Submitted:
28 June 2023
Posted:
29 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
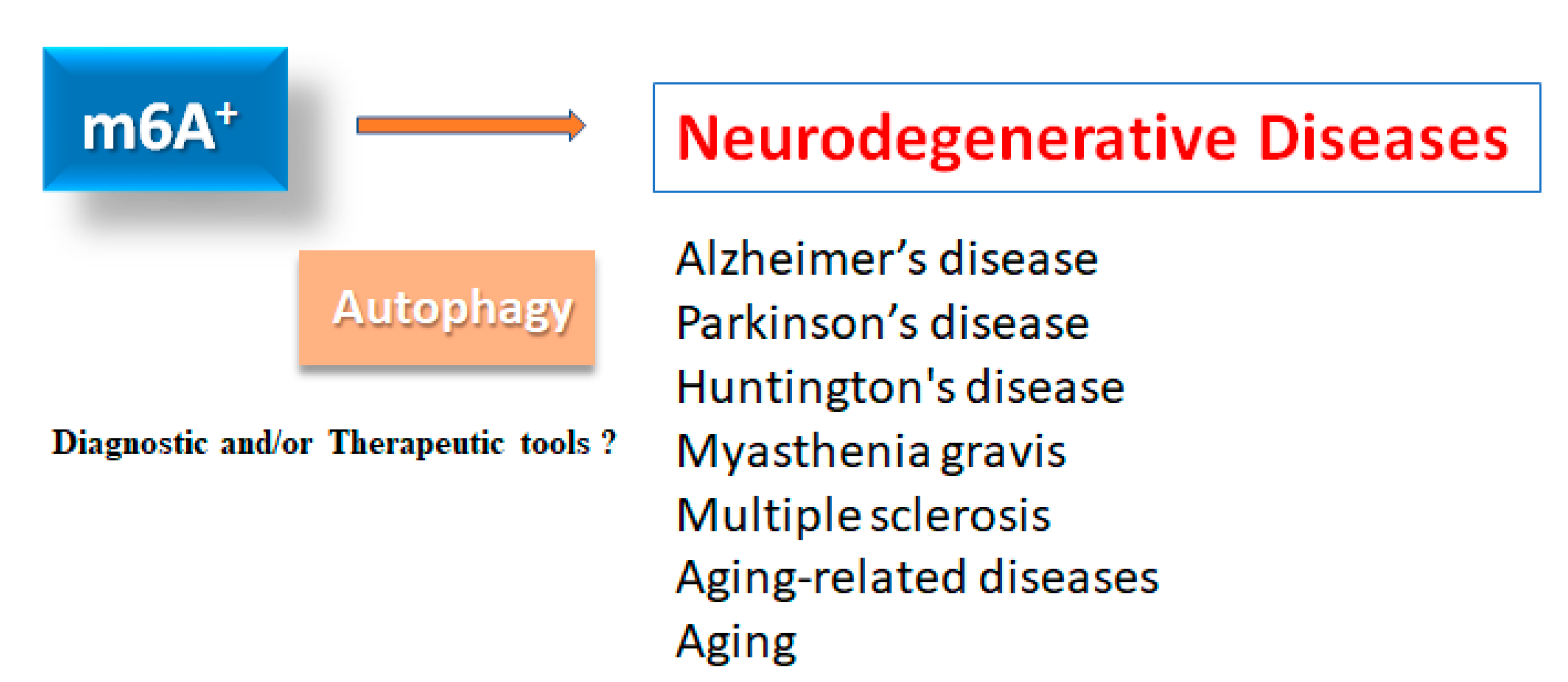
2. m6A methylation of mRNAs in neurodegenerative diseases
2.1. Alzheimer’s disease
2.2. Parkinson’s disease
2.3. Further neurodegenerative disorders and aging
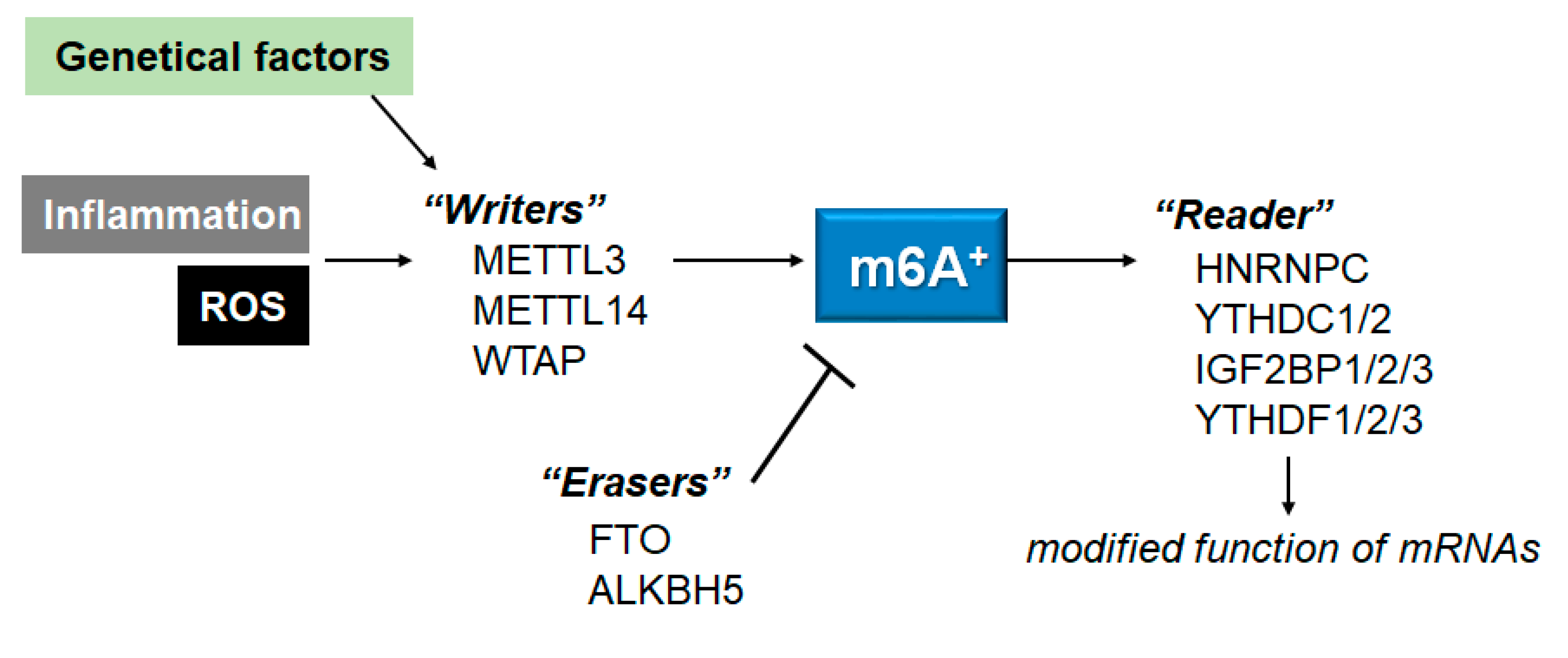
3. ROS, inflammation and m6A mRNAs
4. m6A RNA modifications in autophagy
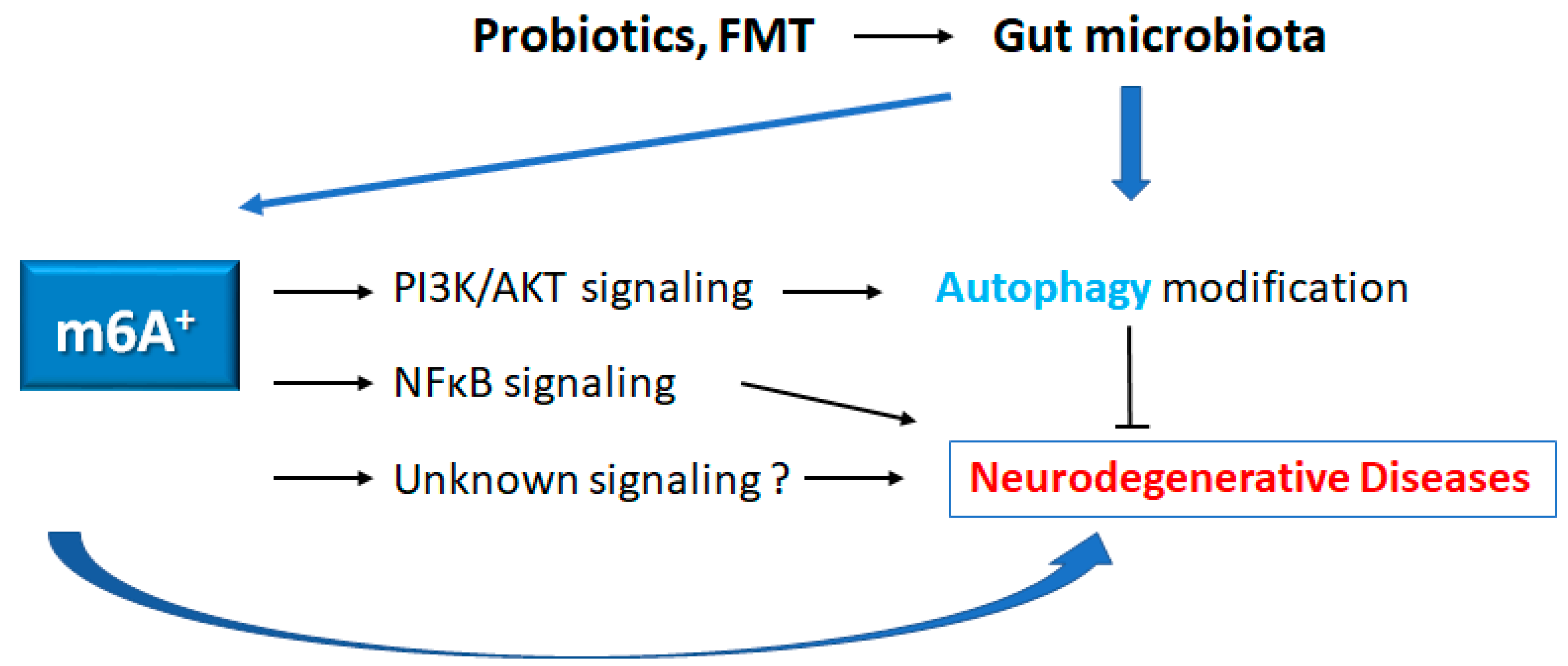
5. Possible therapeutics for neurodegenerative diseases
6. Future perspectives involved in the therapeutics
Author Contributions
Funding
Data Availability Statement
Competing interests statement
Abbreviations
References
- Wang, M.; Pan, W.; Xu, Y.; Zhang, J.; Wan, J.; Jiang, H. Microglia-mediated neuroinflammation: a potential target for the treatment of cardiovascular diseases. J Inflamm Res. 2022, 15, 3083–3094. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martin, P.; Rodriguez-Blazquez, C.; Forjaz, M.J. Quality of life and burden in caregivers for patients with Parkinson’s disease: concepts, assessment and related factors. Expert Rev Pharmacoecon Outcomes Res. 2012, 12, 221–230. [Google Scholar] [CrossRef]
