nat Med 2022, 28, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.G.A.; Krizhanovsky, V. Physiological and Pathological Consequences of Cellular Senescence. Cellular and Molecular Life Sciences 2014, 71, 4373–4386. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Grootaert, M.O.J.; Bennett, M.R. Vascular Smooth Muscle Cells in Atherosclerosis: Time for a Re-Assessment. Cardiovasc Res 2021, 117, 2326–2339. [Google Scholar] [CrossRef]
- Grootaert, M.O.J.; Moulis, M.; Roth, L.; Martinet, W.; Vindis, C.; Bennett, M.R.; de Meyer, G.R.Y. Vascular Smooth Muscle Cell Death, Autophagy and Senescence in Atherosclerosis. Cardiovasc Res 2018, 114, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Basatemur, G.L.; Jørgensen, H.F.; Clarke, M.C.H.; Bennett, M.R.; Mallat, Z. Vascular Smooth Muscle Cells in Atherosclerosis. Nat Rev Cardiol 2019, 16, 727–744. [Google Scholar] [CrossRef]
- Campisi, J.; d’Adda di Fagagna, F. Cellular Senescence: When Bad Things Happen to Good Cells. Nat Rev Mol Cell Biol 2007, 8, 729–740. [Google Scholar] [CrossRef]
- Chi, C.; Li, D.-J.; Jiang, Y.-J.; Tong, J.; Fu, H.; Wu, Y.-H.; Shen, F.-M. Vascular Smooth Muscle Cell Senescence and Age-Related Diseases: State of the Art. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2019, 1865, 1810–1821. [Google Scholar] [CrossRef]
- Ngoi, N.YL.; Liew, A.QX.; Chong, S.J.F.; Davids, M.S.; Clement, M.-V.; Pervaiz, S. The Redox-Senescence Axis and Its Therapeutic Targeting. Redox Biol 2021, 45, 102032. [Google Scholar] [CrossRef]
- Basisty, N.; Kale, A.; Jeon, O.H.; Kuehnemann, C.; Payne, T.; Rao, C.; Holtz, A.; Shah, S.; Sharma, V.; Ferrucci, L.; et al. A Proteomic Atlas of Senescence-Associated Secretomes for Aging Biomarker Development. PLoS Biol 2020, 18, e3000599. [Google Scholar] [CrossRef]
- Stojanović, S.D.; Fuchs, M.; Kunz, M.; Xiao, K.; Just, A.; Pich, A.; Bauersachs, J.; Fiedler, J.; Sedding, D.; Thum, T. Inflammatory Drivers of Cardiovascular Disease: Molecular Characterization of Senescent Coronary Vascular Smooth Muscle Cells. Front Physiol 2020, 11. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Lanigan, F.; Geraghty, J.G.; Bracken, A.P. Transcriptional Regulation of Cellular Senescence. Oncogene 2011, 30, 2901–2911. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat Rev Genet 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular Functions of Long Noncoding RNAs. Nat Cell Biol 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat Rev Genet 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Perry, R.B.-T.; Ulitsky, I. The Functions of Long Noncoding RNAs in Development and Stem Cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef] [PubMed]
