Submitted:
03 July 2023
Posted:
04 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Plant material and Growth
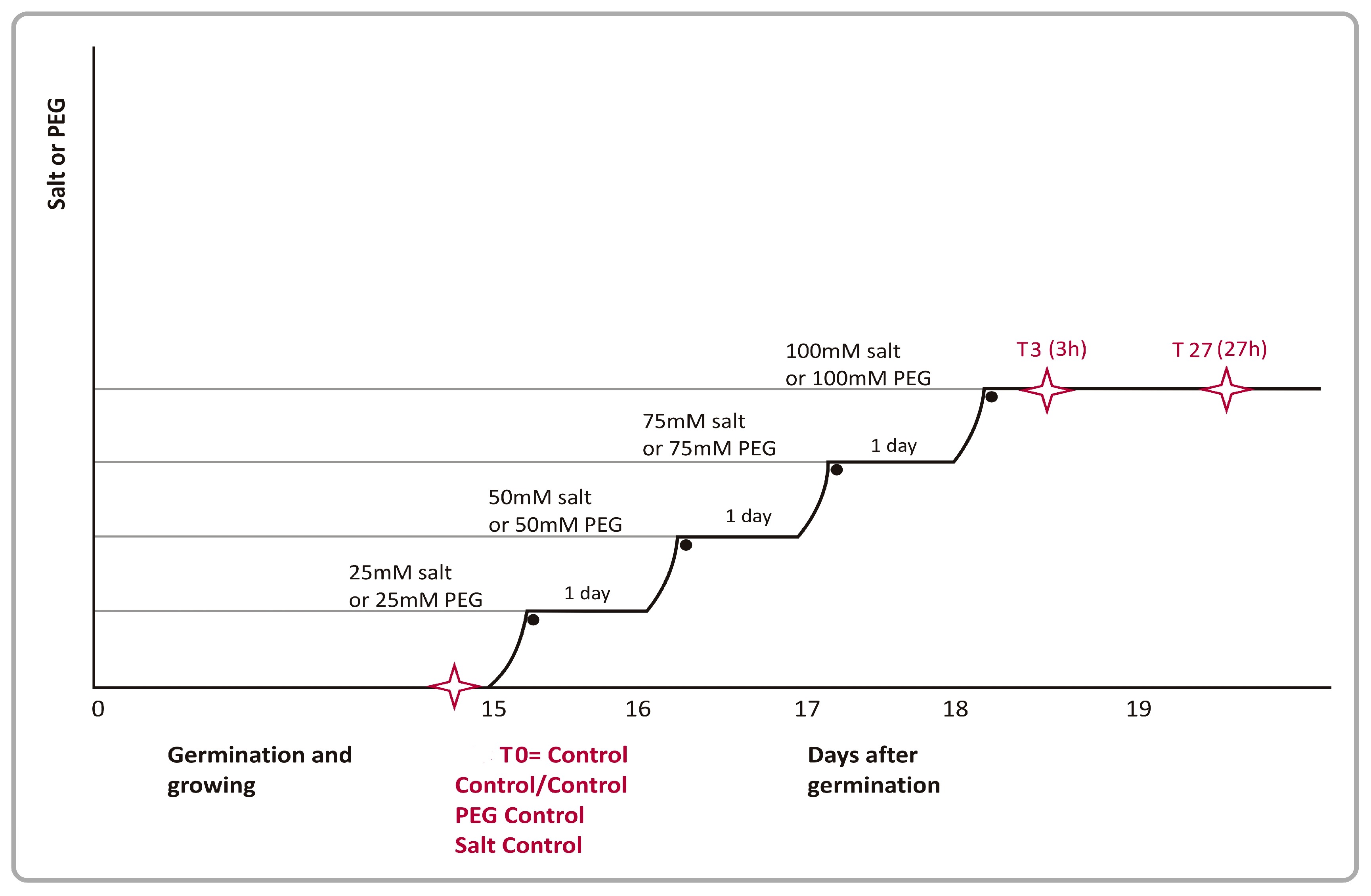
2.2. Stress Applications
2.3. Leaf Water Potential Measurements
2.4. RNA Extractions and Barley Genechip Array Hybridizations
2.5. Microarray Data Processing and Analyzes
2.7. Validation of Microarray Results Using qRT-PCR
3. Results
3.1. Optimizations of Stress Applications
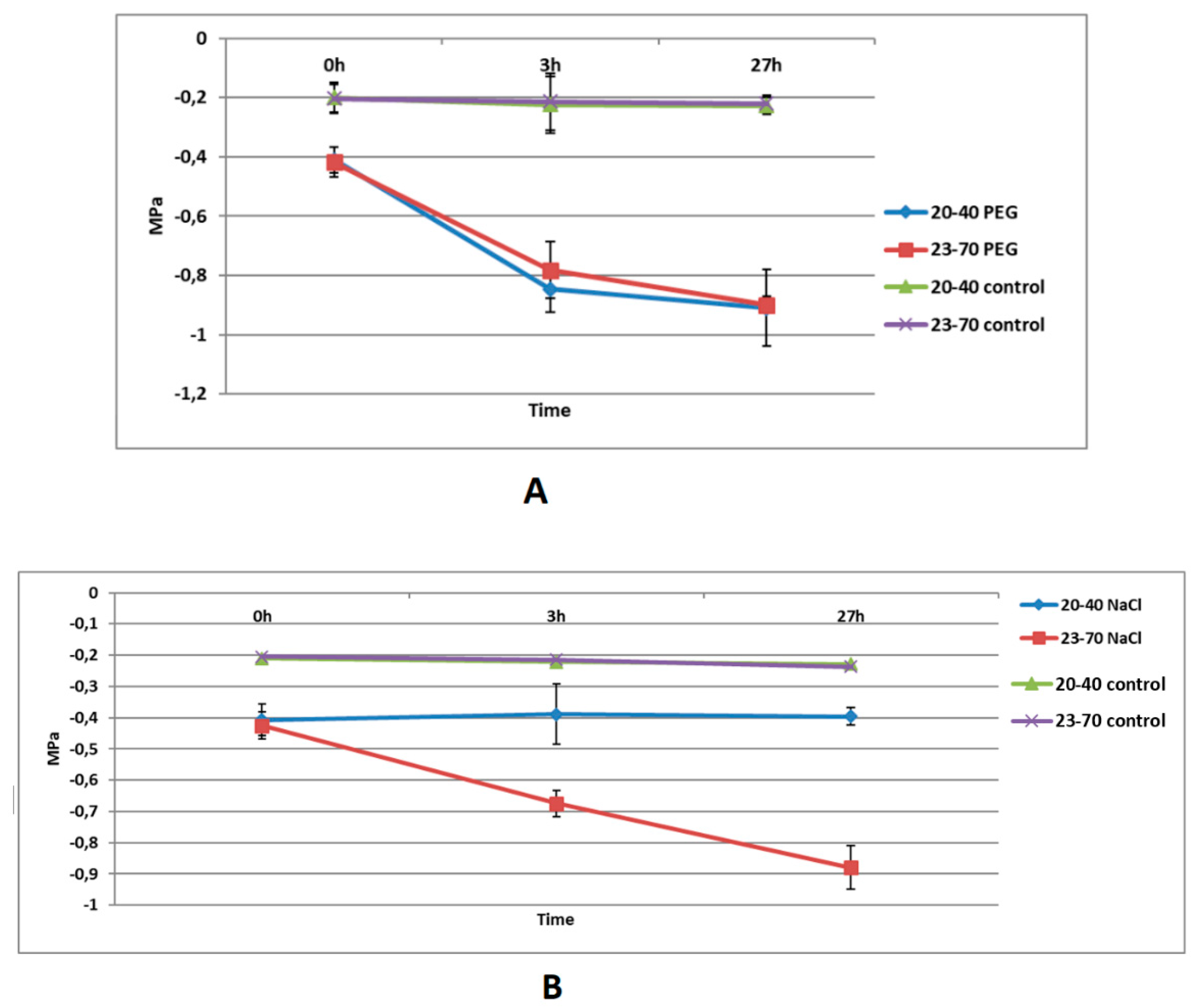
3.2. Leaf Water Potentials of Wild Barley Accessions Under Salt or PEG Stress
3.3. Expression Profiling of Wild Barley Accessions Under Salt and PEG Stress
3.4. Validation of Microarray Results Using qRT-PCR
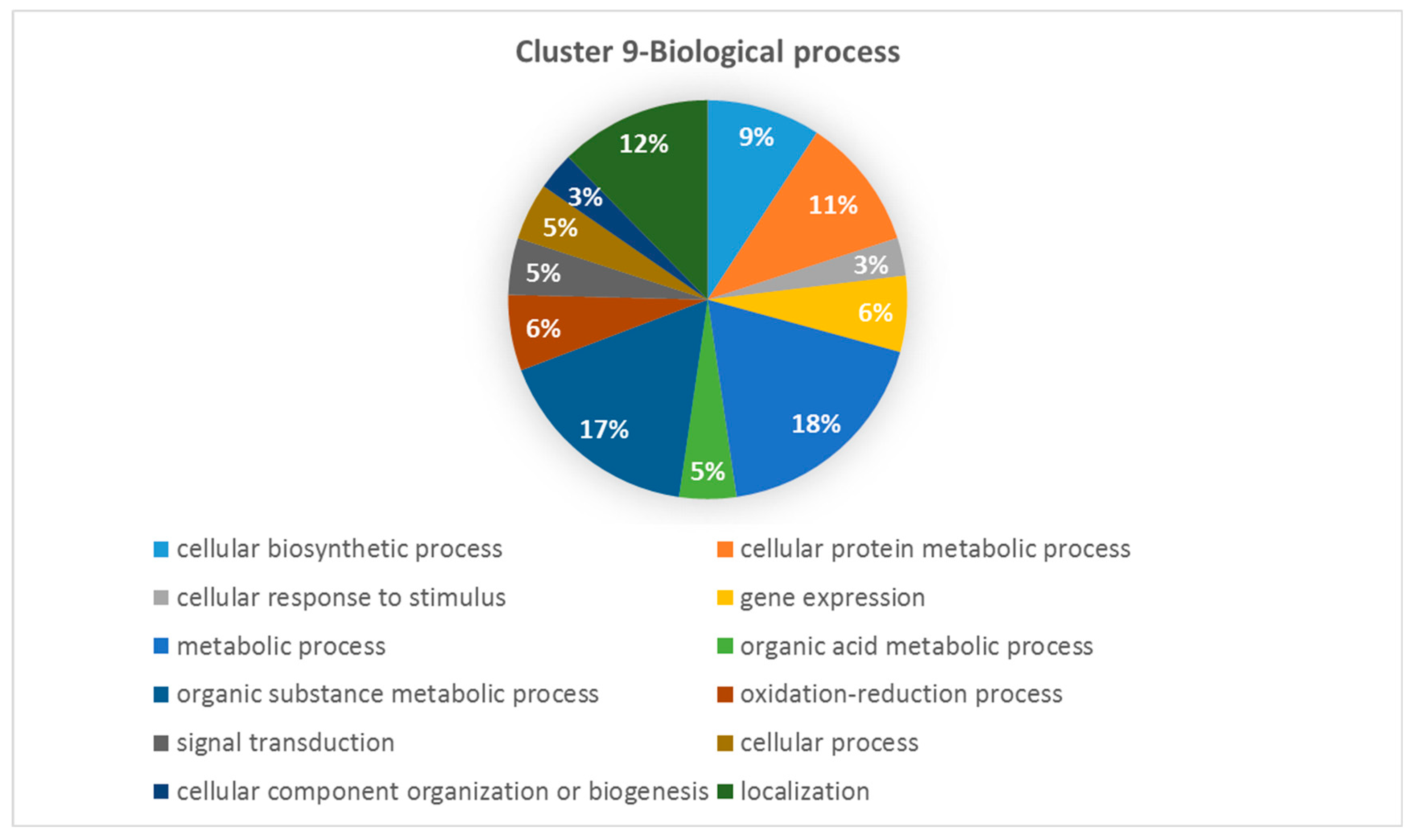
3.5. Biological Processes Impacted by Salinity and Drought Stress in Wild Barley
3.6. Comparison of Transcriptomes Between Genotypes, Tissues and Stress Treatments
4. Discussion
4.1. Leaf Water Potentials of Wild Barley Accessions Under Salt or PEG Stress
4.2. Expression Profiling of Wild Barley Accessions Under Salt and PEG Stress
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tattersall, E.A.; Grimplet, J.; DeLuc, L.; Wheatley, M.D.; Vincent, D.; Osborne, C.; Ergül, A.; Lomen, E.; Blank, R.R.; Schlauch, K.A.; et al. Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct. Integr. Genom. 2007, 7, 317–333. [Google Scholar] [CrossRef]
- Boscaiu, M.; Fita, A. Physiological and molecular characterization of crop resistance to abiotic stresses. Agronomy 2020, 10, 1308. [Google Scholar] [CrossRef]
- Munns, R. Plant Adaptations to Salt and Water Stress: Differences and Commonalities, Adv. Bot. Res. 2011, 57, 2–32. [Google Scholar]
- Shehzad, M.A.; Nawaz, F.; Ahmad, F.; Ahmad, N.; Masood, S. Protective effect of potassium and chitosan supply on growth, physiological processes and antioxidative machinery in sunflower (Helianthus annuus L.) under drought stress. Ecotoxicol Environ. Saf. 2020, 187, 109841. [Google Scholar] [CrossRef]
- Iwuala, E.; Odjegba, V.; Sharma, V.; Alam, A. Drought stress modulates expression of aquaporin gene and photosynthetic efficiency in Pennisetum glaucum (L.) R. Br. genotypes. Curr. Plant Biol 2019, 21, 100131. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought stress in plants: causes, consequences, and tolerance. In Drought Stress Tolerance in Plants; Hossain, M., Wani, S., Bhattacharjee, S., Burritt, D., Tran, LS., Eds.; Springer: Cham, 2016; Vol 1, pp. 1–16. [Google Scholar]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Nabi, R.B.S.; Tayade, R.; Hussain, A.; Kulkarni, K.P.; Imran, Q.M.; Mun, B.G.; Yun, B.W. Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 2019, 161, 120–133. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhao, X. Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS One 2014, 9, e112807. [Google Scholar] [CrossRef]
- Godoy, F.; Olivos-Hernández, K.; Stange, C.; Handford, M. Abiotic stress in crop species: improving tolerance by applying plant metabolites. Plants 2021, 10, 186. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, J.G. Osmotic adjustment and plant adaptation to environmental changes related to drought and salinity. Environ. Rev. 2010, 18, 309–319. [Google Scholar] [CrossRef]
- Bray, E.A. Plant Responses to Water Deficit. Trends Plant Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Hernández, J.A. Salinity tolerance in plants: Trends and perspectives. Int. J. Mol Sci. 2019, 20, 2408. [Google Scholar] [CrossRef]
- Nefissi Ouertani, R.; Arasappan, D.; Ruhlman, T.A.; Ben Chikha, M.; Abid, G.; Mejri, S.; Ghorbel, A.; Jansen, R.K. Effects of salt stress on transcriptional and physiological responses in barley leaves with contrasting salt tolerance. Int. J. Mol. Sci. 2022, 23, 5006. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Naghdyzadegan Jahromi, M.; Razzaghi, F.; Zand-Parsa, S. Strategies to increase barley production and water use efficiency by combining deficit irrigation and nitrogen fertilizer. Irrig. Sci. 2022, 41, 261–275. [Google Scholar] [CrossRef]
- Al-Karaki, G.N. Germination, sodium, and potassium concentrations of barley seeds as influenced by salinity. J. Plant Nutr. 2001, 24, 511–522. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Wahid, A.; Condamine, P.; Cui, X.; Close, T.J. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct. Integr. Genom. 2006, 6, 143–156. [Google Scholar] [CrossRef]
- Samarah, N.H. Effects of drought stress on growth and yield of barley. Agron. Sustain Dev. 2005, 25, 145–149. [Google Scholar] [CrossRef]
- Samarah, N.H.; Alqudah, A.M.; Amayreh, J.A.; McAndrews, G. M. The effect of late-terminal drought stress on yield components of four barley cultivars. J. Agron. Crop Sci. 2009, 195, 427–441. [Google Scholar] [CrossRef]