- Brodaty, H.; Breteler, M.M.; DeKosky, S.T.; Dorenlot, P.; Fratiglioni, L.; Hock, C.; Kenigsberg, P.A.; Scheltens, P.; De Strooper, B. The world of dementia beyond 2020. J Am Geriatr Soc. 2011, 59, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Do, M.D.; Tran, T.N.; Luong, A.B.; Le, L.H.G.; Van Le, T.; Le, K.T.; Van Vo, N.T.; Le, T.N.N.; Vu, H.A.; Mai, T.P. Clinical and genetic analysis of Vietnamese patients diagnosed with early-onset Parkinson's disease. Brain Behav. 2023, 13, e2950. [Google Scholar] [CrossRef] [PubMed]
- Naseri, N.; Sharma, M.; Velinov, M. Autosomal dominant neuronal ceroid lipofuscinosis: Clinical features and molecular basis. Clin Genet. 2021, 99, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2010, 39 (suppl_1), D195-D201.
- Li, Y.; Xiao, J.; Bai, J.; Tian, Y.; Qu, Y.; Chen, X.; Wang, Q.; Li, X.; Zhang, Y.; Xu, J. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. J.Mol Cancer. 2019, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, Y.; Wuermanbieke, S.; Hu, S.; Huang, G. N6-methyladenosine (m6A) methyltransferase METTL3-mediated LINC00680 accelerates osteoarthritis through m6A/SIRT1 manner. Cell Death Discov. 2022, 8, 240. [Google Scholar] [CrossRef]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018, 97, 313–325. [Google Scholar] [CrossRef]
- Widagdo, J.; Anggono, V. The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J Neurochem. 2018, 147, 137–152. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ding, X.; Wang, Y.; Hu, X.; Sun, T.; Wei, M.; Wang, X.; Wu, H. The relationship between the network of non-coding RNAs-molecular targets and N6-methyladenosine modification in colorectal cancer. Front Cell Dev Biol. 2021, 9, 772542. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, X.; Huang, T.; Zhao, X.; Chen, W.; Gu, N.; Zhang, R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020, 48, 6251–6264. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dong, D.; Yu, P.; Chen, T.; Gao, R.; Wei, J.; Mo, Z.; Zhou, H.; Yang, Q.; Yue, C. Prognostic model based on m6A -associated lncRNAs in esophageal cancer. Front Endocrinol (Lausanne). 2022, 13, 947708. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, B.; Sun, H.; Xu, X.; Wang, Y. Epigenetic regulations in neural stem cells and neurological diseases. Stem Cells Int. 2018, 2018. 6087143.