- Puvvula, P.K. LncRNAs Regulatory Networks in Cellular Senescence. Int J Mol Sci 2019, 20, 2615–2634. [Google Scholar] [CrossRef]
- Tan, P.; Guo, Y.-H.; Zhan, J.-K.; Long, L.-M.; Xu, M.-L.; Ye, L.; Ma, X.-Y.; Cui, X.-J.; Wang, H.-Q. LncRNA-ANRIL Inhibits Cell Senescence of Vascular Smooth Muscle Cells by Regulating MiR-181a/Sirt1. Biochemistry and Cell Biology 2019, 97, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wu, J.; Wang, X.; Song, C. Noncoding RNAs in Vascular Aging. Oxid Med Cell Longev 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Damiani, I.; Castiglioni, S.; Sochaj-Gregorczyk, A.; Bonacina, F.; Colombo, I.; Rusconi, V.; Otlewski, J.; Corsini, A.; Bellosta, S. Purification and In Vitro Evaluation of an Anti-HER2 Affibody-Monomethyl Auristatin E Conjugate in HER2-Positive Cancer Cells. Biology (Basel) 2021, 10, 758. [Google Scholar] [CrossRef]
- Piegari, E.; De Angelis, A.; Cappetta, D.; Russo, R.; Esposito, G.; Costantino, S.; Graiani, G.; Frati, C.; Prezioso, L.; Berrino, L.; et al. Doxorubicin Induces Senescence and Impairs Function of Human Cardiac Progenitor Cells. Basic Res Cardiol 2013, 108. [Google Scholar] [CrossRef] [PubMed]
- Filippi-Chiela, E.C.; Oliveira, M.M.; Jurkovski, B.; Callegari-Jacques, S.M.; Silva, V.D. da; Lenz, G. Nuclear Morphometric Analysis (NMA): Screening of Senescence, Apoptosis and Nuclear Irregularities. PLoS One 2012, 7, e42522. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet 2013, 9, e1003368. [Google Scholar] [CrossRef] [PubMed]
- Grammatikakis, I.; Panda, A.C.; Abdelmohsen, K.; Gorospe, M. Long Noncoding RNAs (LncRNAs) and the Molecular Hallmarks of Aging. Aging 2014, 6, 992–1009. [Google Scholar] [CrossRef]
- Montes, M.; Lubas, M.; Arendrup, F.S.; Mentz, B.; Rohatgi, N.; Tumas, S.; Harder, L.M.; Skanderup, A.J.; Andersen, J.S.; Lund, A.H. The Long Non-Coding RNA MIR31HG Regulates the Senescence Associated Secretory Phenotype. Nat Commun 2021, 12, 2459. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Tu, C.; Liu, Y. Role of LncRNAs in Aging and Age-Related Diseases. Aging Medicine 2018, 1, 158–175. [Google Scholar] [CrossRef]
- Li, X.L.; Subramanian, M.; Jones, M.F.; Chaudhary, R.; Singh, D.K.; Zong, X.; Gryder, B.; Sindri, S.; Mo, M.; Schetter, A.; et al. Long Noncoding RNA PURPL Suppresses Basal P53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep 2017, 20, 2408–2423. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome Signature of Cellular Senescence. Nucleic Acids Res 2019, 47, 7294–7305. [Google Scholar] [CrossRef]
- Xu, C.L.; Sang, B.; Liu, G.Z.; Li, J.M.; Zhang, X.D.; Liu, L.X.; Thorne, R.F.; Wu, M. SENEBLOC, a Long Non-Coding RNA Suppresses Senescence via P53-Dependent and Independent Mechanisms. Nucleic Acids Res 2020, 48, 3089–3102. [Google Scholar] [CrossRef]