- Jamshidi, A.; Javanmard, H.R. Evaluation of barley (Hordeum vulgare L.) genotypes for salinity tolerance under field conditions using the stress indices. Ain Shams Eng. 2018, J9, 2093–2099. [Google Scholar] [CrossRef]
- Hamzeh-Kahnoji, Z.; Ebrahimi, A.; Sharifi-Sirchi, G.R.; Majidi-Hervan, E. Monitoring of morphological, biochemical and molecular responses of four contrasting barley genotypes under salinity stress. J. Saudi Soc. Agric. Sci. 2022, 21, 187–196. [Google Scholar] [CrossRef]
- Cramer, G.R.; Epstein, E.; Lauchli, A. Effects of sodium, potassium and calcium on salt-stressed barley. Physiol. Plant 1990, 80, 83–88. [Google Scholar]
- Munns, R.; Rawson, H.M. Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley. Aust. J. Plant Physiol. 1999, 26, 459–464. [Google Scholar] [CrossRef]
- Forster, B.P.; Russell, J.R.; Ellis, R.P.; Handley, L.L.; Robinson, D.; Hackett, C.A.; Nevo, E.; Waugh, R.; Gordon, D.C.; Keıth, R.; et al. Locating genotypes and genes for abiotic stress tolerance in barley: a strategy using maps, markers and the wild species. New Phytol. 1997, 137, 141–147. [Google Scholar] [CrossRef]
- Ueda, A.; Kathiresan, A.; Inada, M.; Narita, Y.; Nakamura, T.; Shi, W.; Takabe, T.; Bennett, C. Osmotic stress in barley regulates expression of a different set of genes than salt stress does. J. Exp. Bot. 2004, 55, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Talame, V.; Ozturk, N.Z.; Bohnert, H.J.; Tuberosa, R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J. Exp. Bot. 2007, 58, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhou, L.; Zhang, Z.; Xian, W.; Han, B. Effect of water stress on physiological and growth charaters of Prunella vulgaris at the vegetative stage. China. J. Chin. Mater. Med. 2009, 34, 1761–1764. [Google Scholar]
- Wang, X.; Chen, Z.H.; Yang, C.; Zhang, X.; Jin, G.; Chen, G.; Wang, Y.; Holford, P.; Nevo, E.; Zhang, G. Genomic adaptation to drought in wild barley is driven by edaphic natural selection at the Tabigha Evolution Slope. PNAS 2018, 115, 5223–5228. [Google Scholar] [CrossRef]
- Nevo, E. Origin, evolution, population genetics and resources for breeding of wild barley, Hordeum spontaneum, in the Fertile Crescent. In: Shewry P (ed) Barley: genetics, biochemistry, molecular biology and biotechnology. CAB International, Wallingford, UK, 1992, pp. 19–43.