- Li, J.; Yang, X.; Qi, Z.; Sang, Y.; Liu, Y.; Xu, B.; Liu, W.; Xu, Z.; Deng, Y. The role of mRNA m6A methylation in the nervous system. Cell Biosci. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Liu, S.; Li, Q.; Chen, K.; Zhang, Q.; Li, G.; Zhuo, L.; Zhai, B.; Sui, X.; Hu, X.; Xie, T. The emerging molecular mechanism of m6A modulators in tumorigenesis and cancer progression. Biomed Pharmacother. 2020, 127, 110098. [Google Scholar] [CrossRef]
- Song, H.; Feng, X.; Zhang, H.; Luo, Y.; Huang, J.; Lin, M.; Jin, J.; Ding, X.; Wu, S.; Huang, H. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019, 15, 1419–1437. [Google Scholar] [CrossRef]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020, 16, 1221–1235. [Google Scholar] [CrossRef]
- Liu, S.; Li, Q.; Li, G.; Zhang, Q.; Zhuo, L.; Han, X.; Zhang, M.; Chen, X.; Pan, T.; Yan, L. The mechanism of m6A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020, 11, 969. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luo, R.; Zhang, W.; He, S.; Wang, B.; Liang, H.; Song, Y.; Ke, W.; Shi, Y.; Feng, X. m6A hypomethylation of DNMT3B regulated by ALKBH5 promotes intervertebral disc degeneration via E4F1 deficiency. Clin Transl Med. 2022, 12, e765. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhan, Y.; Zong, Y. METTL3-mediated LINC00657 promotes osteogenic differentiation of mesenchymal stem cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 2022, 388, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Zhou, X.; Li, Y.-M.; Yang, T.; Zhang, S.-J.; Wang, Y.; Jia, S.-H.; Peng, D.-T. N6-methyladenine-modified DNA was decreased in Alzheimer’s disease patients. World J Clin Cases. 2022, 10, 448. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Fischer, C.; Maralani, P.J.; Black, S.E.; Moody, A.R.; Khademi, A. Alzheimer’s and vascular disease classification using regional texture biomarkers in FLAIR MRI. Neuroimage Clin. 2023, 38, 103385. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Shi, M.; Li, M.; Zeng, J.; He, J. Roles of m6A modification in neurological diseases. Zhong nan da xue xue bao. Yi xue ban = Journal of Central South University. Medical sciences. 2022, 47, 109–115. [Google Scholar]
- Tang, F.; Chen, L.; Gao, H.; Xiao, D.; Li, X. m6A: An emerging role in programmed cell death. Front Cell Dev Biol. 2022, 10, 80. [Google Scholar] [CrossRef]
- Han, L.; Lei, G.; Chen, Z.; Zhang, Y.; Huang, C.; Chen, W. IGF2BP2 regulates MALAT1 by serving as an N6-methyladenosine reader to promote NSCLC proliferation. Front Mol Biosci. 2022, 8, 1328. [Google Scholar] [CrossRef]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. J.Front Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef]
- Pinheiro, L.; Faustino, C. Therapeutic strategies targeting amyloid-β in Alzheimer’s disease. Curr Alzheimer Res. 2019, 16, 418–452. [Google Scholar] [CrossRef]
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 1–19. [Google Scholar] [CrossRef]
- Shao, N.; Ye, T.; Xuan, W.; Zhang, M.; Chen, Q.; Liu, J.; Zhou, P.; Song, H.; Cai, B. The effects of N 6-methyladenosine RNA methylation on the nervous system. Mol Cell Biochem. 2023, 1–13. [Google Scholar] [CrossRef]
- Tang, Z.; Cao, J.; Yao, J.; Fan, X.; Zhao, J.; Zhao, M.; Duan, Q.; Han, B.; Duan, S. KDM1A-mediated upregulation of METTL3 ameliorates Alzheimer's disease via enhancing autophagic clearance of p-Tau through m6A-dependent regulation of STUB1. Free Radic Biol Med. 2023, 195, 343–358. [Google Scholar] [CrossRef]
- Hao, X.; Li, Y.; Huang, G.; Zeng, Y. Role of the N6-methyladenosine regulatory factor in reducing the risk of cardiovascular disease: subtype diagnosis following aerobic exercise-assisted weight loss. Am J Transl Res. 2022, 14, 5363. [Google Scholar] [PubMed]