- Haemmig, S.; Yang, D.; Sun, X.; Das, D.; Ghaffari, S.; Molinaro, R.; Chen, L.; Deng, Y.; Freeman, D.; Moullan, N.; et al. Long Noncoding RNA SNHG12 Integrates a DNA-PK–Mediated DNA Damage Response and Vascular Senescence. Sci Transl Med 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Valieva, Y.; Ivanova, E.; Fayzullin, A.; Kurkov, A.; Igrunkova, A. Senescence-Associated β-Galactosidase Detection in Pathology. Diagnostics 2022, 12, 2309. [Google Scholar] [CrossRef]
- Son, H.N.; Chi, H.N.Q.; Chung, D.C.; Long, L.T. Morphological Changes during Replicative Senescence in Bovine Ovarian Granulosa Cells. Cell Cycle 2019, 18, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Heckenbach, I.; Mkrtchyan, G. v.; Ezra, M. ben; Bakula, D.; Madsen, J.S.; Nielsen, M.H.; Oró, D.; Osborne, B.; Covarrubias, A.J.; Idda, M.L.; et al. Nuclear Morphology Is a Deep Learning Biomarker of Cellular Senescence. Nat Aging 2022, 2, 742–755. [Google Scholar] [CrossRef]
- Machado-Oliveira, G.; Ramos, C.; Marques, A.R.A.; Vieira, O. v. Cell Senescence, Multiple Organelle Dysfunction and Atherosclerosis. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Moriwaki, S.; Sugiyama, Y.; Endo, Y.; Yamazaki, K.; Mori, T.; Takigawa, M.; Inoue, S. Decreased Gene Expression Responsible for Post-Ultraviolet DNA Repair Synthesis in Aging: A Possible Mechanism of Age-Related Reduction in DNA Repair Capacity. Journal of Investigative Dermatology 2005, 124, 435–442. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef]
- Lukášová, E.; Kovařík, A.; Kozubek, S. Consequences of Lamin B1 and Lamin B Receptor Downregulation in Senescence. Cells 2018, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Sofiadis, K.; Josipovic, N.; Nikolic, M.; Kargapolova, Y.; Übelmesser, N.; Varamogianni-Mamatsi, V.; Zirkel, A.; Papadionysiou, I.; Loughran, G.; Keane, J.; et al. HMGB1 Coordinates SASP-related Chromatin Folding and RNA Homeostasis on the Path to Senescence. Mol Syst Biol 2021, 17. [Google Scholar] [CrossRef] [PubMed]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of P53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef] [PubMed]
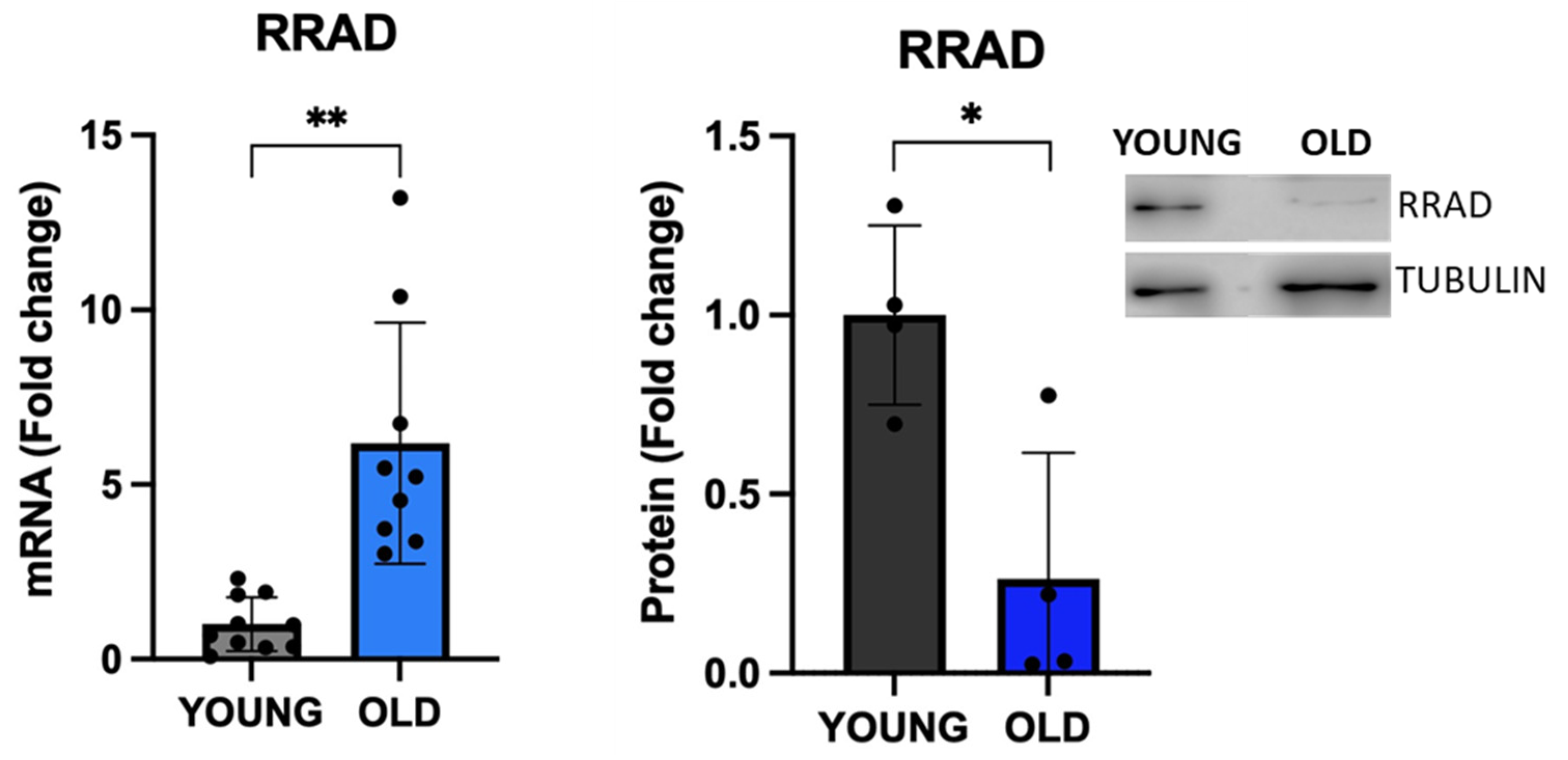
- Wei, Z.; Guo, H.; Qin, J.; Lu, S.; Liu, Q.; Zhang, X.; Zou, Y.; Gong, Y.; Shao, C. Pan-Senescence Transcriptome Analysis Identified RRAD as a Marker and Negative Regulator of Cellular Senescence. Free Radic Biol Med 2019, 130, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Ritschka, B.; Storer, M.; Mas, A.; Heinzmann, F.; Ortells, M.C.; Morton, J.P.; Sansom, O.J.; Zender, L.; Keyes, W.M. The Senescence-Associated Secretory Phenotype Induces Cellular Plasticity and Tissue Regeneration. Genes Dev 2017, 31, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.C.; Banito, A.; Wuestefeld, T.; Georgilis, A.; Janich, P.; Morton, J.P.; Athineos, D.; Kang, T.