- Liang, J.; Chen, X.; Deng, G.; Pan, Z.; Zhang, H.; Li, Q.; Yang, K.; Long, H.; Yu, M. Dehydration induced transcriptomic responses in two Tibetan hulless barley (Hordeum vulgare var. nudum) accessions distinguished by drought tolerance. BMC Genom. 2017, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Gürel, F.; Öztürk, N.Z.; Yörük, E.; Uçarlı, C.; Poyraz, N. Comparison of expression patterns of selected drought-responsive genes in barley (Hordeum vulgare L.) under shock-dehydration and slow drought treatments. Plant Growth Regul. 2016a, 80, 183–193. [Google Scholar] [CrossRef]
- Gürel, F.; Öztürk, N.Z; Uçarlı, C.; Rosellini, D. Barley genes as tools to confer abiotic stress tolerance in crops. Front. Plant Sci. 2016b, 7, 1137. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Collin, A.; Sitko, K.; Janiak, A.; Kalaji, H.M.; Szarejko, I. Genetic and physiological dissection of photosynthesis in barley exposed to drought stress. Int. J. Mol. Sci. 2019, 20, 6341. [Google Scholar] [CrossRef]
- Zhang, M.; Jin, Z.Q.; Zhao, J.; Zhang, G.; Wu, F. Physiological and biochemical responses to drought stress in cultivated and Tibetan wild barley. Plant Growth Regul. 2015, 75, 567–574. [Google Scholar] [CrossRef]
- Ellis, R.P.; Forster, B.P.; Robinson, D.; Handley, L.L.; Gordon, D.C.; Russell, J.R.; Powell, W. Wild barley: a source of genes for crop improvement in the 21st century? J. Exp. Bot. 2000, 51, 9–17. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, H.; Dai, H.; Zhang, G.; Wu, F. Difference in response to drought stress among Tibet wild barley genotypes. Euphytica 2010, 172, 395–403. [Google Scholar] [CrossRef]
- Barati, M.; Majidi, M.M.; Mirlohi, A. Response of cultivated and wild barley germplasm to drought stress at different developmental stages. Crop. Sci. 2015, 55, 2668–2681. [Google Scholar] [CrossRef]
- Barati, M.; Majidi, M.M.; Mostafavi, F.; Mirlohi, A.; Safari, M.; Karami, Z. Evaluation of wild barley species as possible sources of drought tolerance for arid environments. Plant Genet. Resour. 2017, 16, 209–217. [Google Scholar] [CrossRef]
- Cai, K.; Chen, X.; Han, Z.; Wu, X.; Zhang, S.; Li, Q.; Nazir, M.M.; Zhang, G.; Zeng, F. Screening of worldwide barley collection for drought tolerance: The assessment of various physiological measures as the selection criteria. Front. Plant Sci. 2020, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Rensink, W.A.; Buell, C.R. Microarray expression profiling resources for plant genomics. Trends Plant Sci. 2005, 10, 603–609. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Timothy, J. Comparative Transcriptional Profiling of Barley Cultivar Maythorpe and Its Derived Mutant Golden Promise under Salinity Stress. In Plant & Animal Genome XIII International Conference, San Diego, CA. 2005, 234.
- Wu, D.; Cai, S.; Chen, M.; Ye, L.; Chen, Z.; Zhang, H.; Dai, F.; Wu, F.; Zhang, G. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS one 2013, 8, e55431. [Google Scholar] [CrossRef]
- Bedada, G.; Westerbergh, A.; Müller, T.; Galkin, E.; Bdolach, E.; Moshelion, M.; Fridman, E.; Schmid, K.J. Transcriptome sequencing of two wild barley (Hordeum spontaneum L.) ecotypes differentially adapted to drought stress reveals ecotype-specific transcripts. BMC Genom. 2014a, 15, 1–20. [Google Scholar] [CrossRef]
- Bahieldin, A.; Atef, A.; Sabir, J.S.; Gadalla, N.O.; Edris, S.; Alzohairy, A.M.; Radhwan, N.A.; Baeshen, M.N.; Ramadan, A.M.; Eissa, H.F.; et al. RNA-Seq analysis of the wild barley (H. spontaneum) leaf transcriptome under salt stress. CR. Biol. 2015, 338, 285–97. [Google Scholar] [CrossRef]
- Cramer, G.R.; Läuchli, A.; Epstein, E. Effects of NaCl and CaCl2 on ion activities in complex nutrient solutions and root growth of cotton. Plant Physiol. 1986, 81, 792–797. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Beazer-barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.; Schlauch, K.; Keeling, C.I.; Young, S.; Bearfield, J.C.; Blomquist, G.J.; Tittiger, C. Functional genomics of mountain pine beetle (Dendroctonus ponderosae) midguts and fat bodies. BMC Genom. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. 1995, B57, 289–300. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene Ontology annotations and resources. Nucleic Acids Res. 2012, 41, D530–D535. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols, Humana Press, Totowa, N.J. 200, pp. 365–386.
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lavie, B.; Stow, V.; Krugman, T.; Beiles, A.; Nevo, E. Fitness in wild barley from two opposing slopes of a Mediterranean microsite at Mount Carmel, Israel. Barley Genetics Newslett. 1993, 23, 12–14. [Google Scholar]
- Nevo, E.; Beiles, A.; Gutterman, Y.; Storch, N.; Kaplan, D. Genetic resources of wild cereals in Israel and vicinity. I. Phenotypic variation within and between populations of wild wheat, Triticum dicoccoides. Euphytica 1984, 33, 717–735. [Google Scholar] [CrossRef]
- Chen, G.; Krugman, T.; Fahima, T.; Zhang, F.; Korol, A.B.; Nevo, E. Differential patterns of germination and desiccation tolerance of mesic and xeric wild barley (Hordeum spontaneum L.) in Israel. J. Arid Environ. 2004, 56, 95–105. [Google Scholar] [CrossRef]
- Lupu, A.; Nevo, E.; Zamorzaeva, I.; Korol, A. Ecological–genetic feedback in DNA repair in wild barley, Hordeum spontaneum. Genetica 2006, 127, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X. , Ismail, A. M.; Close, T.J. Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ. 2007, 30, 410–421. [Google Scholar] [CrossRef]