- Teng, Y.; Liu, Z.; Chen, X.; Liu, Y.; Geng, F.; Le, W.; Jiang, H.; Yang, L. Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J Cell Mol Med. 2021, 25, 8567–8572. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci. 2019, 10, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Wang, S.; Li, J.; Wen, Y.; Cui, R.; Zhang, K.; Liu, Y.; Yang, X.; Zhang, L.; Xu, B. Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J Hazard Mater. 2022, 426, 128099. [Google Scholar] [CrossRef]
- Ondo, K.; Isono, M.; Nakano, M.; Hashiba, S.; Fukami, T.; Nakajima, M. The N6-methyladenosine modification posttranscriptionally regulates hepatic UGT2B7 expression. Biochem Pharmacol. 2021, 189, 114402. [Google Scholar] [CrossRef]
- Foroud, T.; Siemers, E.; Kleindorfer, D.; Bill, D.J.; Hodes, M.; Norton, J.A.; Conneally, P.M.; Christian, J.C. Cognitive scores in carriers of Huntington's disease gene compared to noncarriers. Ann Neurol. 1995, 37, 657–664. [Google Scholar] [CrossRef]
- Harris, K.L.; Armstrong, M.; Swain, R.; Erzinclioglu, S.; Das, T.; Burgess, N.; Barker, R.A.; Mason, S.L. Huntington's disease patients display progressive deficits in hippocampal-dependent cognition during a task of spatial memory. Cortex. 2019, 119, 417–427. [Google Scholar] [CrossRef]
- Pupak, A.; Singh, A.; Sancho-Balsells, A.; Alcalá-Vida, R.; Espina, M.; Giralt, A.; Martí, E.; Ørom, U.A.V.; Ginés, S.; Brito, V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell Mol Life Sci. 2022, 79, 416. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.M.; Van Den Broeck, J.; Vrolix, K.; Janssen, S.P.; Lemmens, M.A.; Van Der Esch, E.; Duimel, H.; Frederik, P.; Molenaar, P.C.; Martínez-Martínez, P. Antibody effector mechanisms in myasthenia gravis—pathogenesis at the neuromuscular junction. Autoimmunity. 2010, 43 (5-6), 353-370.
- Li, S.; Liu, H.; Ruan, Z.; Guo, R.; Sun, C.; Tang, Y.; Huang, X.; Gao, T.; Hao, S.; Li, H. Landscape analysis of m6A modification regulators related biological functions and immune characteristics in myasthenia gravis. J Transl Med. 2023, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Dalmau, J. Diagnostic and treatment challenges, a new section in N2. Neurol Neuroimmunol Neuroinflamm. 2018, 5, e511. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.-B.; Lei, S.-F.; Qian, Q.-Y.; Guo, Y.-F.; Zhang, Y.-H.; Zhang, H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J Neurol. 2019, 266, 2699–2709. [Google Scholar] [CrossRef] [PubMed]
- Castro-Hernández, R.; Berulava, T.; Metelova, M.; Epple, R.; Peña Centeno, T.; Richter, J.; Kaurani, L.; Pradhan, R.; Sakib, M.S.; Burkhardt, S. Conserved reduction of m6A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts. Proc Natl Acad Sci U S A. 2023, 120, e2204933120. [Google Scholar] [CrossRef]
- Su, X.; Shen, Y.; Jin, Y.; Kim, I.-m.; Weintraub, N.L.; Tang, Y. Aging-associated differences in epitranscriptomic m6A regulation in response to acute cardiac ischemia/reperfusion injury in female mice. Front Pharmacol. 2021, 12, 654316. [Google Scholar] [CrossRef]
- Chen, Y.-s.; Ouyang, X.-p.; Yu, X.-h.; Novák, P.; Zhou, L.; He, P.-p.; Yin, K. N6-Adenosine methylation (m6A) RNA modification: an emerging role in cardiovascular diseases. J Cardiovasc Transl Res. 2021, 14, 857–872. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Luo, J.; Xu, T.; Sun, K. N6-methyladenosine RNA modification in inflammation: roles, mechanisms, and applications. Front Cell Dev Biol. 2021, 9, 670711. [Google Scholar] [CrossRef]