-W.; Lasitschka, F.; Andrulis, M.; et al. A Complex Secretory Program Orchestrated by the Inflammasome Controls Paracrine Senescence. Nat Cell Biol 2013, 15, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, J.; Wen, T.; Zeng, W.; Peng, C.; Yan, S.; Tan, J.; Yang, K.; Liu, S.; Guo, A.; et al. Overexpression of HMGB1 in Melanoma Predicts Patient Survival and Suppression of HMGB1 Induces Cell Cycle Arrest and Senescence in Association with P21 (Waf1/Cip1) up-Regulation via a P53-Independent, Sp1-Dependent Pathway. Oncotarget 2014, 5, 6387–6403. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, Y.; Tan, X.; Liu, W.; He, X.; Pan, D.; Li, E.; Xu, L.; Long, L. Friend or Foe: Regulation, Downstream Effectors of RRAD in Cancer. Biomolecules 2023, 13, 477. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, I.; Kim, S.T.; Lee, J.; Kim, K.-M.; Park, J.O.; Kang, W.K. RRAD Expression in Gastric and Colorectal Cancer with Peritoneal Carcinomatosis. Sci Rep 2019, 9, 19439. [Google Scholar] [CrossRef]
- Belbachir, N.; Portero, V.; al Sayed, Z.R.; Gourraud, J.-B.; Dilasser, F.; Jesel, L.; Guo, H.; Wu, H.; Gaborit, N.; Guilluy, C.; et al. RRAD Mutation Causes Electrical and Cytoskeletal Defects in Cardiomyocytes Derived from a Familial Case of Brugada Syndrome. Eur Heart J 2019, 40, 3081–3094. [Google Scholar] [CrossRef]
- Halter, B.; Gonzalez de Aguilar, J.L.; Rene, F.; Petri, S.; Fricker, B.; Echaniz-Laguna, A.; Dupuis, L.; Larmet, Y.; Loeffler, J.P. Oxidative Stress in Skeletal Muscle Stimulates Early Expression of Rad in a Mouse Model of Amyotrophic Lateral Sclerosis. Free Radic Biol Med 2010, 48, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Buccitelli, C.; Selbach, M. MRNAs, Proteins and the Emerging Principles of Gene Expression Control. Nat Rev Genet 2020, 21, 630–644. [Google Scholar] [CrossRef]
- Han, S.; Li, X.; Wang, K.; Zhu, D.; Meng, B.; Liu, J.; Liang, X.; Jin, Y.; Liu, X.; Wen, Q.; et al. PURPL Represses Autophagic Cell Death to Promote Cutaneous Melanoma by Modulating ULK1 Phosphorylation. Cell Death Dis 2021, 12, 1070. [Google Scholar] [CrossRef]
- Hartford, C.C.R.; Shrestha, R.L.; Pongor, L.; Zhao, Y.; Chen, X.; Fromont, C.; Chaudhary, R.; Li, X.L.; Pasterczyk, K.R.; Kumar, R.; et al. Context-Dependent Function of Long Noncoding RNA PURPL in Transcriptome Regulation during P53 Activation. Mol Cell Biol 2022, 42, e0028922. [Google Scholar] [CrossRef]