- Cramer, G.R.; Ergül, A.; Grimplet, J.; Tillett, R.L.; Tattersall, E.A.R.; Bohlman, M.C.; Vincent, D.; Sonderegger, J.; Evans, C.; Osborne, C.; et al. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct. Integr. Genom. 2007, 7, 111–134. [Google Scholar] [CrossRef]
- Boscai, M.; Donat, P.M.; Llınares, J.; Vicent, P.; Vicente Meana, Ó. Stress-tolerant Wild Plants: a Source of Knowledge and Biotechnological Tools for the Genetic Improvement of Stress Tolerance in Crop Plants. Not. Bot. Hortı. Agrobo. 2012, 40, 323–327. [Google Scholar] [CrossRef]
- Ashoub, A.; Müller, N.; Jiménez-Gómez, J.M.; Brüggemann, W. Prominent alterations of wild barley leaf transcriptome in response to individual and combined drought acclimation and heat shock conditions. Physiol. Plant 2018, 163, 18–29. [Google Scholar]
- Zhang, M.; Fu, M.M.; Qiu, C.W.; Cao, F.; Chen, Z.H.; Zhang, G.; Wu, F. Response of tibetan wild barley genotypes to drought stress and identification of quantitative trait loci by genome-wide association analysis. Int. J. Mol. Sci. 2019, 20, 791. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Bedada, G.; Westerbergh, A.; Nevo, E.; Korol, A.; Schmid, K.J. DNA sequence variation of wild barley Hordeum spontaneum (L.) across environmental gradients in Israel. Heredity 2014b, 112, 646–655. [Google Scholar] [CrossRef]
- Turner, N.C. Turgor maintenance by osmotic adjustment: 40 years of progress. J. Exp. Bot. 2018, 69, 3223–3233. [Google Scholar] [CrossRef] [PubMed]
- Nio, S.A.; Ludong, D.P.M.; Wade, L.J.J.A. Comparison of leaf osmotic adjustment expression in wheat (Triticum aestivum L.) under water deficit between the whole plant and tissue levels. Agric. Nat. Resour. 2018, 52, 33–38. [Google Scholar] [CrossRef]
- Naceur, B.; Paul, R.; Ben Salah, H. Screening of six barley varieties for drought resistance by using leaf water potential, membrane stability, and phosphatasic activity parameters. Prospettive e proposte mediterranee-Rivista di Economia. Agricoltura e Ambiente 1997, 4, 51–54. [Google Scholar]
- González, A.; Martı́n, I.; Ayerbe, L. Barley yield in water-stress conditions.: The influence of precocity, osmotic adjustment and stomatal conductance. Field Crops Res. 1999, 62, 23–34. [Google Scholar] [CrossRef]
- Nayyar, H.; Walia, D.P. Genotypic variation in wheat in response to water stress and abscisic acid-induced accumulation of osmolytes in developing grains. J. Agron. Crop Sci. 2004, 190, 39–45. [Google Scholar] [CrossRef]
- Vysotskaya, L.; Hedley, P.E.; Sharipova, G.; Veselov, D.; Kudoyarova, G.; Morris, J.; Jones, H.G. Effect of salinity on water relations of wild barley plants differing in salt tolerance. AoB Plants 2010, 2010, plq006. [Google Scholar] [CrossRef]
- Ciarmiello, L.F.; Woodrow, P.; Fuggi, A.; Pontecorvo, G.; Carillo, P. Plant Genes for Abiotic Stress, Abiotic Stress in Plants - Mechanisms and Adaptations. In Arun Shanker, B. Venkateswarlu (eds) 2011, 978–953.
- Wang, W.X.; Vinocur, B.; Shoseyov, O.; Altman, A. Biotechnology of plant osmotic stress toelrance and physiological and moleucalr considerations. Acta Hort. 2001, 560, 285–292. [Google Scholar] [CrossRef]
- Ludwig, A.A.; Romeis, T.; Jones, J.D. CDPK-mediated signalling pathways: specificity and cross-talk. J. Exp. Bot. 2004, 55, 181–188. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Frank, W.; Munnik, T.; Kerkmann, K.; Salamini, F.; Bartels, D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 2000, 12, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stevanato, P.; Lv, C.; Li, R.; Geng, G. Comparative physiological and proteomic analysis of two sugar beet genotypes with contrasting salt tolerance. J. Agric. Food Chem. 2019, 67, 6056–6073. [Google Scholar] [CrossRef]
- Görschen, E.; Dunaeva, M.; Hause, B.; Reeh, I.; Wasternack, C.; Parthier, B. Expression of the ribosome-inactivating protein JIP60 from barley in transgenic tobacco leads to an abnormal phenotype and alterations on the level of translation. Planta 1995, 202, 470–478. [Google Scholar]
- Szypulska, E.; Jankowski, K.; Weidner, S. ABA pretreatment can limit salinity-induced proteome changes in growing barley sprouts. Acta Physiol. Plant. 2017, 39, 1–18. [Google Scholar] [CrossRef]
- Barashkova, A.S.; Sadykova, V.S.; Salo, V.A.; Zavriev, S. K.; Rogozhin, E.A. Nigellothionins from black cumin (Nigella sativa l.) seeds demonstrate strong antifungal and cytotoxic activity. Antibiotics 2021, 10, 166. [Google Scholar] [CrossRef]
- Gao, R.; Duan, K.; Guo, G.; Zhizhao, D.; Zhiwei, C.; Liang, L.; Ting, H.; Ruiju, L.; Jianhua, H. Comparative Transcriptional Profiling of Two Contrasting Barley Genotypes under Salinity Stress during the Seedling Stage. Int. J. Genomics 2013, 139, 822–835. [Google Scholar] [CrossRef]
- Lim, J.; Lim, C.W.; Lee, S.C. Core Components of Abscisic Acid Signaling and Their Post-Translational Modification. Front. Plant Sci. 2022, 13, 895698. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the role of abscisic acid insensitive 5 (abi5) and abscisic acid-responsive element binding factors (ABFs) in ABA signaling in different developmental stages in plants. Cells 2021, 10, 1996. [Google Scholar] [CrossRef]
- Vanjildorj, E.; Bae, T.W.; Riu, K.Z.; Kim, S.Y.; Lee, H.Y. Overexpression of Arabidopsis ABF3 gene enhances tolerance to drought and cold in transgenic lettuce (Lactuca sativa). PCTOC 2005, 83, 41–50. [Google Scholar] [CrossRef]