- Mapperley, C.; van de Lagemaat, L.N.; Lawson, H.; Tavosanis, A.; Paris, J.; Campos, J.; Wotherspoon, D.; Durko, J.; Sarapuu, A.; Choe, J. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J Exp Med. 2020, 218, e20200829. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Wu, J.; Li, H.; Luo, C.; Ji, G.; Guan, X.; Liu, J.; Wang, M. METTL3 achieves lipopolysaccharide-induced myocardial injury via m6A -dependent stabilization of Myh3 mRNA. Biochim Biophys Acta Mol Cell Res. 2023, 1870, 119503. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, Q.; Meng, R.; Yi, B.; Xu, Q. METTL 3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J Cell Mol Med. 2018, 22, 2558–2568. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Jin, C.; Wang, Y.; Yu, J.; Wang, R.; Tian, X. FTO promotes the progression of cervical cancer by regulating the N6-methyladenosine modification of ZEB1 and Myc. Mol Carcinog. 2023. [Google Scholar] [CrossRef]
- Luo, J.; Wang, F.; Sun, F.; Yue, T.; Zhou, Q.; Yang, C.; Rong, S.; Yang, P.; Xiong, F.; Yu, Q. Targeted inhibition of FTO demethylase protects mice against LPS-induced septic shock by suppressing NLRP3 inflammasome. Front Immunol. 2021, 12, 663295. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell. 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Deng, Y.; Zhu, H.; Xiao, L.; Liu, C.; Liu, Y.-L.; Gao, W. Identification of the function and mechanism of m6A reader IGF2BP2 in Alzheimer’s disease. Aging (Albany NY). 2021, 13, 24086. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, Y.; Ma, J.; Yu, Y.; Wang, Q.; Gao, P.; Wang, L.; Xu, Z.; Wei, X.; Jing, M. m6A mRNA Methylation Was Associated With Gene Expression and Lipid Metabolism in Liver of Broilers Under Lipopolysaccharide Stimulation. Front Genet. 2022, 13. 818357.
- Qi, L.; Wang, Y.; Hu, H.; Li, P.; Hu, H.; Li, Y.; Wang, K.; Zhao, Y.; Feng, M.; Lyu, H. m6A methyltransferase METTL3 participated in sympathetic neural remodeling post-MI via the TRAF6/NF-κB pathway and ROS production. J Mol Cell Cardiol. 2022, 170, 87–99. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018, 25, 1816–1828. [Google Scholar] [CrossRef]
- Han, Y.-C.; Xie, H.-Z.; Lu, B.; Xiang, R.-L.; Zhang, H.-P.; Li, J.-Y.; Zhang, S.-Y. Lipopolysaccharide alters the m6A epitranscriptomic tagging of RNAs in cardiac tissue. Front Mol Biosci. 2021, 8, 670160. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, Y.; Chen, Z.; Yue, Y.; Huang, H.; Wu, B.; Liu, Y.; Zhou, D.-X.; Zhao, Y. ROS-stimulated protein lysine acetylation is required for crown root development in rice. J Adv Res. 2023, 48, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ni, X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr Drug Targets. 2015, 16, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhuo, R.; Tao, S.; Liang, Y.; Liu, C.; Liu, Q.; Wang, T.; Zhong, X. m6A RNA Methylation Mediates NOD1/NF-kB Signaling Activation in the Liver of Piglets Challenged with Lipopolysaccharide. Antioxidants (Basel). 2022, 11, 1954. [Google Scholar] [CrossRef]
- Shu, F.; Xiao, H.; Li, Q.-N.; Ren, X.-S.; Liu, Z.-G.; Hu, B.-W.; Wang, H.-S.; Wang, H.; Jiang, G.-M. Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct Target Ther. 2023, 8, 32. [Google Scholar] [CrossRef]
- Jin, S.; Zhang, X.; Miao, Y.; Liang, P.; Zhu, K.; She, Y.; Wu, Y.; Liu, D.-A.; Huang, J.; Ren, J. , m6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018, 28, 955–957. [Google Scholar] [CrossRef]
- Liu, J.; Shao, Y.; Li, D.; Li, C. N6-methyladenosine helps Apostichopus japonicus resist Vibrio splendidus infection by targeting coelomocyte autophagy via the AjULK-AjYTHDF/AjEEF-1α axis. Commun Biol. 2023, 6, 547. [Google Scholar] [CrossRef]