- Jiang, L.; Shao, C.; Wu, Q.-J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.-T.; Zhang, Y.; Wang, Y.; et al. NEAT1 Scaffolds RNA-Binding Proteins and the Microprocessor to Globally Enhance Pri-MiRNA Processing. Nat Struct Mol Biol 2017, 24, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K. V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating Gene Expression through RNA Nuclear Retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation from Promoter Region to Paraspeckle Mediates IL8 Expression upon Immune Stimuli. Mol Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Bénard, M.; Fox, A.H.; et al. NEAT1 Long Noncoding RNA Regulates Transcription via Protein Sequestration within Subnuclear Bodies. Mol Biol Cell 2014, 25, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Chen, S.; Wang, X.; Yang, X.; Chen, L.; Huang, T.; Zheng, Y.; Zheng, X.; Wu, X.; Sun, Y.; et al. Exercise Mitigates Endothelial Pyroptosis and Atherosclerosis by Downregulating NEAT1 Through N6-Methyladenosine Modifications. Arterioscler Thromb Vasc Biol 2023, 43, 910–926. [Google Scholar] [CrossRef]
- Idogawa, M.; Ohashi, T.; Sasaki, Y.; Nakase, H.; Tokino, T. Long Non-coding RNA NEAT1 Is a Transcriptional Target of P53 and Modulates P53-induced Transactivation and Tumor-suppressor Function. Int J Cancer 2017, 140, 2785–2791. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. P53 Induces Formation of NEAT1 LncRNA-Containing Paraspeckles That Modulate Replication Stress Response and Chemosensitivity. Nat Med 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 Is a P53-Inducible LincRNA Essential for Transformation Suppression. Genes Dev 2017, 31, 1095–1108. [Google Scholar] [CrossRef]
- Wang, S.; Zuo, H.; Jin, J.; Lv, W.; Xu, Z.; Fan, Y.; Zhang, J.; Zuo, B. Long Noncoding RNA Neat1 Modulates Myogenesis by Recruiting Ezh2. Cell Death Dis 2019, 10, 505. [Google Scholar] [CrossRef]
- Ahmed, A.S.I.; Dong, K.; Liu, J.; Wen, T.; Yu, L.; Xu, F.; Kang, X.; Osman, I.; Hu, G.; Bunting, K.M.; et al. Long Noncoding RNA NEAT1 (Nuclear Paraspeckle Assembly Transcript 1) Is Critical for Phenotypic Switching of Vascular Smooth Muscle Cells. Proceedings of the National Academy of Sciences 2018, 115, E8660–E8667. [Google Scholar] [CrossRef] [PubMed]
- Bielak-Zmijewska, A.; Wnuk, M.; Przybylska, D.; Grabowska, W.; Lewinska, A.; Alster, O.; Korwek, Z.; Cmoch, A.; Myszka, A.; Pikula, S.; et al. A Comparison of Replicative Senescence and Doxorubicin-Induced Premature Senescence of Vascular Smooth Muscle Cells Isolated from Human Aorta. Biogerontology 2014, 15, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Katsuumi, G.; Shimizu, I.; Yoshida, Y.; Minamino, T. Vascular Senescence in Cardiovascular and Metabolic Diseases. Front Cardiovasc Med 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Uryga, A.K.; Bennett, M.R. Ageing Induced Vascular Smooth Muscle Cell Senescence in Atherosclerosis. J Physiol 2016, 594, 2115–2124. [Google Scholar] [CrossRef]