- Wang, Z.; Su, G.; Li, M.; Ke, Q.; Soo Young, K.; Hongbing, L.; Jin, H.; Bingcheng, X.; Xi-Ping, D.; Sang-Soo, K. Overexpressing Arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol. Biochem. 2016, 109, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Binott, J.J.; Owuoche, J.O.; Bartels, D. Physiological and molecular characterization of Kenyan barley (Hordeum vulgare L.) seedlings for salinity and drought tolerance. Euphytica 2017, 213, 1–23. [Google Scholar] [CrossRef]
- Suprunova, T.; Krugman, T.; Fahima, T.; Chen, G.; Shams, I.; Korol, A.; Nevo, E. Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit. Plant Cell Environ. 2004, 27, 1297–1308. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Wheat and barley dehydrins under cold, drought, and salinity–what can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 5, 343. [Google Scholar] [CrossRef]
- Sewelam, N.; Kazan, K.; Schenk, P.M. Global Plant Stress Signaling: Reactive Oxygen Species at the Cross-Road. Front. Plant Sci. 2016, 23, 187. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kim, H.S.; Park, S.C.; Ji, C.Y.; Yang, J.W.; Lee, H.U.; Kwak, S.S. Downregulation of swpa4 peroxidase expression in transgenic sweet potato plants decreases abiotic stress tolerance and reduces stress-related peroxidase expression. Plant Biotechnol. Rep. 2021, 15, 69–76. [Google Scholar] [CrossRef]
- Harb, A.; Simpson, C.; Guo, W.; Govindan, G.; Kakani, V.G.; Sunkar, R. The effect of drought on transcriptome and hormonal profiles in barley genotypes with contrasting drought tolerance. Front. Plant Sci. 2020, 11, 618491. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Ge, T.; Suı, F-G. ; Baı, L. et al. Effects of Water Stress on the Protective Enzyme Activities and Lipid Peroxidation in Roots and Leaves of Summer Maize. Agric. Sci. China 2006, 5, 291–298. [Google Scholar] [CrossRef]
- Anum, J.; O’Shea, C.; Zeeshan Hyder, M.; Farrukh, S.; Skriver, K.; Malik, S.I.; Yasmin, T. Germin like protein genes exhibit modular expression during salt and drought stress in elite rice cultivars. Mol. Biol. Rep. 2022, 49, 293–302. [Google Scholar] [CrossRef]
- Hurkman, W.; Tanaka, C.K. Efect of salt stress on germin gene expression in barley roots. Plant Physiol. 1996, 110, 971–7. [Google Scholar] [CrossRef]
- Valentovičová, K.; Halušková, Ľ.; Huttová, J.; Mistrík, I.; Tamás, L. Efect of heavy metals and temperature on the oxalate oxidase activity and lignifcation of metaxylem vessels in barley roots. Environ. Exp. Bot. 2009, 66, 457–62. [Google Scholar] [CrossRef]
- Voothuluru, P.; Sharp, R.E. Apoplastic hydrogen peroxide in the growth zone of the maize primary root under water stress. I. Increased levels are specific to the apical region of growth maintenance. J. Exp. Bot. 2013, 64, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Shi, W.; Nakamura, T.; Takabe, T. Analysis of salt-inducible genes in barley roots by differential display. J. Plant Res. 2002, 115, 0119–0130. [Google Scholar] [CrossRef] [PubMed]
- Chmielewska, K.; Rodziewicz, P.; Swarcewicz, B.; et al. Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front. Plant Sci. 2016, 7, 1108. [Google Scholar] [CrossRef]
- Anders, J.V.; Davis, G.D. Abiotic stress alters transcript profiles and activity of glutathione S-transferase, glutathione peroxidase, and glutathione reductase in Euphorbia esula. Physiol. Plant 2004, 120, 421–433. [Google Scholar] [CrossRef]
- Rezaei, M.K.; Shobbar, Z.S.; Shahbazi, M.; Abedini, R.; Zare, S. Glutathione S-transferase (GST) family in barley: Identification of members, enzyme activity, and gene expression pattern. J. Plant Physiol. 2013, 170, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, Y.; Duan, R.; Fan, J.; Jiao, P.; Sun, H.; Guan, S.; Liu, S. Overexpression of Maize Glutathione S-Transferase ZmGST26 Decreases Drought Resistance of Arabidopsis. Agronomy 2022, 12, 2948. [Google Scholar] [CrossRef]
- Chen, J.H.; Jiang, H.W.; Hsieh, E.J.; Chen, H. Y.; Chien, C.T.; Hsieh, H.L.; Lin, T.P. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012, 158, 340–351. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, N.; Liu, Z.; Liu, S.; Liu, C.; Lin, J.; Yang, H.; Li, S.; Yukawa, Y. The AtGSTU7 gene influences glutathione-dependent seed germination under ABA and osmotic stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 528, 538–544. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.J.; Zeng, Q.Y. Overexpression of three orthologous glutathione S-transferases from Populus increased salt and drought resistance in Arabidopsis. Biochem. Syst. Ecol. 2019, 83, 57–61. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Yoon, H.W.; Kim, M.C.; Shin, P.G.; et al. Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Mol. Gen. Genet. 1997, 255, 359–371. [Google Scholar] [CrossRef]
- Zinati, Z.; Sazegari, S. Identification of important genes involved in priming induced drought tolerance in barley through transcriptomic data mining. Crop Pasture Sci. 2022, 73, 1011–1025. [Google Scholar] [CrossRef]
- Cui, L.; Yang, G.; Yan, J.; Pan, Y.; Nie, X. Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genom. 2019, 20, 750. [Google Scholar] [CrossRef]
- Dudziak, K.; Zapalska, M.; Börner, A.; Szczerba, H.; Kowalczyk, K.; Nowak, M. Analysis of wheat gene expression related to the oxidative stress response and signal transduction under short-term osmotic stress. Sci. Rep. 2019, 9, 1–14. [Google Scholar]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef]