- Yu, T.; Qi, X.; Zhang, L.; Ning, W.; Gao, D.; Xu, T.; Ma, Y.; Knott, J.G.; Sathanawongs, A.; Cao, Z. Dynamic reprogramming and function of RNA N6-methyladenosine modification during porcine early embryonic development. Zygote. 2021, 29, 417–426. [Google Scholar] [CrossRef]
- Fang, Z.M.; Zhang, S.M.; Luo, H.; Jiang, D.S.; Huo, B.; Zhong, X.; Feng, X.; Cheng, W.; Chen, Y.; Feng, G. Methyltransferase-like 3 suppresses phenotypic switching of vascular smooth muscle cells by activating autophagosome formation. Cell Prolif. 2023, 56, e13386. [Google Scholar] [CrossRef]
- Liang, J.; Sun, J.; Zhang, W.; Wang, X.; Xu, Y.; Peng, Y.; Zhang, L.; Xiong, W.; Liu, Y.; Liu, H. Novel Insights into The Roles of N6-methyladenosine (m6A) Modification and Autophagy in Human Diseases. Int J Biol Sci. 2023, 19, 705. [Google Scholar] [CrossRef]
- Wang, F.; Liao, Y.; Zhang, M.; Zhu, Y.; Wang, W.; Cai, H.; Liang, J.; Song, F.; Hou, C.; Huang, S. N6-methyladenosine demethyltransferase FTO-mediated autophagy in malignant development of oral squamous cell carcinoma. Oncogene. 2021, 40, 3885–3898. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, S.; Zhang, Y.; Zhao, M. Rat bone marrow mesenchymal stem cells (BMSCs) inhibit liver fibrosis by activating GSK3β and inhibiting the Wnt3a/β-catenin pathway. Infect Agent Cancer. 2022, 17, 1–10. [Google Scholar] [CrossRef]
- Huang, P.; Liu, M.; Zhang, J.; Zhong, X.; Zhong, C. The Potential Role of m6A in the Regulation of TBI-Induced BGA Dysfunction. Antioxidants (Basel). 2022, 11, 1521. [Google Scholar] [CrossRef]
- Yu, H.; Zhong, H.; Sun, J.; Li, N.; Chen, J.; Shen, B.; Huang, P.; Shen, X.; Huang, S.; Zhong, Y. Molecular signaling from microglia impacts macroglia autophagy and neurons survival in glaucoma. iScience. 2023, 26. 106839.
- Chen, S.-j.; Bao, L.; Keefer, K.; Shanmughapriya, S.; Chen, L.; Lee, J.; Wang, J.; Zhang, X.-Q.; Hirschler-Laszkiewicz, I. ; Merali, S Transient receptor potential ion channel TRPM2 promotes AML proliferation and survival through modulation of mitochondrial function, ROS, and autophagy. Cell Death Dis. 2020, 11, 247. [Google Scholar] [CrossRef]
- Hao, W.; Dian, M.; Zhou, Y.; Zhong, Q.; Pang, W.; Li, Z.; Zhao, Y.; Ma, J.; Lin, X.; Luo, R. Autophagy induction promoted by m6A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat Commun. 2022, 13, 5845. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Dian, M.; Wang, J.; Sun, Y.; Xiao, D. Epitranscriptomic turbo for autophagy boost: m6A reader YTHDF3. Autophagy. 2022, 19, 1882–1884. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wang, J.; Wang, Q.; Liu, P.; Zhao, H. , Interaction between N6-methyladenosine and autophagy in the regulation of bone and tissue degeneration. Front Bioeng Biotechnol. 2022, 10, 978283. [Google Scholar] [CrossRef] [PubMed]
- Almasi, S.; Crawford Parks, T.E.; Ravel-Chapuis, A.; MacKenzie, A.; Côté, J.; Cowan, K.N.; Jasmin, B.J. Differential regulation of autophagy by STAU1 in alveolar rhabdomyosarcoma and non-transformed skeletal muscle cells. Cell Oncol (Dordr). 2021, 44, 851–870. [Google Scholar] [CrossRef]
- Singh, S.B.; Lin, H.C. Autophagy counters LPS-mediated suppression of lysozyme. Innate Immun. 2017, 23, 537–545. [Google Scholar] [CrossRef]
- Sánchez-Vidaña, D.I.; Li, J.; Abokyi, S.; Chan, J.N.-M.; Ngai, S.P.-C.; Lau, B.W.-M. In vitro methods in autophagy research: Applications in neurodegenerative diseases and mood disorders. Front Mol Neurosci. 2023, 16, 130. [Google Scholar] [CrossRef]
- Nagase, N.; Ikeda, Y.; Tsuji, A.; Kitagishi, Y.; Matsuda, S. Efficacy of probiotics on the modulation of gut microbiota in the treatment of diabetic nephropathy. World J Diabetes. 2022, 13, 150. [Google Scholar] [CrossRef]
- Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Tsuji, A.; Matsuda, S. Promising role of D-amino acids in irritable bowel syndrome. World J Gastroenterol. 2022, 28, 4471. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jang, S. RNA m6A methyltransferase Mettl3 regulates spatial neural patterning in Xenopus laevis. Mol Cell Biol. 2021, 41, e00104–e00121. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, Y.; Lu, M.; Song, M.; Yu, Z.; Wang, J.; Wang, S.; Ren, J.; Yang, Y.-G.; Liu, G.-H. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020, 48, 11083–11096. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, T.; Liu, Y.; Fu, J.; Wei, X.; Liu, D.; Ma, W.; Gu, H.; Yuan, Z. Nuclear factor IC disrupts cellular homeostasis between autophagy and apoptosis via miR-200b-Ambra1 in neural tube defects. Cell Death Dis. 2021, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Rinaldi, L.; Boccia, G.; Chianese, A.; Sasso, F.C.; De Caro, F.; Franci, G.; Galdiero, M. Regulation of m6A methylation as a new therapeutic option against COVID-19. Pharmaceuticals (Basel). 2021, 14, 1135. [Google Scholar] [CrossRef]
- Chen, B.; Ye, F.; Yu, L.; Jia, G.; Huang, X.; Zhang, X.; Peng, S.; Chen, K.; Wang, M.; Gong, S.; et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012, 134, 17963–17971. [Google Scholar] [CrossRef]
- Shen, C.; Zhang, Z.; Xie, T.; Ji, J.; Xu, J.; Lin, L.; Yan, J.; Kang, A.; Dai, Q.; Dong, Y. Rhein suppresses lung inflammatory injury induced by human respiratory syncytial virus through inhibiting NLRP3 inflammasome activation via NF-κB pathway in mice. Front Pharmacol. 2020, 10, 1600. [Google Scholar] [CrossRef]
- Jabs, S.; Biton, A.; Bécavin, C.; Nahori, M.-A.; Ghozlane, A.; Pagliuso, A.; Spanò, G.; Guérineau, V.; Touboul, D. ; Giai Gianetto, Q Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat Commun. 2020, 11, 1344. [Google Scholar] [CrossRef]
- Koh, Y.-C.; Ho, C.-T.; Pan, M.-H. , Recent advances in cancer chemoprevention with phytochemicals. J Food Drug Anal. 2020, 28, 14–37. [Google Scholar] [CrossRef] [PubMed]
- Medina-Larqué, A.-S.; Rodríguez-Daza, M.-C.; Roquim, M.; Dudonné, S.; Pilon, G.; Levy, É.; Marette, A.; Roy, D.; Jacques, H.; Desjardins, Y. Cranberry polyphenols and agave agavins impact gut immune response and microbiota composition while improving gut barrier function, inflammation, and glucose metabolism in mice fed an obesogenic diet. Front Immunol. 2022, 13, 871080. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, A.; Yoshikawa, S.; Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Morikawa, S.; Nakashima, M.; Asai, T.; Matsuda, S. Tactics with prebiotics for the treatment of metabolic dysfunction-associated fatty liver disease via the improvement of mitophagy. Int J Mol Sci. 2023, 24, 5465. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Ikeda, Y.; Tsuji, A.; Matsuda, S. A New Concept of Associations between Gut Microbiota, Immunity and Central Nervous System for the Innovative Treatment of Neurodegenerative Disorders. Metabolites. 2022, 12, 1052. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Fu, J.; Xiao, X.; Wang, F.; Jin, M.; Fang, W.; Wang, Y.; Zong, X. Faecal microbiota transplantation-mediated jejunal microbiota changes halt high-fat diet-induced obesity in mice via retarding intestinal fat absorption. Microb Biotechnol. 2022, 15, 337–352. [Google Scholar] [CrossRef]
- Sawamura, H.; Taniguchi, K.; Ikeda, Y.; Tsuji, A.; Kitagishi, Y.; Matsuda, S. , Gut microbiota could modulate the effects of neuro-immune responses and memory traces via the gut-brain-immune axis in schizophrenia. Explor. Neuroprot. Ther. 2022, 2, 74–86. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