- Sweeney, M.; Cook, S.A.; Gil, J. Therapeutic Opportunities for Senolysis in Cardiovascular Disease. FEBS J 2023, 290, 1235–1255. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, J.; Deng, L.; Ishimwe, N.; Pauli, J.; Wu, W.; Shan, S.; Kempf, W.; Ballantyne, M.D.; Kim, D.; et al. INKILN Is a Novel Long Noncoding RNA Promoting Vascular Smooth Muscle Inflammation via Scaffolding MKL1 and USP10. Circulation 2023, 148, 47–67. [Google Scholar] [CrossRef]
- Josefs, T.; Boon, R.A. The Long Non-Coding Road to Atherosclerosis. Curr Atheroscler Rep 2020, 22, 55. [Google Scholar] [CrossRef]
- Cohn, R.L.; Gasek, N.S.; Kuchel, G.A.; Xu, M. The Heterogeneity of Cellular Senescence: Insights at the Single-Cell Level. Trends Cell Biol 2023, 33, 9–17. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of P16Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
| lncRNA symbol | lncRNA name | Expression in cellular senescence | Function/Mechanism of action | References |
|---|---|---|---|---|
| CDKN2B-AS1 | CDKN2B antisense RNA 1 | Up/Down | Suppresses the expression of CDKN2A/p16 and CDKN2B/p15 by recruiting the repressive Polycomb complexes | [18] |
| H19 | H19 imprinted maternally expressed transcript | Up/Down | Inhibits p53, CDKN1C, IL6/STAT3/p21 pathway (anti-senescence function). Derepresses β-catenin (pro-senescence function). | [18] |
| HOTAIR | HOX transcript antisense RNA | Up | Activates NF-κB/IL6 and p53/p21 pathways through DNA damage response. | [18] |
| GAS5 | growth arrest specific 5 | - | Sponges miR-223 which inhibits the anti-senescence NAMPT enzyme. | [20] |
| MALAT1 | metastasis associated lung adenocarcinoma transcript 1 | Down | Controls cell cycle progression by regulating p53 activity and the expression of B-MYB transcription factor. | [24] |
| MEG3 | maternally expressed 3 | Up | Enhances p53 transcription and reduces p53 degradation. Promotes p53 binding to target promoters. | [18,25] |
| MIR31HG | MIR31 host gene | Up | Activates the expression and secretion of SASP components. | [26] |
| NEAT1 | nuclear paraspeckle assembly transcript 1 | - | Facilitates the expression of IL8. | [27] |
| PURPL | p53 upregulated regulator of p53 levels | Up | Negatively controls p53 levels by interfering with p53-MYBBP1A complex. | [28,29] |
| SBLC | Senebloc | Down | Promotes p53 degradation. Mediates epigenetic silencing of p21 through regulation HDAC5. | [30] |
| SNHG12 | small nucleolar RNA host gene 12 | - | Inhibits p16, p21 and γH2AX expression. Regulates DNA damage response via interaction with DNA-PK kinase. | [31] |
| TUG1 | taurine up regulated 1 | Up | Growth arrestor induced by p53 upon DNA damage. Inhibits the pro-proliferation HOXB7 transcription factor | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