- Zhou, M.; Cheng, H.; Chiang, V.L.; Li, W.; Yang, C.; Wang, C. PtrbZIP3 transcription factor regulates drought tolerance of Populus trichocarpa. Environ. Exp. Bot. 2023, 208, 105231. [Google Scholar] [CrossRef]
- Oh, S.J.; Song, S.I.; Kim, Y.S.; et al. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Xu, Y.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Tian, S.; Dong, H.; Guo, C. Pleiotropic effects of TaMYB3R1 on plant development and response to osmotic stress in transgenic Arabidopsis. Gene 2015, 558, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Shan, Z.; Zhou, S.; Yang, Q.; Gai, J.; Li, Y. GmDNAJC7 from Soybean Is Involved in Plant Tolerance to Alkaline-Salt, Salt, and Drought Stresses. Agronomy 2022, 12, 1419. [Google Scholar] [CrossRef]
- Zhichang, Z.; Wanrong, Z.; Jinping, Y.; Jianjun, Z.; Xufeng, L.Z.L.; Yang, Y. Over-expression of Arabidopsis DnaJ (Hsp40) contributes to NaCl-stress tolerance. Afr. J. Biotechnol. 2010, 9, 972–978. [Google Scholar]
- Sugino, M.; Hibino, T.; Tanaka, Y.; Nii, N.; Takabe, T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica acquires resistance to salt stress in transgenic tobacco plants. Plant Sci. 1999, 146, 81–88. [Google Scholar] [CrossRef]
- Isayenkov, S.; Hilo, A.; Rizzo, P.; Tandron Moya, Y.A.; Rolletschek, H.; Borisjuk, L.; Radchuk, V. Adaptation strategies of halophytic barley Hordeum marinum ssp. marinum to high salinity and osmotic stress. Int. J. Mol. Sci. 2020, 21, 9019. [Google Scholar] [CrossRef]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, Z.; Wang, A.; Zhu, J. Cold-regulated gene LeCOR413PM2 confers cold stress tolerance in tomato plants. Gene 2021, 764, 145097. [Google Scholar] [CrossRef]
- Lee, K.W.; Choi, G.J.; Kim, K.Y.; Ji, H.C.; Zaman, R.; Lee, S. H. Identification of drought induced differentially expressed genes in barley leaves using the annealing control-primer-based Gene Fishing technique. Aust. J. Crop Sci. 2011, 5, 1364–1369. [Google Scholar]
- Das, R.R.; Pradhan, S.; Parida, A. De-novo transcriptome analysis unveils differentially expressed genes regulating drought and salt stress response in Panicum sumatrense. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]


| Up-regulated genes | ||||||
|---|---|---|---|---|---|---|
| Probe Set ID | Accession No | E-valuec | Annotation | Genotype | Tissue | Time Point |
| Contig1568_x_at* | S22515 | 2,00E-65 | Thionin precursor | 23-70 | Leaf | 3h-27h |
| Contig3501_at* | Q00531 | e-108 | 60 kDa jasmonate-induced protein | 23-70 | Leaf | 3h |
| 20-40 | Leaf | 27h | ||||
| HV06O23u_at | NP_567835.1 | mRNA cleavage factor subunit - like protein | 23-70 | Leaf | 3h-27h | |
| 23-70 | Root | 3h-27h | ||||
| 20-40 | Leaf | 3h-27h | ||||
| 20-40 | Root | 3h-27h | ||||
| Contig13483_at | NP_188806.1 | 5,00E-70 | Integral membrane protein, putative | 23-70 | Leaf | 3h-27h |
| Contig21604_at* | CAC83006.1 | 1,00E-42 | Putative glutathione synthetase | 23-70 | Leaf | 3h-27h |
| Contig1865_at | AAM08517.1 | 3,00E-90 | Putative peroxidase | 23-70 | Leaf | 3h |
| Contig1718_s_at* | AAD02260.1 | 3,00E-30 | Dehydrin 9 | 23-70 | Root | 27h |
| 20-40 | Leaf | 3h-27h | ||||
| 20-40 | Root | 3h-27h | ||||
| Contig8961_at* | T02663 | 9,00E-17 | Abscisic acid- and stress-induced protein | 20-40 | Leaf | 3h-27h |
| Contig9774_s_at | Unknown | 20-40 | Leaf | 3h-27h | ||
| Contig4281_s_at* | CAC12881.1 | 2,00E-77 | Cold-regulated protein | 23-70 | Leaf | 3h |
| 23-70 | Root | 27h | ||||
| 20-40 | Leaf | 3h-27h | ||||
| Contig3112_at* | T06810 | 2,00E-87 | Cold acclimation protein WCOR413 | 20-40 | Leaf | 3h-27h |
| 20-40 | Root | 3h | ||||
| Contig15982_at | NP_567949.1 | 1,00E-31 | Abscisic acid responsive elements-binding factor(ABF3) | 20-40 | Leaf | 3h |
| HR01N22u_s_at | BAB62578.1 | 1,00E-15 | Putative protein kinase SPK-3 | 20-40 | Leaf | 3h-27h |
| Contig9239_s_at | Unknown | 23-70 | Leaf | 27h | ||
| 20-40 | Leaf | 27h | ||||
| Contig6546_at* | T06480 | e-101 | Safener-induced In2.1-like protein | 23-70 | Root | 3h |
| Contig3017_at* | CAA74595.1 | e-126 | Oxalate oxidase | 23-70 | Root | 3h-27 |
| 20-40 | Root | 27h | ||||
| Contig4213_at | CAB85629.1 | 1,00E-50 | Putative ripening-related protein | 23-70 | Root | 3h-27 |
| Contig5838_at | AAG40562.1 | 4,00E-88 | Glutathione-S-transferase 2 | 23-70 | Root | 3h |
| Contig17190_at* | AAL83988.1 | 2,00E-36 | Putative heat shock protein | 23-70 | Root | 27h |
| Contig3783_s_at* | AAC31615.1 | 2,00E-17 | Hypersensitive-induced reaction protein 3 | 20-40 | Root | 3h-27 |
| Contig15282_at | AAG34845.1 | 6,00E-41 | Glutathione S-transferase GST 37 | 23-70 | Root | 3h |
| 20-40 | Root | 3h | ||||
| Down-regulated genes | ||||||
| rbah13p07_s_at* | AAM76682.1 | 2,00E-24 | Peroxidase | 23-70 | Leaf | 3h-27h |
| 23-70 | Root | 3h | ||||
| 20-40 | Leaf | 3h-27h | ||||
| 20-40 | Root | 3h | ||||
| Contig25448_at | BAC20673.1 | 3,00E+38 | Serine/threonine kinase-like protein | 23-70 | Leaf | 3h-27h |
| Contig15729_at* | BAB63804.1 | 7,00E-30 | Putative I-box binding factor | 23-70 | Leaf | 3h |
| 23-70 | Root | 3h-27h | ||||
| 20-40 | Root | 3h | ||||
| Contig15489_at* | AAK73104.1 | 3,00E-33 | MAP kinase kinase | 23-70 | Leaf | 3h |
| 20-40 | Root | 3h | ||||
| Contig2112_at | S61406 | e-103 | Peroxidase 2 precursor | 23-70 | Leaf | 3h |
| 23-70 | Root | 3h | ||||
| 20-40 | Root | 3h | ||||
| Contig17190_at* | AAL83988.1 | 2,00E-36 | Putative heat shock protein | 23-70 | Leaf | 3h |
| Contig3783_at* | AAC31615.1 | 2,00E-17 | Physical impedance induced protein | 20-40 | Leaf | 3h-27h |
| Contig13932_at | AAL34131.1 | 6,00E-27 | Unknown | 23-70 | Leaf | 3h-27h |
| 20-40 | Leaf | 3h-27h | ||||
| Contig3636_at | CAC37639.1 | 8,00E-60 | SERK2 protein | 20-40 | Leaf | 27h |
| Contig6546_at* | T064480 | e-101 | Safener-induced In2.1-like protein | 20-40 | Leaf | 27h |
| Contig3875_s_at* | NP_566088.2 | 1,00E-23 | MYB-related transcription factor | 20-40 | Leaf | 3h |
| Contig21604_at* | CAC83006.1 | 1,00E-42 | Putative glutathione synthetase | 23-70 | Root | 3h-27h |
| 20-40 | Root | 3h-27h | ||||
| Contig3216_at | BAC10287.1 | 1,00E-20 | Defensin | 23-70 | Root | 3h-27h |
| Contig18873_at | P17801 | 4,00E-53 | Putative receptor protein kinase ZMPK1 precursor | 23-70 | Root | 3h-27h |
| 20-40 | Root | 3h | ||||
| EBro03_SQ007_C24_at | AAL73531.1 | 1,00E-18 | TNP2-like protein | 23-70 | Root | 3h |
| Contig8961_at* | T02663 | 9,00E-17 | Abscisic acid- and stress-induced protein | 23-70 | Root | 3h-27h |
| Contig10179_s_at | Unknown | 20-40 | Root | 3h | ||
| Contig25015_at* | NP_680449.1 | 2,00E-08 | Unknown | 23-70 | Root | 3h |
| 20-40 | Root | 3h | ||||
| HC109A09_T3_at | NP_079900.1 | 6,00E-12 | Unknown | 23-70 | Root | 3h-27 |
| 20-40 | Root | 3h-27 | ||||
| Contig3156_s_at | T05956 | 7,00E-36 | Germin-like protein | 23-70 | Leaf | 27h |
| 23-70 | Root | 3h | ||||
| 20-40 | Root | 27h |
| Up-regulated genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Probe Set ID | Accession No | E-valuec | Annotation | Genotype | Tissue | Time Point | ||
| Contig3501_at* | Q00531 | e-108 | 60 kDa jasmonate-induced protein | 23-70 | Leaf | 3h | ||
| 23-70 | Root | 3h | ||||||
| 20-40 | Leaf | 3h-27h | ||||||
| Contig6004_x_at | NP_564147.1 | 2,00E-10 | Unknown | 23-70 | Leaf | 3h-27h | ||
| Contig4281_s_at* | CAC12881.1 | 2,00E-77 | Cold-regulated protein | 23-70 | Leaf | 27h | ||
| Contig10808_at | BAB02515.1 | 8,00E-45 | Transporter-like protein | 23-70 | Leaf | 27h | ||
| 23-70 | Root | 3h-27h | ||||||
| 20-40 | Root | 3h-27h | ||||||
| Contig1568_x_at* | S22515 | 2,00E-65 | Thionin precursor | 23-70 | Leaf | 3h-27h | ||
| Contig3967_at | AAK62365.1 | 5,00E-23 | Unknown | 23-70 | Root | 27h | ||
| 20-40 | Leaf | 3h | ||||||
| Contig8961_at* | T02663 | 9,00E-17 | Abscisic acid- and stress-induced protein | 20-40 | Leaf | 3h | ||
| Contig1718_s_at* | AAD02260.1 | 3,00E-30 | Dehydrin 9 | 20-40 | Leaf | 3h | ||
| 20-40 | Root | 3h-27h | ||||||
| Contig24926_at | BAC10698.1 | 4,00E-13 | Putative leucine rich repeat containing protein kinase | 20-40 | Leaf | 27h | ||
| Contig6546_at* | T06480 | e-101 | Safener-induced In2.1-like protein | 23-70 | Root | 3h | ||
| 20-40 | Root | 3h | ||||||
| Contig21640_at | AAG40562.1 | 1,00E-61 | Glutathione-S-transferase 2 | 23-70 | Root | 3h | ||
| 20-40 | Root | 3h | ||||||
| Contig3017_at* | CAA74595.1 | e-126 | Oxalate oxidase | 23-70 | Root | 3h-27 | ||
| 20-40 | Root | 3h-27h | ||||||
| HW06F04u_s_at | Unknown | 23-70 | Root | 3h | ||||
| Contig3783_at* | AAC31615.1 | 2,00E-17 | Physical impedance induced protein | 20-40 | Root | 3h-27h | ||
| Contig3783_s_at* | AAN17464.1 | e-126 | Hypersensitive-induced reaction protein 3 | 23-70 | Root | 3h-27h | ||
| 20-40 | Root | 3h-27h | ||||||
| Contig3112_at* | T06810 | 2,00E-87 | Cold acclimation protein WCOR413 | 20-40 | Root | 3h | ||
| Down-regulated genes | ||||||||
| Contig ID | Accession No | E-valuec | Annotation | Genotype | Tissue | Time Point | ||
| rbah13p07_s_at* | AAM76682.1 | 2,00E-24 | Peroxidase | 23-70 | Leaf | 3h | ||
| 20-40 | Leaf | 3h-27h | ||||||
| Contig15729_at* | BAB63804.1 | 7,00E-30 | Putative I-box binding factor | 23-70 | Leaf | 3h | ||
| 23-70 | Root | 3h-27h | ||||||
| 20-40 | Root | 3h | ||||||
| Contig6953_at | Unknown | 23-70 | Leaf | 3h-27h | ||||
| 20-40 | Leaf | 27h | ||||||
| Contig15489_at* | AAK73104.1 | 3,00E-33 | MAP kinase kinase | 23-70 | Leaf | 3h | ||
| 20-40 | Root | 3h-27h | ||||||
| Contig11487_at | AAM63624.1 | 2,00E-33 | DnaJ protein, putative | 23-70 | Leaf | 3h | ||
| 23-70 | Root | 3h | ||||||
| Contig17190_at* | AAL83988.1 | 2,00E-36 | Putative heat shock protein | 23-70 | Leaf | 3h | ||
| 23-70 | Root | 3h | ||||||
| Contig3875_s_at* | NP_566088.2 | 1,00E-23 | MYB-related transcription factor | 20-40 | Leaf | 3h | ||
| Contig3783_at* | AAC31615.1 | 2,00E-17 | Physical impedance induced protein | 20-40 | Leaf | 3h-27h | ||
| Contig18873_at* | P17801 | 4,00E-53 | Putative receptor protein kinase ZMPK1 precursor | 23-70 | Root | 3h-27h | ||
| Contig9042_s_at | Unknown | 23-70 | Root | 3h | ||||
| 20-40 | Root | 3h | ||||||
| HV05A09u_s_at | P49970 | 9,00E-23 | Signal Recognition Particle 54 KD Protein3 (SRP54) | 23-70 | Root | 3h | ||
| Contig8961_at* | T02663 | 9,00E-17 | Abscisic acid- and stress-induced protein | 23-70 | Root | 3h | ||
| 20-40 | Root | 3h | ||||||
| EBro03_SQ007_C24_at | AAL73531.1 | 1,00E-18 | TNP2-like protein | 23-70 | Root | 3h | ||
| Contig3216_at | BAC10287.1 | 1,00E-20 | Defensin | 23-70 | Root | 3h-27h | ||
| Contig25015_at* | NP_680449.1 | 2,00E-08 | Unknown | 23-70 | Root | 3h-27h | ||
| 20-40 | Root | 3h-27h | ||||||
| Contig21604_at* | CAC83006.1 | 1,00E-42 | Putative glutathione synthetase | 23-70 | Root | 3h | ||
| 20-40 | Root | 3h-27h | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





